Introduction
Eukaryotic chromosomes are composed of tandemly
repetitive telomere sequences that protect the ends from damage and
rearrangements. Telomere repeats are synthesized by telomerase, a
ribonucleic acid (RNA)-protein complex (1). Telomerase enables cancer cells to
achieve replicative immortality, which is one of the hallmarks of
cancer (2). Telomerase activity is
currently the most general molecular marker for the identification
of human cancer and can be detected in 85–90% of all tumors
(3). Human telomerase reverse
transcriptase (hTERT), which confers the catalytic activity of
telomerase, is the restricting factor for telomerase activity
(4). Investigations have shown that
hTERT has a significant role in cancer tumorigenesis, growth,
migration and invasion (5).
Clinical studies have also proven that the overexpression of hTERT
is associated with cancer progression and poor outcomes, although
the underlying mechanism remains unclear (6). The majority of studies have focused on
the co-regulation of hTERT via transcriptional regulation, the
presence or absence of various activators and repressors, as well
as the epigenetic pathways of DNA methylation and histone
modifications (7). Few studies
aimed to examine the regulation of hTERT via
post-transcription.
microRNAs (miRNAs) are thought to control gene
expression at the post-transcriptional level by degrading or
repressing target messenger RNAs (mRNAs) (8). miRNAs are a class of newly identified
eukaryotes, highly conserved, with a length of ~18–24 nt endogenous
non-coding single-stranded RNA. They have crucial regulatory
functions in cell differentiation, proliferation, and apoptosis
(9). Each miRNA has multiple target
genes, with well over one third of human genes appearing to be
conserved miRNA targets (10).
Recent findings have shown that the altered expression pattern of
miRNAs is involved in many forms of cancer as oncogenes and tumor
suppressors, playing an important role in cancer occurrence,
development and progression, especially in colorectal cancer (CRC)
(11).
Three different miRNAs (miR138 in human anaplastic
thyroid carcinoma cell lines, miR-1207 and miR1266 in gastric
cancer) have been identified that may influence hTERT expression
(12,13). Since a miRNA can affect hundreds of
target gene regulation, a gene can also be affected by molecular
regulation of multiple miRNAs. Therefore, other miRNAs may be
involved in the post-transcriptional regulation of hTERT.
Through retrieval of microRNA microarray data and
biological information technology analysis, the expression of
miRNAs with the potential to directly regulate hTERT was
downregulated. Reverse-transcriptase quantitative polymerase chain
reaction (RT-qPCR) was used to verify the expression of these
miRNAs in the CRC and corresponding normal tissues. Subsequently,
immunohistochemistry (IHC) was used in the detection of hTERT
protein expression in the same samples. A preliminary validation of
the miRNAs with the potential to regulate hTERT was obtained by
identifying whether there is a negative relationship between
expression of miRNAs and the hTERT IHC data.
Materials and methods
Patients, tissue samples and
follow-up
The CRC and corresponding normal tissues were
retrospectively recruited from 84 patients with CRC at the Tumor
Hospital of Guangxi Medical University between 2007 and 2010. The
present study was approved by the Institutional Review Board of the
Guangxi Medical University. None of the CRC patients had undergone
neo-adjuvant radiotherapy, chemotherapy or other treatment prior to
surgery. Pathological diagnosis was also standardised and reviewed
by committee criteria, chaired by a senior academic pathologist.
Patient clinicopathological information and follow-up data of CRC
were available from the documents of the hospital. The
clinicopathological characteristics of patients are shown in
Table I.
 | Table IThe correlation between miR-138-5p and
miR-422a expression and clinicopathological parameters in 84
colorectal cancer patients. |
Table I
The correlation between miR-138-5p and
miR-422a expression and clinicopathological parameters in 84
colorectal cancer patients.
| Parameters | Total (n=84) | miR-138-5p
| P-value | miR-422a
| P-value |
|---|
| Low (n=43) | High (n=41) | Low (n=42) | High (n=42) |
|---|
| Age (years) | | | | 0.166 | | | 0.450 |
| <50 | 21 | 8 | 13 | | 9 | 12 | |
| ≥50 | 63 | 35 | 28 | | 33 | 30 | |
| Gender | | | | 0.497 | | | 0.381 |
| Male | 46 | 22 | 24 | | 21 | 25 | |
| Female | 38 | 21 | 17 | | 21 | 17 | |
| Tumor size (cm) | | | | 0.130 | | | 0.378 |
| <5 | 36 | 15 | 21 | | 20 | 16 | |
| ≥5 | 48 | 28 | 20 | | 22 | 26 | |
| Tumor site | | | | 0.257 | | | 0.659 |
| Colon | 48 | 22 | 26 | | 23 | 25 | |
| Rectum | 36 | 21 | 15 | | 19 | 17 | |
| Differentiation | | | | 0.071 | | | 0.825 |
| G1+G2 | 49 | 21 | 28 | | 24 | 25 | |
| G3+G4 | 35 | 22 | 13 | | 18 | 17 | |
| T | | | | 0.238 | | | 0.242 |
| T1+T2 | 14 | 9 | 5 | | 9 | 5 | |
| T3+T4 | 70 | 34 | 36 | | 33 | 37 | |
| N | | | | 0.126 | | | 0.023a |
| Negative | 30 | 12 | 18 | | 10 | 20 | |
| Positive | 54 | 31 | 23 | | 32 | 22 | |
| M | | | | 0.000a | | | 0.287 |
| Negative | 66 | 27 | 39 | | 31 | 35 | |
| Positive | 18 | 16 | 2 | | 11 | 7 | |
| TNM stage | | | | 0.123 | | | 0.064 |
| I+II | 28 | 11 | 17 | | 10 | 18 | |
| III+IV | 56 | 32 | 24 | | 32 | 24 | |
Retrieval of microRNA microarray
data
Gene Expression Omnibus (GEO) is a public functional
genomics data repository supporting MIAME compliant data
submissions. The GEO datasets were searched for relevant studies
using the terms: (colon or rectum or rectal or colorectal) and
(cancer or tumor or neoplasm). The study type was limited to
‘non-coding RNA profiling by array’, the organism was ‘homo
sapiens’ and the attribute name was ‘tissue’. The dataset was
included in our analysis when the certain criteria were met, i.e.,
the dataset was required to i) be microarray data expression of
miRNAs in CRC; ii) be controlled studies in the CRC and
corresponding normal tissue; iii) include ≥3 samples and iv) be
available for download.
