Introduction
Radiotherapy is the most commonly used therapeutic
approach for managing cancer and acts mainly through the induction
of DNA damage (1). Impairment in
the DNA repair proteins, which physiologically protect cells from
persistent DNA injury, can affect the efficacy of cancer therapy
(2). The DNA damage repair (DDR)
response is an intricate signal transduction pathway activated upon
DNA damage. Cancer cells often show significant alterations at the
level of the DDR response and develop resistance to DNA
damage-inducing agents (3,4).
Recently, increasing evidence suggests that
microRNAs (miRNAs) take an active part in the regulation of the DNA
damage/repair network (5,6). miRNAs are endogenous short non-coding
molecules able to regulate gene expression at the
post-transcriptional level (7). In
the last decade, miRNAs, a new class of molecules able to
post-transcriptionally regulate gene expression, have emerged to be
involved in several fundamental physiological and pathological
biomolecular and cellular mechanisms (8,9). miRNA
profiles have been analyzed and several differentially expressed
miRNAs involved in many cellular functions such as apoptosis, cell
cycle control, and DNA damage/repair were identified in response to
different radiation doses (10).
miRNAs are non-encoding small RNAs existing
extensively in plants, animals and viruses, at an approximate
length of 21–23 nt and are highly conserved. They bind to a
specific mRNA 3′UTR and regulate gene transcription. Mature miRNAs
together with other proteins form into RNA-induced silencing
complex, resulting in the degradation or translation suppression of
target mRNAs when binding to a target mRNA 3′UTR (11).
The analysis of miRNA-modulated gene regulation in
the DDR and its involvement in cancer pathogenesis and progression
will help to understand and define the impact of these small
molecules in DNA damage/repair as well as chemoresistant and
radioresistant mechanisms (12,13).
This knowledge will expand the characterization of molecules and
networks involved in pathways activated upon DNA damage and the
subsequent alterations at the level of fundamental processes such
as cell cycle control and apoptosis (14). The identification of miRNA-modulated
genes and the effects of deregulated functions of miRNAs will make
it possible to acquire information concerning the prognosis,
chemoresistance or radioresistance, and then the response to
therapeutic treatments in cancer. However, the implications of
miRNA-mRNA interactions in genotoxic mechanisms remain unknown
(15).
In the present study, we investigated the global
miRNA expression in an established radioresistant lung cancer cell
line from our previous study and analyzed the identified miRNA-mRNA
interaction to radiotherapy. The aim of this study was to clarify
the mechanisms of resistance in lung cancer cells and to also
identified specific miRNAs in lung cancer cells following radiation
treatment.
Materials and methods
Cells and cell culture
The human lung cancer cell line A549 was purchased
from the American Type Culture Collection (ATCC) and cultured in
RPMI-1640 with Dulbecco’s modified Eagle’s medium (Sigma-Aldrich,
St. Louis, MO, USA) supplemented with 20% fetal calf serum, 0.05%
L-glutamine, 150 UI/ml penicillin and 50 μg/ml streptomycin
in a humidified atmosphere with 5% CO2 at 37°C. The
cultures were split every second day by dilution at a concentration
of 2×105 cells/ml. The cell counts were performed with a
hemocytometer; the cell membrane integrity was determined using the
trypan blue exclusion technique. Cell lines in the maximal range of
up to 20 passages were used for the present study. All cell lines
tested negative for mycoplasma contamination by polymer chain
reaction (PCR) methods (16). Cell
lines were authenticated using the short tandem repeats (STR)
testing.
Development of acquired radioresistant
cells
A549 cells (1×106) were plated in
75-cm2 culture flasks and irradiated with 4 Gy of γ-rays
using a Theratron Cobalt-60 treatment unit (Siemens, Concord, CA,
USA) at a dose rate of 1 Gy/min when the cells were at ~60%
confluency in the culture flask. Immediately following irradiation,
the culture medium was renewed, and the cells were returned to the
incubator. When the A549 cells reached ~90% confluency, they were
trypsinized, counted and passaged into new culture flasks. Again,
the cells were treated with 4-Gy γ-rays when they reached ~60%
confluency. The irradiation was performed 13 times for a total dose
of 60 Gy (irradiated with 2 Gy of γ-rays at the final irradiation)
over 5 months. The parental cells were trypsinized, counted and
passaged under the same conditions without irradiation.
In vitro X-ray irradiation
The final cell density was adjusted to
1×106 cells/ml, and the samples were placed at 37°C in a
5% CO2 incubator. X-irradiation was performed using a
Theratron Cobalt-60 treatment unit (Siemens) at a dose rate of 2
Gy/min. Non-irradiated cells were treated in similar way, but at a
zero radiation dose.
Colony formation assay
The cells were trypsinized and resuspended in T25
flasks, followed by γ-ray exposure at room temperature at single
doses of 0, 4, 8, and 12 Gy. After irradiation, the cells were
seeded onto 6-cm dishes and incubated for 14 days without
disturbance. The seeded cell number was increased as the radiation
dose increased. Formed colonies were visualized by staining with
0.02% crystal violet solution (w/v in 75% ethanol). The plating
efficiency was determined as the ratio of the number of colonies
divided by the number of cells seeded. The surviving fraction was
determined from the ratio of plating efficiencies of irradiated
cells compared to the no irradiated control. Each data value
represents the mean of three independent experiments ± SEM.
Total RNA preparation
The samples were ground by using liquid nitrogen.
Total RNA was extracted from lung tissues using an animal tissue
RNA purification kit (Norgen Biotek, Thorold, ON, USA) as
recommended by the manufacturer. RNA samples were measured using
Bioanalyzer to determine RNA integrity number.
miRNA microarray chip analysis
miRNA expression profiling was performed using the
TaqMan Rodent miRNA Array Card A and Card B (Applied Biosystems,
Foster City, CA, USA). Each chip contained multiple quality control
probes and employed dual-color chip to examine miRNA expression
profiling in mouse lung tissues. Probes were synthesized in
situ with photosensitive PGR. The sequence consisted of 2
fragments, namely, a chemically modified oligonucleotide encoding
fragment complementary to target miRNA or other target RNA, and an
extension arm at a certain distance to the connected encoding
sequence, which lessened the hybridization spatial impairment. The
temper of probe hybridization was balanced by the chemical
modification. Cy3- and Cy5-specific fluorescent labels and Axon
GenePix 4000B microarray scanner were used to capture hybridization
images. ArrayPro (Media Cybernetics, Bethesda, MD, USA) was
utilized to complete the digital transformation.
Quantitative real-time PCR (qRT-PCR)
An approximate length of 21–23 nt of miRNA resulted
in difficulties in conventional PCR test. We used
TaqMan® microRNA assays (Applied Biosystems) to examine
the miRNA differential expression profiling in PTC as recommended
by the manufacturer. Sample RNA (10 ng) was reversely transcribed
into cDNA by using specific stem-loop primers and
TaqMan® microRNA reverse transcription kit. With cDNA as
the template, TaqMan microRNA assay and TaqMan®
Universal PCR Master Mix were used for the serial real-time PCR.
RNU48 was used as an internal control to minimize the variation
among reverse transcription, PCR and samples. Data were collected,
analyzed, and normalized using the Applied Biosystems analysis
software to determine the differential expression profiles of the
miRNAs. All of the experiments were conducted in triplicate.
Expression levels were calculated using the relative quantification
method (ΔΔCT) in the ABI PRISM 7500 sequence detection system
(Applied Biosystems), according to the manufacturer’s
instructions.
miRNA overexpression and knockdown
miR-1323 was overexpressed by reverse transfection
of 100 nM pre-miR precursor molecule using siPORT NeoFX
transfection reagent as per the manufacturer’s instructions and
compared with the pre-miR negative control (miR-control) (all from
Life Technologies/Ambion). Inhibition of miR-1323 was achieved by
reverse transfection of 30 nM anti-miR-1323 inhibitor (Life
Technologies/Ambion) using siPORT NeoFX transfection reagent and
compared with the anti-miR negative control.
Luciferase constructs and assays
Full-length human PRKDC 3′UTR sequences were
amplified by PCR and cloned into the Spe1/HindIII
restriction sites of the pMIR Report Luciferase plasmid (Life
Technologies/Invitrogen, Carlsbad, CA, USA). The putative miR-1323
target sites in the 3′UTR of PRKDC were mutated using the
QuikChange XL Site-Directed Mutagenesis kit (Stratagene, La Jolla,
CA, USA). Resistant cells were transfected with 750 ng wild-type or
mutant PRKDC 3′UTR luciferase reporter constructs and stable cell
lines generated by puromycin selection. Stable cell lines were
reverse transfected with 10 nM miR-1323 or miR-control. Cells were
incubated for a further 24 h, and luciferase activity was assessed
using the Dual-Luciferase assay system (Promega, Fitchburg, WI,
USA).
Western blot analysis
Proteins were resolved on 12% polyacrylamide gels,
transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA,
USA) and blocked with 5% non-fat dairy milk in Tris-buffered saline
(20 mM Tris and 150 mM NaCl, pH 7.4) with 0.1% Tween-20.
Statistical and bioinformatic analysis of
the microarray data
In the process and analysis of data, the background