Biological information technology
The common analysis methods of extract
differentially regulated miRNAs from the dataset included
previously were: significance analysis of microarrays (SAM), CyberT
and rank products (RP), of which CyberT is the most commonly used
analytical tool of computational biology, and bioinformatics
analysis as its algorithm can be completely implemented in the
linear models for the microarray data (Limma) package (14). In this study, the data were analyzed
by using language R (3.2 versions). To test for differential
expression, CyberT algorithm and the bayesian adjusted t-statistics
from the Limma package statistical methods were used (15,16). A
multiple-testing correction based on the false discovery rate (FDR)
was performed.
The miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/) is
a publically available comprehensive resource, hosting the
predicted as well as the experimentally validated miRNA-target
interaction pairs (17). miRWalk
online software was used to predict miRNAs with the potential to
interact with hTERT. The candidate hTERT-targeting miRNAs were
selected for analysis in the subsequent experiments.
Reverse transcriptase quantitative
polymerase chain reaction
Reverse transcriptase quantitative polymerase chain
reaction (RT-qPCR) was performed to detect the included miRNAs
expression. Total RNA was extracted from tissue samples using a
Qiagen miRNeasy Mini kit (Qiagen GmbH, Hilden, Germany) according
to the manufacturer’s instructions. The total RNA was then eluted
in a 30-μl volume of elution buffer. RNAs were quantified by
Nanodrop 2000 (PeqLab Biotechnology GmbH, Erlangen, Germany). RNA
samples were converted to cDNA using miScript II RT kit (Qiagen).
qPCR was performed in a total volume of 20 μl reaction
mixture containing cDNA product, specific primers for each miRNA
(Invitrogen, Carlsbad, CA, USA), miScript Universal primer
(Qiagen), SYBR-Green qPCR Master Mix (Thermo Fisher Scientific,
Waltham, MA, USA) and nuclease-free water. The U6 snRNA gene was
used as an internal control. The sequences of the specific forward
primers are shown in Table II. PCR
reactions were conducted at 95°C for 7 min, followed by 40 cycles
of 95°C for 10 sec, and 60°C for 30 sec in a Mx3000P Real-Time
Quantitative PCR system (Agilent Technologies, Inc., Santa Clara,
USA). To minimize data variation in separate runs, paired cancer
and corresponding normal tissues from the same patient were
detected on the same runs. The Ct value was calculated using
MxPro-Mx3000P software (Agilent Technologies) using the automatic
threshold setting. The reactions were run in triplicate. Results
were presented as the levels of expression following normalization
to U6 using the 2−Δ∆Ct method (18).
 | Table IIPCR primer sequences for miRNAs and
U6. |
Table II
PCR primer sequences for miRNAs and
U6.
| Genes | Forward primers
(5′–3′) |
|---|
|
hsa-miR-133a-3p |
TTTGGTCCCCTTCAACCAGCTG |
| hsa-miR-133b |
TTTGGTCCCCTTCAACCAGCTA |
| hsa-miR-422a |
ACTGGACTTAGGGTCAGAAGG |
| hsa-miR-29c-3p |
GCTAGCACCATTTGAAATCGGTTA |
| hsa-miR-124-3p |
TAAGGCACGCGGTGAATG |
| hsa-miR-138-5p |
GCAGCTGGTGTTGTGAATCA |
| hsa-miR-150-5p |
CTCCCAACCCTTGTACCAGTG |
|
hsa-miR-378a-3p |
ACTGGACTTGGAGTCAGAAGGC |
| U6 |
CGCAAGGATGACACGCAAATTCGT |
Immunohistochemistry
Sections (4-μm) were cut from the selected
paraffin blocks and dried overnight at 37°C. The sections were
dewaxed and rehydrated, exposed to 3% H2O2
solution for 10 min to block endogenous peroxidase and subjected to
antigen retrieval in Tris-EDTA (pH 9.0). Subsequently, the slides
were incubated with the rabbit monoclonal antibody against hTERT
(1:100; Abcam, Cambridge, MA, USA), overnight at 4°C. Following
three washes in 0.01 mol/l phosphate-buffered saline (PBS, pH=7.4),
labeling was detected by adding a secondary antibody for 15 min at
room temperature and diaminobenzidine (both from Maxim-Bio, Fuzhou,
China) after washing in PBS again. The sections were then
counterstained with hematoxylin, dehydrated, and mounted. Negative
controls were conducted by replacing the primary antibody with
PBS.