was initially excluded. Any ‘bad spot’ that showed a signaling
value deviation above 50% of the average of repeated spots and/or a
spot CV above 0.5 was deleted preceding the calculation of the mean
and standard deviation for repeated spots. The normalization was
completed by locally weighted regression. For the double-labeling
experiments, the ratio of 2 sets of detected signals (log2
transformed) and P-value for the Student-t test were calculated. A
P-value <0.05 was considered to be statistically significant.
Significantly differential miRNAs were defined as those showing a
logarithm (log2) for the ratio of the treatment group signal to
control signal ≥1 or ≤−1. Multiple target gene prediction software
including miRanda (http://www.mocrorna.org), TargetScan (http://www.targetscan.org), mirTarget2 (http://mirdb.org/miRDB), RNA22 (http://cbcsrv.watson.ibm.com/rna22.html), and
microTv3.0 (http://diana.cslab.ece.ntua.gr/microT) were used to
forecast some potential target genes showing a sequential
differential expression profiling. The genes identified by at least
3 software, were taken as target genes to minimize the false
positivity. In order to further characterize the molecular
functions of forecasted target genes, MAS database (http://bioinfo.capitalbio.com/mas3/) and GDM
database (http://gdm.fmrp.usp.br/) were used to
make a preliminary analysis of forecasted target genes.
Results
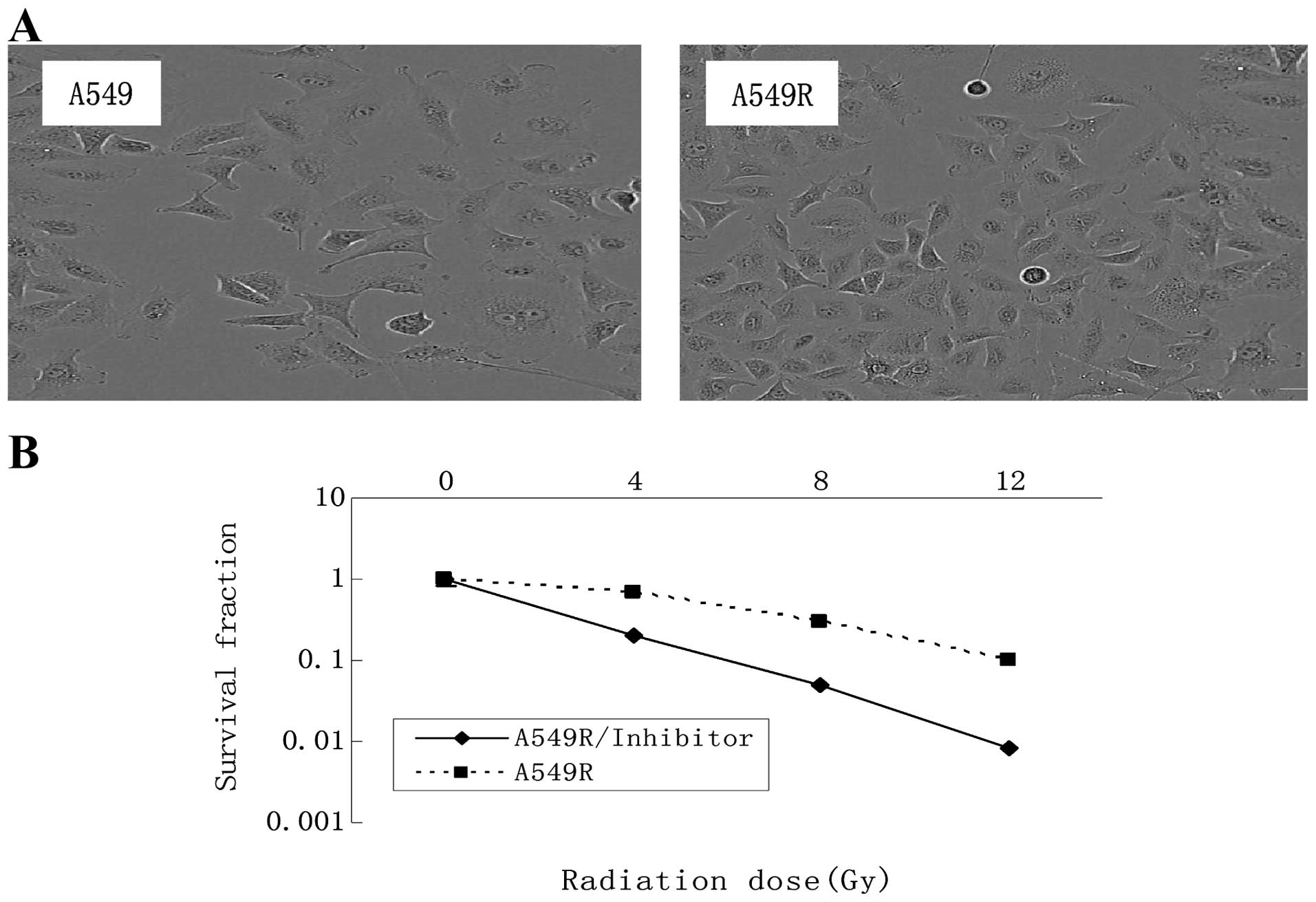
Morphology and radiosensitivity of the
A549R cells
The radioresistant cells designated as A549R, were
obtained by subjecting A549 cells to 5 months of fractionated
irradiation with a total dose of 60 Gy and 10 additional passages
without irradiation. No obvious change in the cell morphology was
observed following irradiation (Fig.
1A). The radiosensitivity of the A549 and A549R cells was
compared using a colony formation assay (Fig. 1B). Each point on the survival curve
represents the mean surviving fraction from triplicate experiments.
As expected, the A549R cells had a higher survival rate than the
A549 cells, indicating that the A549R cells were more
radioresistant than the A549 cells.
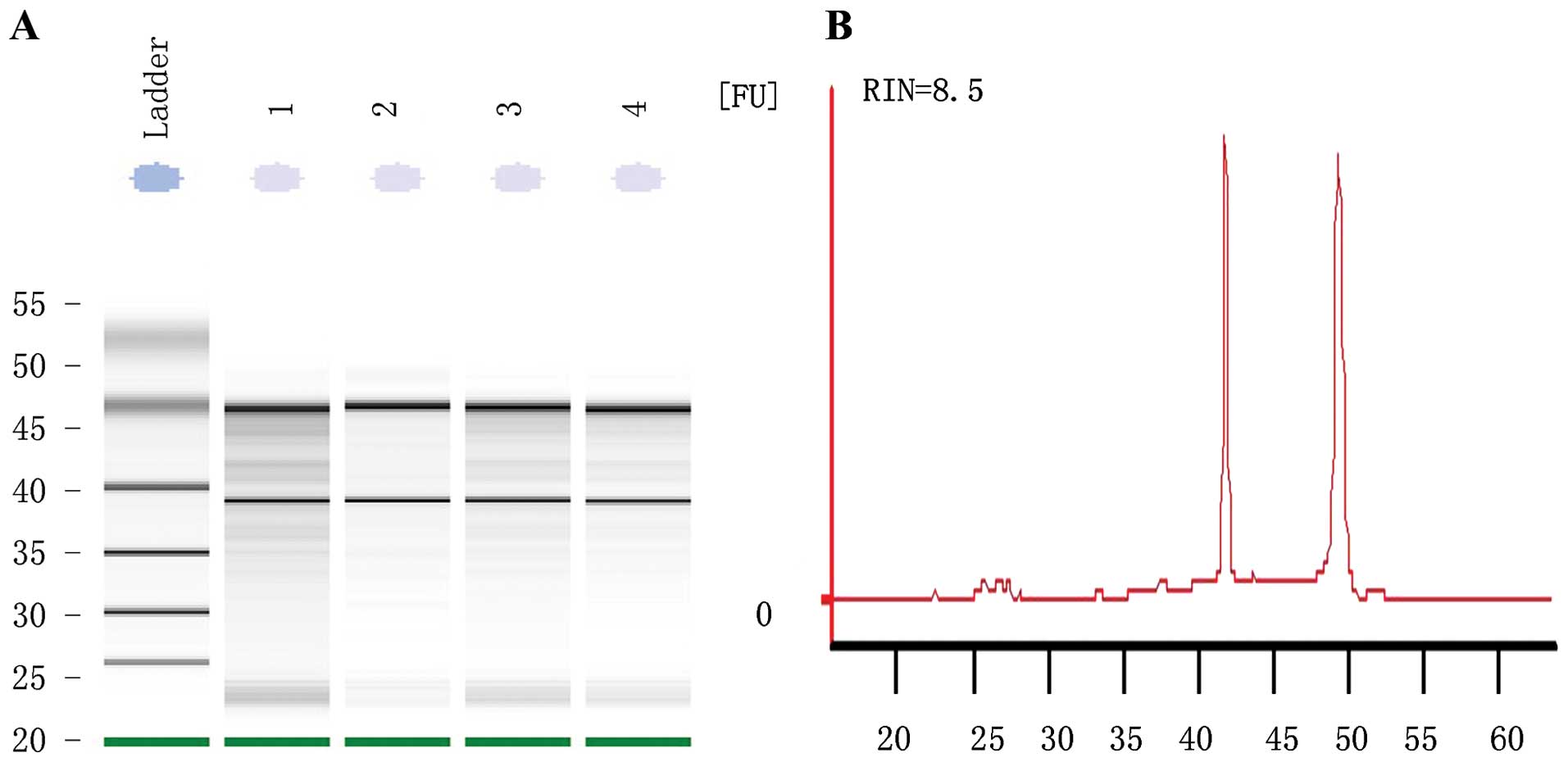
Purity and integrity of RNA
RNA extracted from the parental and resistant cells
showed an RIN number >8, suggesting the high purity of total RNA
without any protein or DNA residual (Fig. 2). The formaldehyde denaturing
agarose gel electrophoresis image showed that the 28S:18S band
neared 2:1 in a clear and trailing-free manner, without any
non-specific band, suggesting the integrity of degradation-free
RNA. The quality control experiment confirmed the suitability of
the RNA sample for further miRNA microarray and reverse
transcription PCR.
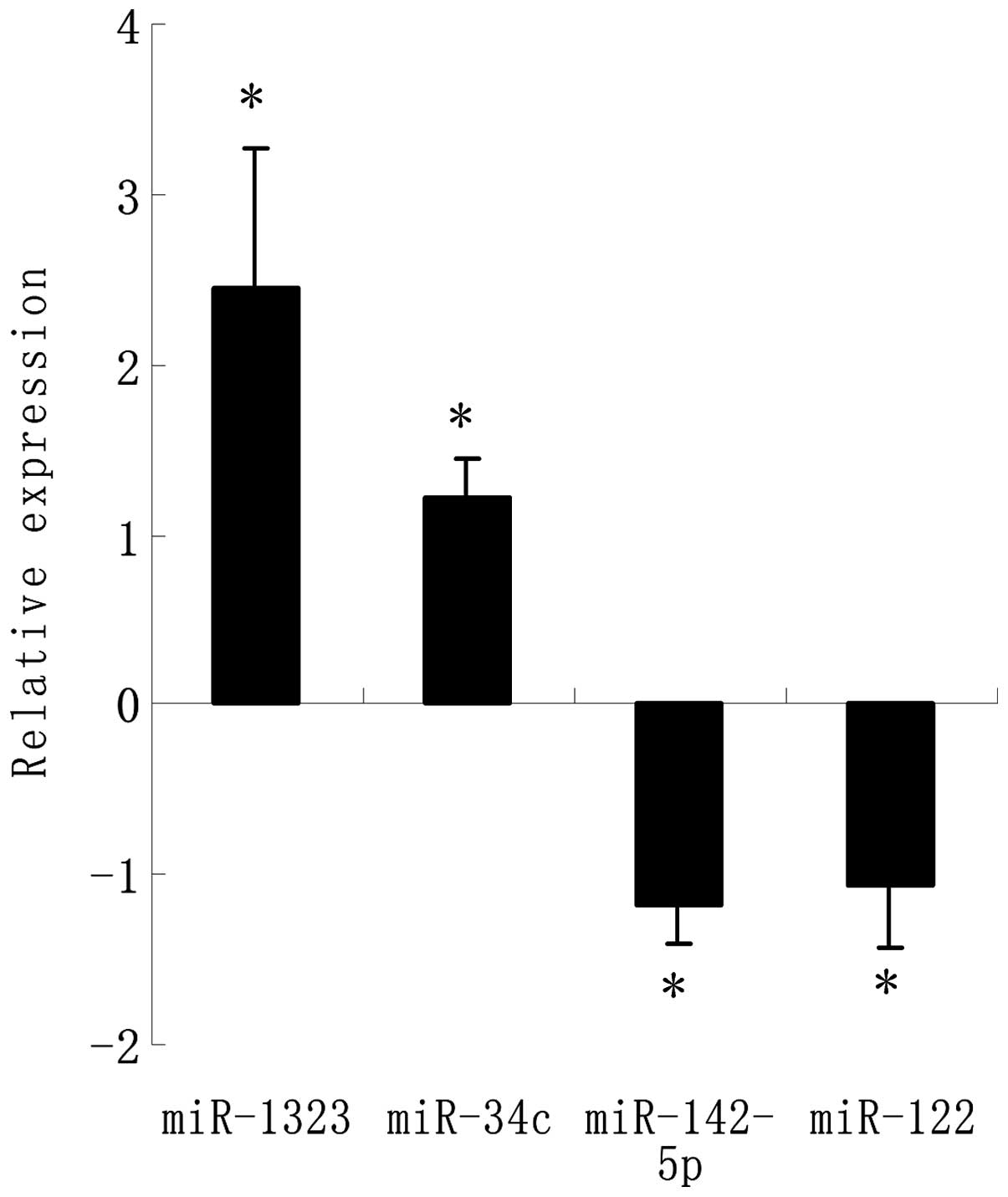
Expression of miRNAs and qRT-PCR
verification between the control and acquired radioresistant
cells
A microarray platform optimized for the analysis of
a panel of human miRNAs was used to analyze and compare the pattern
of miRNA expression between the parental A549 cell line and its
counterpart resistant cell line. The expression profile of 43
miRNAs significantly changed (2.0- to 14.0-fold) including 25
upregulated miRNAs and 18 downregulated miRNAs in the resistant
cells as compared to the parental cells (Table I). The four most significantly
changed miRNA were chosen to be verified by RT-PCR which showed the
same results as the array analysis (Fig. 3). Among these, miR-1323 had the
highest fold-change in the exposure tissues. Compared to those in
the parental cells, miR-1323 was significantly upregulated and
miR-142-5p was significantly downregulated (P<0.05),
respectively, in the resistance exposure group.
 | Table IDifferential expression of miRNAs in
the resistant subclone and parental cells
(resistance/parental). |
Table I
Differential expression of miRNAs in
the resistant subclone and parental cells
(resistance/parental).