The sections were blindly and independently assessed
microscopically by two well-trained pathologists. For the
assessment of hTERT, five high-power fields in each specimen were
randomly selected, and yellow or brown staining of the cytoplasm or
nuclear was considered positive staining. The hTERT immunostaining
score was calculated with a semi-quantitative scoring system as the
intensity (0, no staining; 1, weak staining; 2, moderate staining;
3, strong staining) and the percentage (extent staining) of tumor
cells that were stained (0, <10% of tumor cells stained; 1,
10–50% of positive cells; 2, >50 and <75% of positive cells;
3, >75% of positive cells). An overall score was obtained as the
product of the intensity and distribution of positive staining
(19). Cases with 0 points were
considered to be negative (0), cases with a final score of 1–3 as
weakly positive (1+), cases with a final score of 4–7 as moderately
positive (2+) and cases with a final score of >7 as strongly
positive (3+).
Statistical analysis
Statistical analysis was performed using Statistical
Program for Social Sciences (SPSS) software, version 16.0 (SPSS
Inc., Chicago, IL, USA). RT-qPCR data were expressed as medians
(IQR, interquartile range). Possible differences between the CRC
and corresponding normal tissue groups were analyzed using the
Wilcoxon’s signed-rank test. Correlations between the expression of
hTERT proteins (immunohistochemical scores) with the expression of
miRNAs were evaluated using the Spearman’s rank-order correlation
coefficient. Survival curves were obtained by the Kaplan-Meier
method. A comparison between curves was made using the log-rank
test. The tests were performed as two-tailed and the level of
significance was set as P<0.05.
Results
Identification of downregulated miRNAs
through retrieval of microRNA microarray data and language R
analysis
A total of eight datasets fulfilled the inclusion
criteria for our analysis: GSE10259, GSE33127, GSE35602, GSE38389,
GSE39845, GSE49246, GSE18392 and GSE35982, respectively.
Subsequently, we used language R to analyze the eight included
data. First, 173 significantly downregulated miRNAs were selected
with log2FC<−0.5 and a P-value of <0.05 by
analyzing language R. Second, according to the number of chip data
source arrangement (highest score, 8 points and lowest score 1
point), 45 significantly downregulated miRNAs with a frequency of
≥2 were identified. An intersection of the 45 miRNAs and
downregulated miRNAs provided from the published literature
corresponding to the eight microarray data was performed.
Thirty-two significantly downregulated miRNAs in CRC tissue were
found.
Identification of candidate
hTERT-targeting miRNAs through biological information
technology
Initially, 359 miRNAs were found by using miRWalk
online software. Points ≥2 were selected, duplicate values were
removed, and ultimately 125 hTERT regulation-related miRNAs were
obtained according to the integral arrangement (results of the 1
point represent only one software prediction, with a maximum of 6
points and a minimum of 1 point). miRNAs were obtained by the
intersection of the 32 significantly downregulated miRNAs and the
125 miRNAs associated with the regulation of hTERT. Eight miRNAs
with the potential to interact with hTERT were predicted:
miR-29c-3p, miR-124-3p, miR-133a-3p, miR-133b, miR-138-5p,
miR-150-5p, miR-378a-3p and miR-422a, respectively.
Validation of the 8 selected miRNAs by
RT-qPCR
To validate the relative expression levels of 8
selected miRNAs in CRC and corresponding normal tissues, we
performed qPCR experiments. The results revealed that the
expression level in CRC tissues of miR-124-3p, miR-133a-3p,
miR-133b, miR-138-5p, miR-150-5p, miR-378a-3p and miR-422a
(P<0.0001 for all) were statistically significantly
downregulated when compared with the corresponding normal tissues.