| No. | miRNA | Fold changea (log2) | P-value | No. | miRNA | Fold change
(log2) | P-value |
|---|
| 1 | miR-1323 | 13.234 | 0.0032 | 23 | miR-375 | 2.344 | 0.0008 |
| 2 | miR-34c | 11.422 | 0.0051 | 24 | miR-744 | 2.123 | 0.0013 |
| 3 | miR-181 | 11.233 | 0.0178 | 25 | miR-140-3p | 2.098 | 0.0340 |
| 4 | miR-214 | 9.966 | 0.0020 | 26 | miR-142-5p | −14.098 | 0.0043 |
| 5 | miR-31 | 8.434 | 0.0411 | 27 | miR-122 | −9.856 | 0.0237 |
| 6 | miR-148a | 8.234 | 0.0069 | 28 | miR-7 | −9.678 | 0.0198 |
| 7 | miR-299-5p | 8.012 | 0.0260 | 29 | miR-146a | −7.345 | 0.0260 |
| 8 | miR-127 | 6.987 | 0.0145 | 30 | miR-126 | −5.345 | 0.0103 |
| 9 | miR-660 | 6.782 | 0.0071 | 31 | miR-424 | −5.231 | 0.0071 |
| 10 | miR-22 | 6.340 | 0.0124 | 32 | miR-494 | −5.013 | 0.0342 |
| 11 | miR-24-1 | 5.233 | 0.0048 | 33 | miR-224 | −4.875 | 0.0304 |
| 12 | miR-766 | 5.123 | 0.0021 | 34 | miR-155 | −4.456 | 0.0102 |
| 13 | miR-762 | 5.076 | 0.0161 | 35 | miR-638 | −4.123 | 0.0051 |
| 14 | miR-143 | 4.671 | 0.0052 | 36 | miR-483-3p | −4.091 | 0.0178 |
| 15 | miR-376a | 4.234 | 0.0225 | 37 | miR-92a | −3.987 | 0.0003 |
| 16 | miR-99a | 4.123 | 0.0146 | 38 | miR-505 | −3.675 | 0.0311 |
| 17 | miR-132 | 4.098 | 0.0301 | 39 | miR-125-3p | −3.102 | 0.0254 |
| 18 | miR-145 | 3.567 | 0.0169 | 40 | miR-572 | −2.514 | 0.0260 |
| 19 | miR-125b | 3.123 | 0.0024 | 41 | miR-100 | −2.287 | 0.0175 |
| 20 | miR-130a | 2.987 | 0.0071 | 42 | miR-720 | −2.173 | 0.0091 |
| 21 | miR-423-5p | 2.871 | 0.0176 | 43 | let-7e | −2.098 | 0.0105 |
| 22 | miR-497 | 2.456 | 0.0248 | | | | |
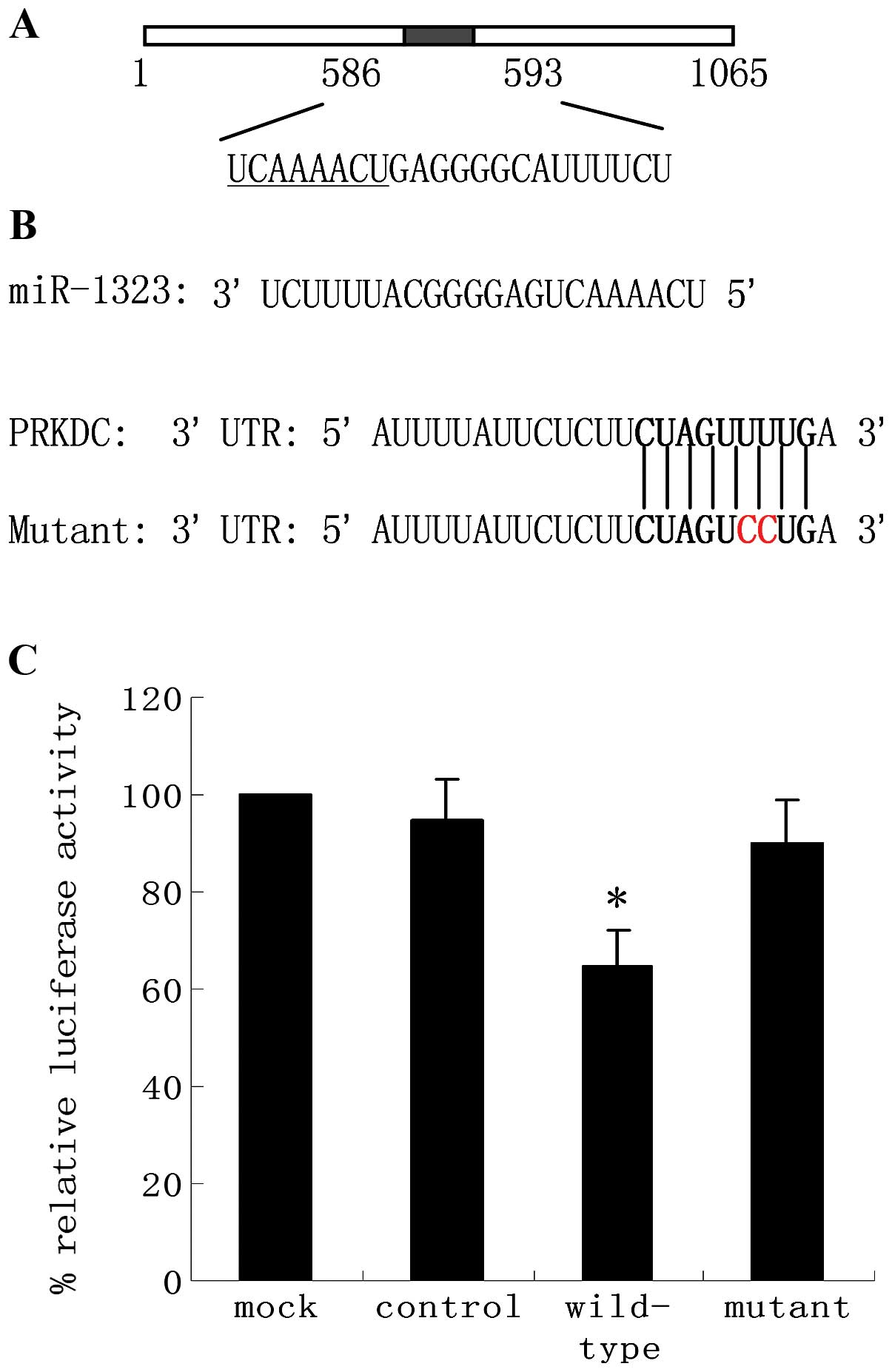
miR-1323 directly suppresses expression
of the luciferase reporter containing the PRKDC 3′UTR
To identify miRNAs that regulate human PRKDC
expression, we used the web-based algorithms TargetScan, miRanda
and PITA. All these tools predicted a putative binding site for
miR-1323 in the PRKDC 3′UTR (Fig.
4A). To validate the interaction between miR-1323 and the PRKDC
3′UTR, we cloned full-length wild-type PRKDC 3′UTR into the
reporter vector to serve as the 3′UTR of the luciferase coding
region. Acquired resistant cells were used to generate stable
wild-type PRKDC-luciferase cell lines. These cells were then
transfected with miR-1323 or miR-control, as a negative control,
and luciferase activity was measured. miR-1323 overexpression
increased luciferase activity driven by PRKDC 3′UTRs. To determine
whether this effect is direct, the predicted miR-1323-binding sites
in the 3′UTR of luciferase-PRKDC plasmids were mutated generating
luciferase PRKDC-mutant constructs (Fig. 4B). Co-transfection of lung cells
with the parental luciferase construct without the PRKDC 3′UTR did
not significantly change the expression of the reporter. However,
when the miR-1323 target site from the PRKDC 3′UTR was inserted
into the luciferase construct, expression of luciferase was
strongly increased when co-transfected with miR-1323 (Fig. 4C). This enhanced expression was
suppressed by a single-base mutation in the binding site. Taken
together, these data indicate that miR-1323 interacts with specific
elements in the 3′UTR of PRKDC.
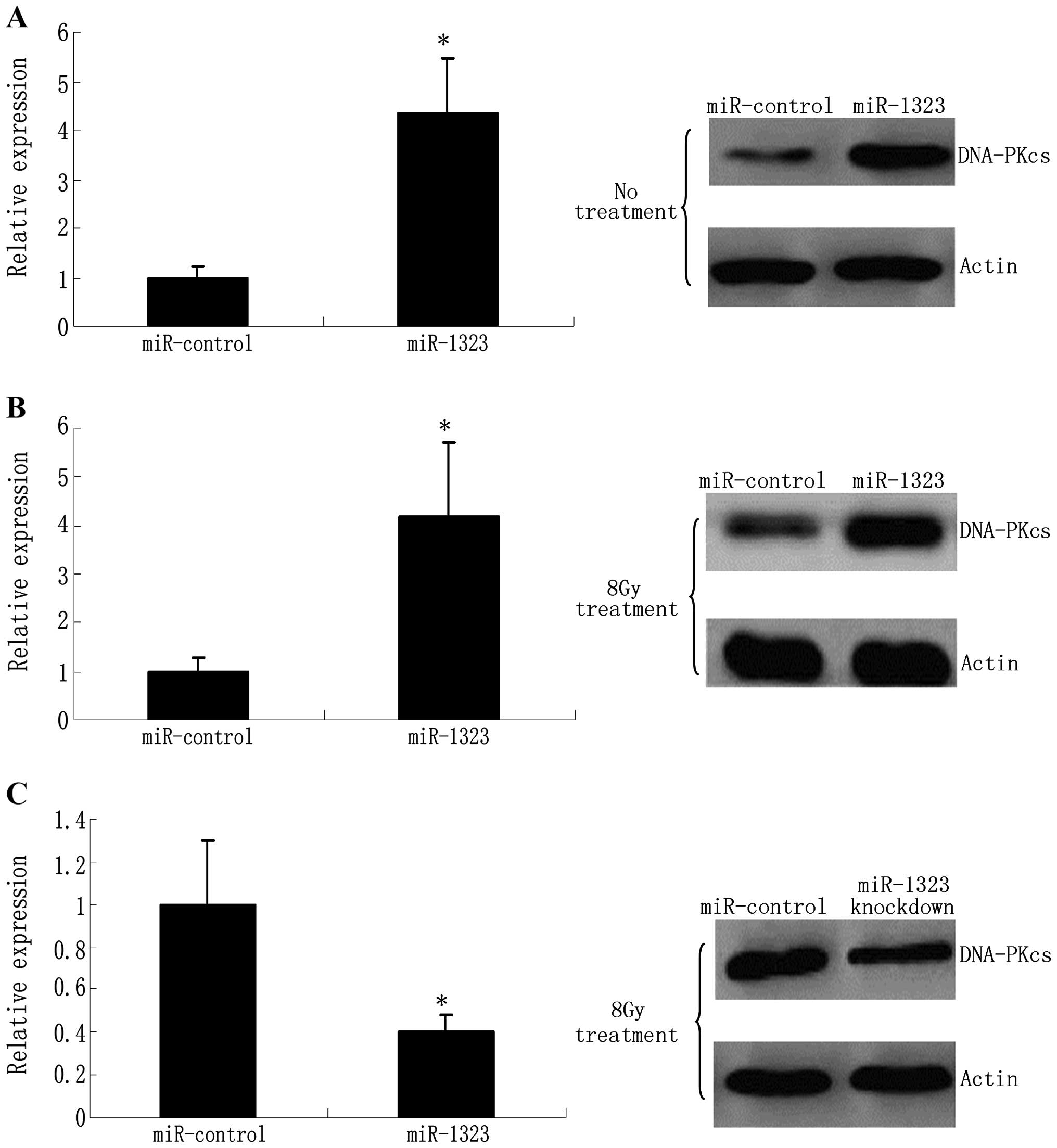
miR-1323 promotes the production of
PRKDC
To investigate whether miR-1323 modulates endogenous
PRKDC expression, we examined the effects of miR-1323
overexpression on PRKDC protein levels in human primary lung cell
lines. Upon transfection, miR-1323 was overexpressed 5-fold in the
resistant cells as determined by real-time quantitative PCR
(RT-qPCR) (Fig. 5A). Nevertheless,
miR-1323 overexpression caused an increase in the DNA-PKcs protein
levels compared with the miR-control in the cell lines (Fig. 5A).
miR-1323 knockdown effects cellular
response to radiation
Owing to the role of DNA-dependent protein kinases
(DNA-PKcs) in response to genotoxic insults, we explored the
regulation of miR-1323 and DNA-PKcs following genotoxic stress
stimuli. Firstly, resistant cells were treated with 8-Gy radiation.
Radiation-induced accumulation of DNA-PKcs protein was found to
correlate with a concomitant increase in miR-1323 levels (Fig. 5B). These data suggest that an
increase in miR-1323 and accumulation of DNA-PKcs are a general
response to DNA double-break DNA adducts caused by radiation. To
understand the role of miR-1323 in response to DNA damage, we
knocked down miR-1323 in resistant cells that were then treated
with 8 Gy. Unlike the control cells, miR-1323-knockdown cells
failed to recruit the DNA-PKcs protein (Fig. 5C).