However, there was no significant difference in the levels of
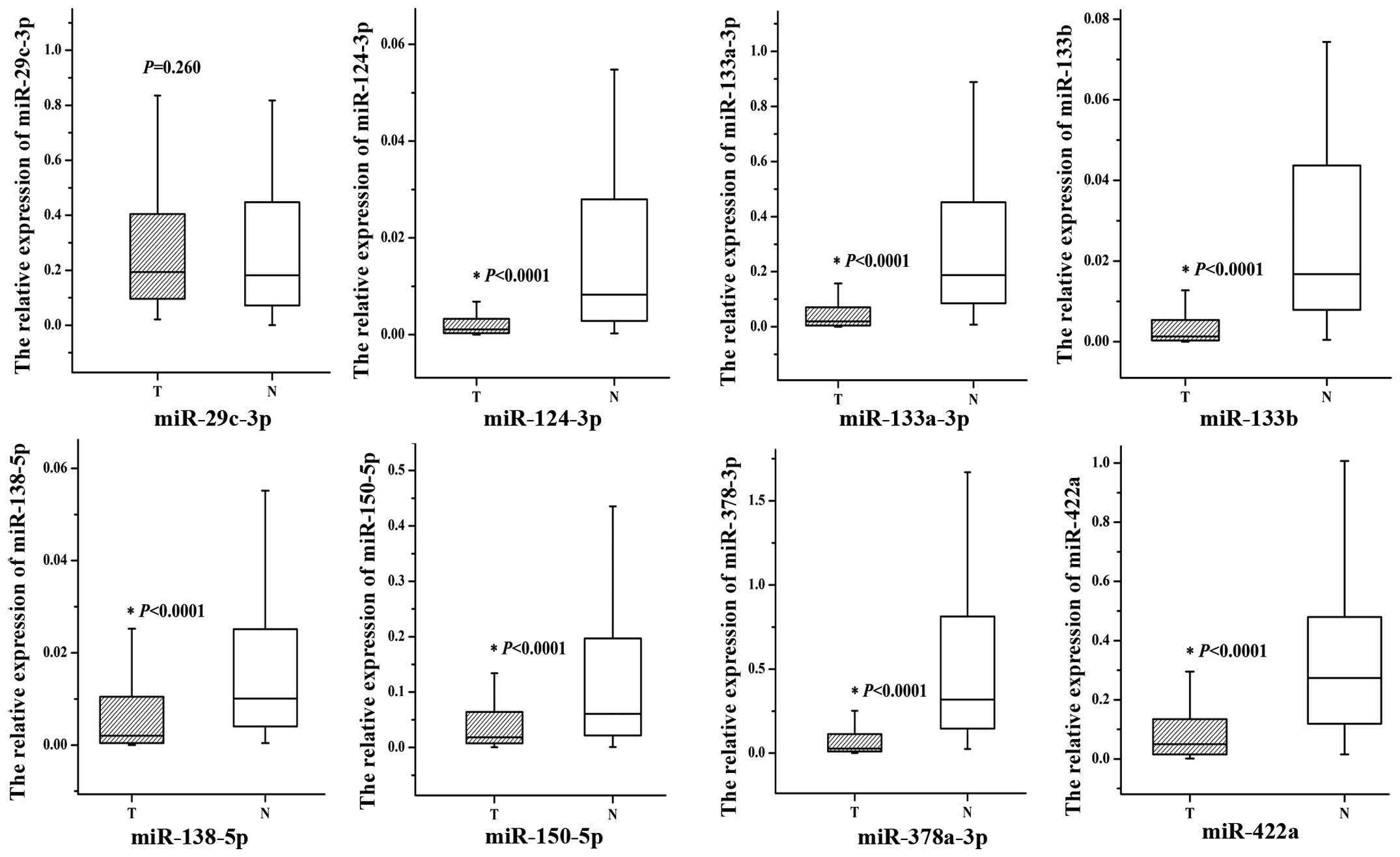
miR-29c-3p (P=0.260, Fig. 1 and
Table III). The amplification
curves and the melting curves of 8 miRNAs and U6 in the qPCR phase
are shown in Fig. 2.
 | Table IIImiRNAs levels of colorectal cancer
and corresponding normal tissue groups using RT-qPCR. |
Table III
miRNAs levels of colorectal cancer
and corresponding normal tissue groups using RT-qPCR.
| microRNA | Median (25–75th)
| P-value |
|---|
| Cancer group | Normal group |
|---|
| miR-29c-3p | 0.193822
(0.096222–0.405425) | 0.181904
(0.070936–0.449074) | 0.0260 |
| miR-124-3p | 0.001082
(0.000298–0.003321) | 0.008278
(0.002707–0.028471) | <0.0001 |
| miR-133a-3p | 0.019915
(0.004420–0.073774) | 0.187506
(0.083645–0.456176) | <0.0001 |
| miR-133b | 0.001285
(0.000296–0.005439) | 0.016779
(0.007922–0.043813) | <0.0001 |
| miR-138-5p | 0.002022
(0.00380–0.010544) | 0.010097
(0.004016–0.025342) | <0.0001 |
| miR-150-5p | 0.018194
(0.007202–0.064146) | 0.060581
(0.020490–0.197861) | <0.0001 |
| miR-378a-3p | 0.027426
(0.009510–0.114644) | 0.318763
(0.145602–0.820874) | <0.0001 |
| miR-422a | 0.050255
(0.014860–0.134671) | 0.273810
(0.117263–0.488233) | <0.0001 |
Immunohistochemical expression of hTERT
protein in paraffin sections and the correlations with expression