Discussion
In an effort to better understand the mechanisms
underlying radioresistance, different radioresistant cell models,
including the glioma MGR2R (17),
glioblastoma U251 (18), colon
adenocarcinoma WiDr (19) and small
cell lung cancer cell line H69 (20), have been generated using distinct IR
exposure methods. Repeated low-dose IR exposure and sublethal IR
exposure are the most frequently adopted strategies that are used
to establish radioresistant cancer cell models. In our opinion, the
gradually increasing doses of IR exposure decreased the potential
for cellular contamination after repeated low-dose IR exposure and
avoided sudden cell death during sublethal IR exposure. However,
the manner in which IR exposure can maximally enhance the
radioresistant capability remains to be investigated in the future
(10).
miRNAs are considered to be involved in multiple
malignant cell behaviors including radioresistance (21,22).
Previous studies have used miRNA arrays to identify different miRNA
expression profiles that are associated with cancer initiation and
progression (23–25). In the present study, high-throughput
sequencing technology was applied to discover novel miRNAs
associated with radioresistance. Our results demonstrated that 18
known miRNAs were downregulated and 25 known miRNAs were
upregulated. Some of the previously identified miRNAs have been
reported to regulate the proliferation, apoptosis, migration and
invasion of cancer cells and have prognostic value (10).
Among these, miR-1323 exhibited the highest
fold-change in the radioresistant cells. Further functional study
revealed its binding site on PRKDC (NM_001081640). This gene
encodes the catalytic subunit of the DNA-PK. It functions with the
Ku70/Ku80 heterodimer protein in DNA double-strand break repair and
recombination. The protein encoded is a member of the PI3/PI4
kinase family. miR-1323 is located on chromosome 19. This sequence
was identified as an miRNA candidate by Berezikov et al
(26), and was confirmed later by
cloning (27). A previous study
reported that it was overexpressed in radioresistant NPC cancer
cells and it had significantly lower expression in skeletal muscle
from aged persons (28). All of
these implied its possible critical role in the DNA repair
pathway.
Studies have identified some novel miRNAs which are
involved in the DNA repair pathway and regulate the DNA damage
response to radiation (29–31). Yan et al (32) identified miR-101 as a molecule able
to sensitize cancer cells to IR by targeting the 3′UTR of DNA-PKcs
and ATM transcripts. The authors demonstrated that miR-101
overexpression could be used for rendering tumor cells more
sensitive to radiation in in vitro and in vivo
models. miR-18a was able to affect DNA damage response mechanisms
through ATM downregulation (33).
miR-18a was overexpressed in breast cancer cell lines and tumors
and its ectopic expression downregulated ATM by direct interaction
with the 3′UTR of the gene. ATM siRNA and miR-18a overexpression
caused reduction of homologous recombination and DNA repair in
breast cancer cells, making them more sensitive to ionizing
radiation. Conversely, inhibition of miR-18a led to an increase in
homologous recombination and DNA repair efficiency, thus reducing
cellular radiosensitivity. Several miRNAs able to inhibit
γH2AX foci formation were identified. miR-138
specifically targeted the H2AX 3′UTR, reducing its
expression and inducing chromosomal instability after DNA damage.
miR-138 overexpression inhibited homologous recombination and
increased sensitivity to DNA damaging agents. Chang et al
(34) showed that miR-3928 was
induced by ionizing radiation in HeLa cells and targeted the
endoribonuclease Dicer. miR-3928 overexpression promoted ATR
activation and Chk1 phosphorylation. miR-3928 overexpression was
also able to downregulate several miRNAs, including miR-185,
miR-300, and miR-663. Additionally, Wang et al (35) demonstrated that miR-185, whose
expression is reduced after ionizing radiation exposure in renal
cell carcinoma (RCC), targeted ATR. miR-185 expression sensitized
RCC cells to X-rays both in vivo and in vitro and
enhanced radiation-induced apoptosis as well as inhibition of
proliferation by repressing the ATR pathway. Therefore, miR-185
could be potentially used to radiosensitize cancer cells.
In conclusion, miRNAs involved in the regulation of
DNA damage/repair mechanisms can be considered as markers to
predict the response to radiotherapy and can be utilized hereafter
to define personalized treatments (36). The identification of miRNAs that are
associated with radioresistance may lead to more individualized and
efficient treatments for cancer patients. In this regard,
expression levels of miRNAs could be evaluated in serum and/or
tumor specimens to predict radiosensitivity and optimal radiation
dose, in order to make the treatment more effective and to limit
both side effects and normal tissue injury. In addition, expression
and activity of miRNAs able to affect the response to chemotherapy
or radiotherapy could be specifically modified and modulated to
enhance the expected therapeutic effects. The use of artificial
miRNAs with sequences able to target genes already known to have
important roles in DNA damage/repair mechanisms could be of great
impact in regulating mechanisms able to render cancer cells more
sensitive to DNA damaging agents.
Acknowledgments
We acknowledge the Guizhou Province Programs for
Science and Technology Development.
References
|
1
|
Begg AC, Stewart FA and Vens C: Strategies
to improve radiotherapy with targeted drugs. Nat Rev Cancer.
11:239–253. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kastan MB: DNA damage responses:
mechanisms and roles in human disease: 2007 G.H.A. Clowes Memorial
Award Lecture. Mol Cancer Res. 6:517–524. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hosoya N and Miyagawa K: Targeting DNA
damage response in cancer therapy. Cancer Sci. 105:370–388.
2014.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lomax ME, Folkes LK and O’Neill P:
Biological consequences of radiation-induced DNA damage: relevance
to radiotherapy. Clin Oncol (R Coll Radiol). 25:578–585. 2013.
View Article : Google Scholar
|
|
5
|
Czochor JR and Glazer PM: microRNAs in
cancer cell response to ionizing radiation. Antioxid Redox Signal.