levels of the 7 verified miRNAs
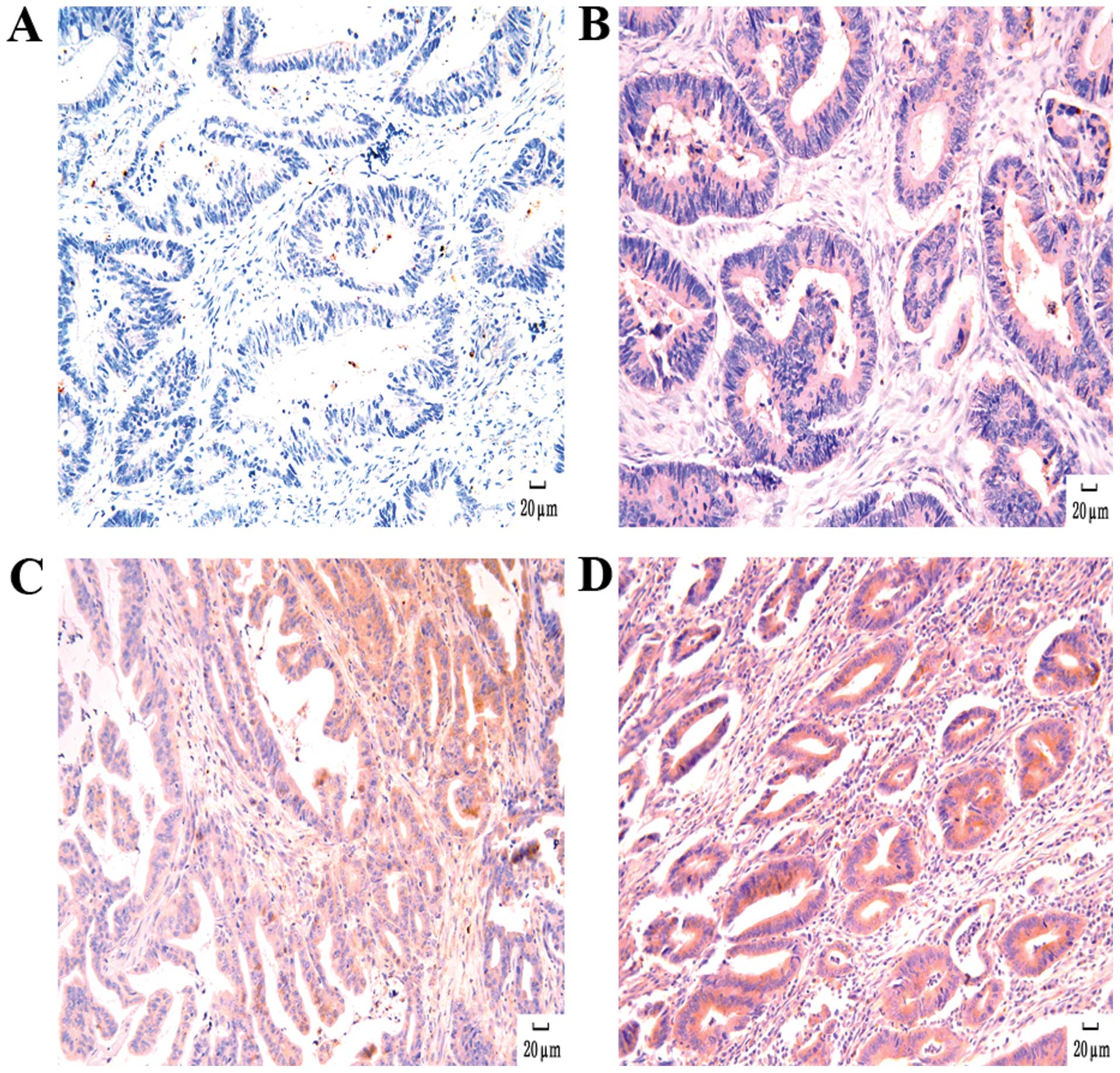
Positive hTERT inmunoreactivity was observed in
cancer tissue of 60 patients, while in 24 patients it was negative.
As shown in Fig. 3, the positive
expression of hTERT was localized to the cytoplasm or nuclei in
colorectal tumor cells. Spearman’s rank-order correlation
coefficient between miRNAs and hTERT protein expression was
calculated (r). As the frequency distribution for miRNAs in the
group was non-parametric, the median (non-parametric distribution)
was used as the cut-off level of expression to divide patients into
the high- and low-expression groups for statistical testing. Our
results suggested that hTERT protein showed a significant negative
correlation with the expression levels of miR-138-5p (r=−0.362,
P=0.001) and miR422a (−0.306, P=0.005), while the correlation
between hTERT and other miRNAs (miR-124-3p, miR-133a-3p, miR-133b,
miR-150-5p and miR-378a-3p) revealed no significant negative
correlation (Table IV). Therefore,
the downregulated expression of miR-138-5p and miR-422a potentially
inhibited hTERT expression.
 | Table IVThe relationship between the
expression of miRNAs and hTERT protein expression in colorectal
cancer tissues. |
Table IV
The relationship between the
expression of miRNAs and hTERT protein expression in colorectal
cancer tissues.
| Groups | Total (84) | hTERT
| r | P-value |
|---|
| − (24) | + (34) | ++ (16) | +++ (10) |
|---|
| miR-124-3p | | | | | | −0.147 | 0.183 |
| Low | 42 | 8 | 21 | 6 | 7 | | |
| High | 42 | 16 | 13 | 10 | 3 | | |
| miR-133a-3p | | | | | | −0.208 | 0.058 |
| Low | 42 | 9 | 17 | 8 | 8 | | |
| High | 42 | 15 | 17 | 8 | 2 | | |
| miR-133b | | | | | | −0.018 | 0.874 |
| Low | 42 | 11 | 19 | 6 | 6 | | |
| High | 42 | 13 | 15 | 10 | 4 | | |
| miR-138-5p | | | | | | −0.362 | 0.001a |
| Low | 43 | 6 | 19 | 9 | 9 | | |
| High | 41 | 18 | 15 | 7 | 1 | | |
| miR-150-5p | | | | | | −0.185 | 0.092 |
| Low | 42 | 8 | 19 | 9 | 6 | | |
| High | 42 | 16 | 15 | 7 | 4 | | |
| miR-378a-3p | | | | | | −0.064 | 0.562 |
| Low | 42 | 9 | 21 | 7 | 5 | | |
| High | 42 | 15 | 13 | 9 | 5 | | |
| miR-422a | | | | | | −0.306 | 0.005a |
| Low | 42 | 7 | 17 | 11 | 7 | | |
| High | 42 | 17 | 17 | 5 | 3 | | |
Relationship between miR-138-5p and
miR-422a expression and clinicopathological factors in CRC
patients
For statistical testing, we primarily divided
patients into the high- and low-expression groups according to the
median value of each miRNA. The expression levels of miR-138-5p and
miR-422a were compared between different cohorts depending on
various clinicopathological characteristics (Table I). There were no significant
variations in miR-138-5p between the subgroups regarding age,
gender, tumor size, tumor site, differentiation, the depth of
invasion, lymph-node metastasis and TNM stage. However, a
statistically significant difference in miR-138-5p expression was
observed with regard to distant metastasis (P<0.000). A
significant difference in miR-422a expression was identified
between the subgroups according to lymph-node metastasis (P=0.023).
However, the data showed no significant association between
miR-422a relative expression levels and other parameters.
Downregulated miR-422a associated with
shorter overall survival (OS)
The survival analysis of the 84 studied patients was
implemented using information available from the clinical
follow-ups. The median follow-up was 58 months (range, 8–86
months). At the end of follow-up, 62 patients succumbed, 22
patients remained alive, and the follow-up rate was 100%. Results
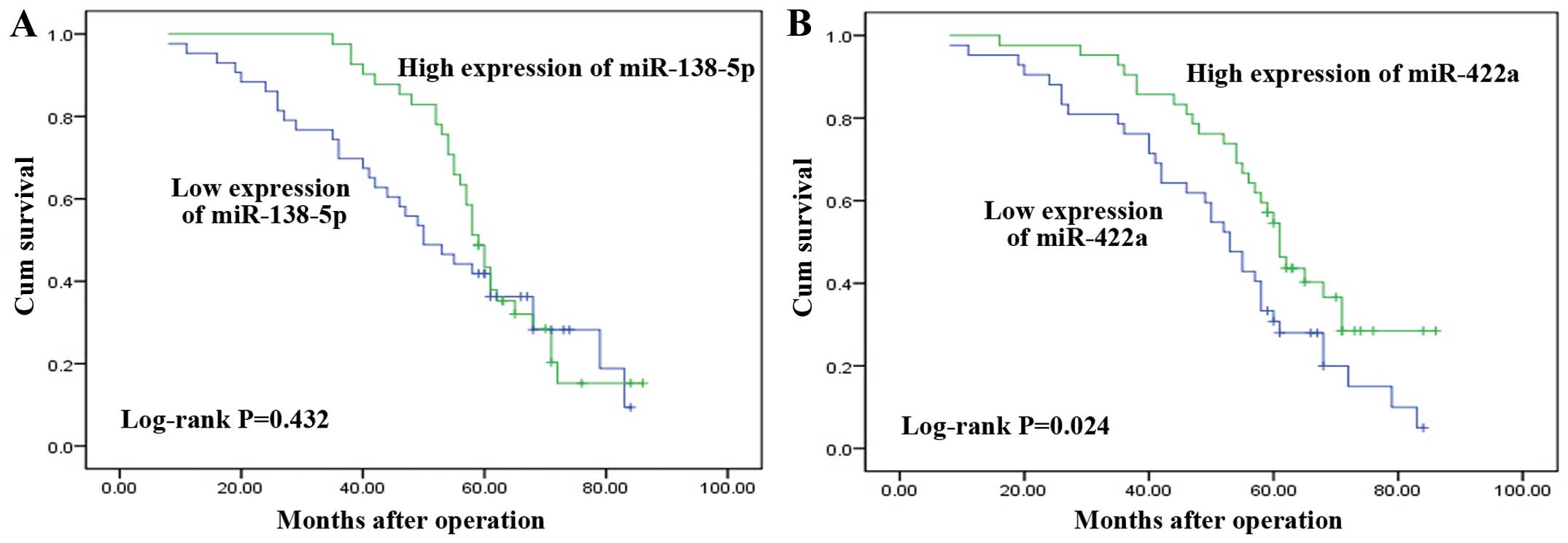
of the Kaplan-Meier method and log-rank test showed that the OS of
CRC patients with a high-expression miR-138-5p was longer than that
of patients with low-expression miR-138-5p (the estimated median OS
time was 59 and 50 months, respectively), albeit the difference was
not statistically significant (P=0.432, Fig. 4A). However, patients with
low-expression miR-422a had significantly poorer OS (P=0.024,
Fig. 4B). The estimated median OS
time was 61 months in the miR-422a high-expression group but 53
months in the miR-422a low-expression group. Our result suggested
that a lower expression of miR-422a was associated with reduced OS
in CRC patients.