21:293–312. 2014.Epub ahead of print. View Article : Google Scholar
|
|
6
|
Metheetrairut C and Slack FJ: MicroRNAs in
the ionizing radiation response and in radiotherapy. Curr Opin
Genet Dev. 23:12–19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Meltzer PS: Cancer genomics: Small RNAs
with big impacts. Nature. 435:745–746. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Trang P, Weidhaas JB and Slack FJ:
MicroRNAs as potential cancer therapeutics. Oncogene. 27(Suppl 2):
S52–S57. 2008. View Article : Google Scholar
|
|
9
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar :
|
|
10
|
Li G, Qiu Y, Su Z, Ren S, Liu C, Tian Y
and Liu Y: Genome-wide analyses of radioresistance-associated miRNA
expression profile in nasopharyngeal carcinoma using next
generation deep sequencing. PLoS One. 8:e844862013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weidhaas JB, Babar I, Nallur SM, Trang P,
Roush S, Boehm M, Gillespie E and Slack FJ: MicroRNAs as potential
agents to alter resistance to cytotoxic anticancer therapy. Cancer
Res. 67:11111–11116. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu ZL, Wang H, Liu J and Wang ZX:
MicroRNA-21 (miR-21) expression promotes growth, metastasis, and
chemo- or radioresistance in non-small cell lung cancer cells by
targeting PTEN. Mol Cell Biochem. 372:35–45. 2013. View Article : Google Scholar
|
|
14
|
Wouters MD, van Gent DC, Hoeijmakers JH
and Pothof J: MicroRNAs, the DNA damage response and cancer. Mutat
Res. 717:54–66. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tessitore A, Cicciarelli G, Del Vecchio F,
Gaggiano A, Verzella D, Fischietti M, Vecchiotti D, Capece D,
Zazzeroni F and Alesse E: MicroRNAs in the DNA damage/repair
network and cancer. Int J Genomics. 2014:8202482014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Uphoff CC and Drexler HG: Detecting
mycoplasma contamination in cell cultures by polymerase chain
reaction. Methods Mol Biol. 731:93–103. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng JJ, Hu Z, Xia YF and Chen ZP:
Radioresistant subline of human glioma cell line MGR2R induced by
repeated high dose X-ray irradiation. Ai Zheng. 25:45–50. 2006.In
Chinese. PubMed/NCBI
|
|
18
|
Lee HC, Kim DW, Jung KY, Park IC, Park MJ,
Kim MS, Woo SH, Rhee CH, Yoo H, Lee SH, et al: Increased expression
of antioxidant enzymes in radioresistant variant from U251 human
glioblastoma cell line. Int J Mol Med. 13:883–887. 2004.PubMed/NCBI
|
|
19
|
Virsik-Köpp P, Hofman-Hüther H, Rave-Fränk
M and Schmidberger H: The effect of wortmannin on radiation-induced
chromosome aberration formation in the radioresistant tumor cell
line WiDr. Radiat Res. 164:148–156. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Henness S, Davey MW, Harvie RM and Davey
RA: Fractionated irradiation of H69 small-cell lung cancer cells
causes stable radiation and drug resistance with increased MRP1,
MRP2, and topoisomerase IIalpha expression. Int J Radiat Oncol Biol
Phys. 54:895–902. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Su H, Jin X, Zhang X, Xue S, Deng X, Shen
L, Fang Y and Xie C: Identification of microRNAs involved in the
radioresistance of esophageal cancer cells. Cell Biol Int.
38:318–325. 2014. View Article : Google Scholar
|
|
22
|
Wang P, Zhang J, Zhang L, Zhu Z, Fan J,
Chen L, Zhuang L, Luo J, Chen H, Liu L, et al: MicroRNA 23b
regulates autophagy associated with radioresistance of pancreatic
cancer cells. Gastroenterology. 145:1133–1143. e122013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang XC, Wang W, Zhang ZB, Zhao J, Tan XG
and Luo JC: Overexpression of miRNA-21 promotes
radiation-resistance of non-small cell lung cancer. Radiat Oncol.
8:1462013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Besse A, Sana J, Fadrus P and Slaby O:
MicroRNAs involved in chemo- and radioresistance of high-grade
gliomas. Tumour Biol. 34:1969–1978. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin J, Liu C, Gao F, Mitchel RE, Zhao L,
Yang Y, Lei J and Cai J: miR-200c enhances radiosensitivity of
human breast cancer cells. J Cell Biochem. 114:606–615. 2013.
View Article : Google Scholar
|
|
26
|
Berezikov E, van Tetering G, Verheul M,
van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M,
Guryev V, Takada S, et al: Many novel mammalian microRNA candidates
identified by extensive cloning and RAKE analysis. Genome Res.
16:1289–1298. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Afanasyeva EA, Hotz-Wagenblatt A, Glatting
KH and Westermann F: New miRNAs cloned from neuroblastoma. BMC
Genomics. 9:522008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mercken EM, Majounie E, Ding J, Guo R, Kim
J, Bernier M, Mattison J, Cookson MR, Gorospe M, de Cabo R, et al:
Age-associated miRNA alterations in skeletal muscle from rhesus
monkeys reversed by caloric restriction. Aging (Albany, NY).
5:692–703. 2013.
|
|
29
|
Gasparini P, Lovat F, Fassan M, Casadei L,
Cascione L, Jacob NK, Carasi S, et al: Protective role of miR-155
in breast cancer through RAD51 targeting impairs homologous
recombination after irradiation. Proc Natl Acad Sci USA.
111:4536–4541. 2014.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo P, Lan J, Ge J, Nie Q, Guo L, Qiu Y
and Mao Q: miR-26a enhances the radiosensitivity of glioblastoma
multiforme cells through targeting of ataxia-telangiectasia
mutated. Exp Cell Res. 320:200–208. 2014. View Article : Google Scholar
|
|
31
|
Liu YJ, Lin YF, Chen YF, Luo EC, Sher YP,
Tsai MH, Chuang EY and Lai LC: MicroRNA-449a enhances
radiosensitivity in CL1–0 lung adenocarcinoma cells. PLoS One.
8:e623832013. View Article : Google Scholar
|
|
32
|
Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo
YY, Mao H, Hao C, Olson JJ, Curran WJ, et al: Targeting DNA-PKcs
and ATM with miR-101 sensitizes tumors to radiation. PLoS One.
5:e113972010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song L, Lin C, Wu Z, Gong H, Zeng Y, Wu J,
Li M and Li J: miR-18a impairs DNA damage response through
downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS
One. 6:e254542011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chang L, Hu W, Ye C, Yao B, Song L, Wu X,
Ding N, Wang J and Zhou G: miR-3928 activates ATR pathway by
targeting Dicer. RNA Biol. 9:1247–1254. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang J, He J, Su F, Ding N, Hu W, Yao B,
Wang W and Zhou G: Repression of ATR pathway by miR-185 enhances
radiation-induced apoptosis and proliferation inhibition. Cell
Death Dis. 4:e6992013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao L, Bode AM, Cao Y and Dong Z:
Regulatory mechanisms and clinical perspectives of miRNA in tumor
radiosensitivity. Carcinogenesis. 33:2220–2227. 2012. View Article : Google Scholar : PubMed/NCBI
|