Discussion
CRC is the third most common cancer in humans and
the mortality of CRC accounts for ~9% of all cancer deaths
(20). Therefore, gaining a better
understanding of the biological behavior of the tumor is critical
to improve treatment strategies and patient outcomes. The
dysregulation of miRNAs and hTERT have been suggested to play a
crucial role in the regulation of tumorigenesis and metastases of
various types of cancer. The finding that miRNAs influence cell
proliferation, apoptosis, metabolism, and transformation in many
subtypes of cancer at a post-transcriptional level, has led to
great attention being focused on novel tumor-suppressor
oncogene-targeting miRNAs.
In this study, we initially identified 8 miRNAs that
were potentially involved in the regulation of hTERT by using a
combination of retrieving and analysing microRNA microarray data
and bioinformatics analysis. The expression levels of the 7 miRNAs
(miR-124-3p, miR-133a-3p, miR-133b, miR-138-5p, miR-150-5p and
miR-378a-3p, miR-422a) were found to be significantly decreased in
84 pairs of the CRC tissues when compared with their matched
corresponding normal tissues using RT-qPCR. Our results suggest
that hTERT protein showed a significant negative correlation with
the expression levels of miR-138-5p (r=−0.362, P=0.001) and miR422a
(−0.306, P=0.005). Thus, miR-138-5p and miR-422a may be more likely
to inhibit hTERT expression potentially compared to the remaining 5
miRNAs.
To the best of our knowledge, this is the first
study to screen and validate preliminarily miRNAs with regulation
of hTERT in CRC. Other studies have identified 3 different miRNAs
that influence hTERT expression. miR-138 was the first demonstrated
hTERT-targeting miRNA. Mitomo et al (13) have shown that the overexpression of
miR-138 induced a reduction in hTERT protein expression in human
anaplastic thyroid carcinoma. Additionally, using luciferase
reporter assay those authors confirmed target specificity between
miR-138 and the hTERT 3′-untranslated region. Chen et al
(12) determined that miR-1207-5p
and miR-1266 interact with the 3′UTR of hTERT and suppress gastric
cancer growth and invasion by targeting hTERT. However, in addition
to the main mechanism described above, miRNAs can also regulate the
expression of hTERT through influence of other transcription
factors. Wang et al (21)
found that miR-21 regulates hTERT expression mediated by STAT3,
thereby controlling glioblastoma cell growth. Moreover, the scope
of functional miRNA-mRNA interactions have been expanded from RNA
3′UTRs to include the coding regions of the targeted RNAs (22). These mechanisms described above
offered possible interpretations of the observed negative
statistical association between the downregulated miR-138-5p and
miR-422 and the overexpression of hTERT protein.
In contrast to hTERT promoting tumor metastasis,
miR-138 and miR-422a have been found to be potential tumor
suppressors in certain types of cancer. It has been shown that
miR-138 may have an effect on tumor metastasis by targeting SOX4
and HIF1a in ovarian cancer and targeting MMP2/MMP9 in
cholangiocarcinoma (23,24). Downregulation of miR-138 promotes
metastasis by directly targeting TWIST2 and is associated with
lymph-node metastasis, distant metastasis, and predicted poor
prognosis in CRC (25). Previous
findings suggested that miR-422a may play a protective role against
CRC, which was shown by its decreased expression in CRC when
compared to normal tissue (26).
Furthermore, miR-422a can suppress tumor cell proliferation by
inhibiting related pathways in osteosarcoma (27). Notably, miR-138-5p or miR-422a may
have a negative connection with hTERT in tumor cell proliferation
and metastasis. Thus, combined with the results of our study, the
probability of miR-138-5p and miR-422a potentially inhibiting hTERT
expression in CRC is valuable. However, this results remains to be
confirmed in future studies
Similar to previous studies showing the
downregulation of miR-138-5p and miR422a in cancer tissues, in our
study, we investigated whether or not the decreased levels of
miR-138-5p and miR-422a in CRC were associated with the
clinicopathology and survival of patients. A statistically
significant difference in miR-138-5p expression was observed with
regard to distant metastasis (P<0.000) while a significant
difference in miR-422a expression was also noted between subgroups
according to lymph-node metastasis (P=0.023). In addition, we found
that the high- vs. low-expression group of miR-422a showed a highly
significant difference in CRC patients (P=0.024), which suggests
that the downregulation of miR-422a was associated with a poorer
prognosis.
The present study had some limitations. First,
immunohistochemistry was used to detect hTERT protein expression in
CRC instead of western blotting. Second, in the present analysis,
elevated levels of miR-422a expression were found to have a
prognostic role in CRC, but it was not possible to confirm miR-422a
as an independent predictive factor. Third, validation of miRNAs
with the regulation of hTERT in CRC requires a cell function test,
which is to be conducted in future studies.
In conclusion, our results confirm that miR-124-3p,
miR-133a-3p, miR-133b, miR-138-5p, miR-150-5p, miR-378a-3p and
miR-422a expression levels were downregulated in CRC and that
miR-138-5p and miR-422a were found to potentially interact with
hTERT. Investigation of the suppression of malignant behavior of
these miRNAs in CRC may be useful as a diagnostic or prognostic
tool at least, and may contribute to the development of a new
effective treatment for CRC.
Acknowledgments
This study was funded by the scientific research
fund of Guangxi Zhuang Autonomous Region Education Department (no.
302304).
References
|
1
|
Feng J, Funk WD, Wang SS, Weinrich SL,
Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, et al:
The RNA component of human telomerase. Science. 269:1236–1241.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cukusić A, Skrobot Vidacek N, Sopta M and
Rubelj I: Telomerase regulation at the crossroads of cell fate.
Cytogenet Genome Res. 122:263–272. 2008. View Article : Google Scholar
|
|
4
|
Poole JC, Andrews LG and Tollefsbol TO:
Activity, function, and gene regulation of the catalytic subunit of
telomerase (hTERT). Gene. 269:1–12. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, Chen X, Xu X, Wang X, Wang X,
Yuan G, Sun D, Ka W, He D, Wen Z, et al: Knockdown of hTERT alters
biophysical properties of K562 cells resulting in decreased
migration rate in vitro. Cell Biochem Biophys. 61:595–603. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bertorelle R, Briarava M, Rampazzo E,
Biasini L, Agostini M, Maretto I, Lonardi S, Friso ML, Mescoli C,
Zagonel V, et al: Telomerase is an independent prognostic marker of
overall survival in patients with colorectal cancer. Br J Cancer.
108:278–284. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Daniel M, Peek GW and Tollefsbol TO:
Regulation of the human catalytic subunit of telomerase (hTERT).
Gene. 498:135–146. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schetter AJ, Okayama H and Harris CC: The
role of microRNAs in colorectal cancer. Cancer J. 18:244–252. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen L, Lü MH, Zhang D, Hao NB, Fan YH, Wu
YY, Wang SM, Xie R, Fang DC, Zhang H, et al: miR-1207-5p and
miR-1266 suppress gastric cancer growth and invasion by targeting
telomerase reverse transcriptase. Cell Death Dis. 5:e10342014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mitomo S, Maesawa C, Ogasawara S, Iwaya T,
Shibazaki M, Yashima-Abo A, Kotani K, Oikawa H, Sakurai E, Izutsu
N, et al: Downregulation of miR-138 is associated with
overexpression of human telomerase reverse transcriptase protein in
human anaplastic thyroid carcinoma cell lines. Cancer Sci.
99:280–286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ritchie ME, Silver J, Oshlack A, Holmes M,
Diyagama D, Holloway A and Smyth GK: A comparison of background
correction methods for two-colour microarrays. Bioinformatics.
23:2700–2707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kayala MA and Baldi P: Cyber-T web server:
differential analysis of high-throughput data. Nucleic Acids Res.
40(W1): W553–W559. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baldi P and Long AD: A Bayesian framework
for the analysis of microarray expression data: regularized t-test
and statistical inferences of gene changes. Bioinformatics.
17:509–519. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dweep H, Gretz N and Sticht C: miRWalk
database for miRNA-target interactions. Methods Mol Biol.
1182:289–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Li J, Cao X, Fang Y, Liao ZE, Liu YY,
Huang BD and Han YJ: Overexpression of hTERT in potentially
malignant colorectal laterally spreading tumors. Mol Med Rep.
7:1409–1412. 2013.PubMed/NCBI
|
|
20
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang YY, Sun G, Luo H, Wang XF, Lan FM,
Yue X, Fu LS, Pu PY, Kang CS, Liu N, et al: miR-21 modulates hTERT
through a STAT3-dependent manner on glioblastoma cell growth. CNS
Neurosci Ther. 18:722–728. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chi SW, Zang JB, Mele A and Darnell RB:
Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature.
460:479–486. 2009.PubMed/NCBI
|
|
23
|
Yeh YM, Chuang CM, Chao KC and Wang LH:
MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis
by targeting SOX4 and HIF-1α. Int J Cancer. 133:867–878. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Q, Tang H, Yin S and Dong C:
Downregulation of microRNA-138 enhances the proliferation,
migration and invasion of cholangiocarcinoma cells through the
upregulation of RhoC/p-ERK/MMP-2/MMP-9. Oncol Rep. 29:2046–2052.
2013.PubMed/NCBI
|
|
25
|
Long L, Huang G, Zhu H, Guo Y, Liu Y and
Huo J: Down-regulation of miR-138 promotes colorectal cancer
metastasis via directly targeting TWIST2. J Transl Med. 11:2752013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Faltejskova P, Svoboda M, Srutova K,
Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K,
Kiss I, et al: Identification and functional screening of microRNAs
highly deregulated in colorectal cancer. J Cell Mol Med.
16:2655–2666. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gougelet A, Pissaloux D, Besse A, Perez J,
Duc A, Dutour A, Blay JY and Alberti L: Micro-RNA profiles in
osteosarcoma as a predictive tool for ifosfamide response. Int J
Cancer. 129:680–690. 2011. View Article : Google Scholar
|