Introduction
Ovarian cancer is the leading cause of mortality
from gynecologic malignancies in the world (1). Ovarian cancer is often diagnosed in
advanced stages, resulting in a poor survival rate (2). The 5-year survival rate of patients
with advanced-stage disease is merely about 5–30% (3). Recent investigations to characterize
genetic alterations in ovarian cancer have discovered extensive
cytogenetic and molecular alterations in these tumors (4–9).
However, only limited knowledge has been obtained regarding the
basic molecular mechanisms that deregulate the growth of the
ovarian epithelium and lead to the invasive and metastatic behavior
of these tumors.
TMEM45A (also called DERP7, DNAPTP4 or FLJ10134)
belongs to the large family of genes encoding predicted
transmembrane (TMEM) proteins. Recently, there are a few studies
concerning the expression and functions of TMEM45A in cancers
(10–13). TMEM45A was reported as an
epigenetically regulated gene in MCF-7 breast cancer cells
(14). Overexpression of TMEM45A
has been shown to favor chemoresistance of liver and breast cancer
cells under hypoxic conditions (11). It has been reported that high
expression of TMEM45A is associated with poor prognosis in patients
with breast (11), bladder
(12) and ovarian cancer (10). However, there remains a lack of
in-depth research on the expression pattern and biological
functions of THEM45A in ovarian cancers.
To investigate the role of TMEM45A in ovarian
cancer, we compared its expression between ovarian cancer and
normal tissues. The effects of TMEM45A silencing on the
proliferation, adhesion and invasion of ovarian cancer cells were
then assessed. The possible involved mechanisms were also explored.
Our study provides evidence that TMEM45A is overexpressed in
ovarian cancer and it may be an effective therapeutic target for
this disease.
Materials and methods
Bioinformatic analysis
The Cancer Genome Atlas (TCGA) RNA-sequencing and
corresponding clinical data were downloaded from the TCGA website
https://tcga-data.nci.nih.gov/tcga/following approval
of this project by the consortium. Data from 568 ovarian cancers
and 8 adjacent normal tissues were used for RNA-seq analysis. To
further investigate the biological pathways involved in ovarian
cancer pathogenesis through the TMEM45A pathway, we performed a
gene set enrichment analysis (GSEA) (15) by using GSEA version 2.0 from the
Broad Institute at MIT. The data in question were analyzed in terms
of their differential enrichment in a predefined biological set of
genes (representing pathways). In this study, GSEA firstly
generated an ordered list of all genes according to their
correlation with TMEM45A expression, and then a predefined gene set
(signature of gene expression upon perturbation of certain
cancer-related gene) receives an enrichment score (ES), which is a
measure of statistical evidence rejecting the null hypothesis that
its members are randomly distributed in the ordered list. The
expression level of TMEM45A was used as a phenotype label, and
‘metric for ranking genes’ was set to Pearson correlation. All
other basic and advanced fields were set to default. The KEGG gene
sets of the biological process database (c2.KEGG.v4.0) from the
Molecular Signatures Database (MSig DB) (http://www.broad.mit.edu/gsea/msigdb/index.jsp)
were used for enrichment analysis.
Cancer specimens
Tumor tissues and paired non-cancerous tissues were
collected from 25 patients diagnosed with epithelial ovarian serous
adenocarcinoma, who were admitted to the Department of Gynecology
and Obstetrics, Yangpu Hospital, Tongji University School of
Medicine (Shanghai, China) between 2010 and 2013. Informed consent
was obtained from all patients. This study was approved by the
Ethics Committee of Yangpu Hospital, Shanghai, China.
Cell lines
OVCAR3, A2780, HO-8910, CAOV3, SK-OV-3 and HEK293T
cells were obtained from the American Type Culture Collection
(ATCC; Rockville, MD, USA). All culture media were supplemented
with 10% fetal bovine serum (FBS), 100 mg/ml penicillin G, and 50
μg/ml streptomycin (all from Invitrogen Life Technologies,
Carlsbad, CA, USA). The ovarian cancer cell lines, OVCAR3, A2780
and HO-8910, were cultured in RPMI-1640, and the CAOV3, SK-OV-3 and
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium
(DMEM) (all from Invitrogen Life Technologies). All cells were
maintained at 37°C in 5% CO2.
RNA extraction and real-time PCR
Total RNA was extracted using TRIzol reagent
(Invitrogen Life Technologies) according to the manufacturer’s
instructions. mRNA contained in 2 μg total RNA was reverse
transcribed using a cDNA synthesis kit (Thermo Fisher Scientific,
Rockford, IL, USA) according to the manufacturer’s instructions.
Real-time PCR was performed to detect mRNA levels of the indicated
genes. GAPDH served as an internal control. The primers used were:
TMEM45A, 5′-ACCAAGTTGGATCATGGGGA-3′ and 5′-AGCCATGCCAGTTAAAGCCA-3′;
GAPDH, 5′-AATCCCATCACCATCTTC-3′ and 5′-AGGCTGTTGTCATACTTC-3′;
TGF-β1, 5′-GACTACTACGCCAAGGAGGTC-3′ and 5′-GAG AGCAACACGGGTTCAG-3′;
TGF-β2, 5′-AGAGCAGAAGGCGAATGG-3′ and 5′-AAAGTGCAGCAGGGACAG-3′;
RhoA, 5′-TAGTCCACGGTCTGGTCTTC-3′ and 5′-CTTTCCACAGGCTCCATCAC-3′;
ROCK2, 5′-CAGCAATGGTAAGCGTAAAG-3′ and 5′-GTAGAGACGGAGTTTCACTATG-3′.
All reactions were conducted on an ABI 7300 real-time PCR machine
(Applied Biosystems, Foster City, CA, USA) using the following
cycling parameters: 95°C for 10 min, followed by 40 cycles of 95°C
for 15 sec, and 60°C for 45 sec. The gene expression was calculated
using the ΔΔCt method. All data represent the average of 3
replicates.
RNA interference and construction of
stable cell lines
Three shRNAs targeting position 713–733
(GAGGCCTTTATCTTC TACAAC; named TMEM45A-Ri-1), position 821–841
(GAGT TCCTTGTTCGGAACAAT; named TMEM45A-Ri-2) and position 1248–1268
(TAAGTGTACTGTTTGCATTTC; named TMEM45A-Ri-3) of human TMEM45A mRNA
were cloned into a lentiviral vector (PLKO.1). A non-specific
scramble shRNA sequence (CCTAAGGTTAAGTCGCCC TCG) was used as a
negative control. The constructs were then transfected into HEK293T
cells with lentiviral packaging vectors by using Lipofectamine 2000
(Invitrogen) according to the manufacturer’s instructions. Viruses
were collected 48 h after transfection and used to infect the A2780
and HO-8910 cells.
Western blotting
The cells were lysed with radioimmunoprecipitation
assay buffer (50 mmol/l Tris-HCl, 150 mmol/l NaCl, 1% Triton X-100,
0.1% SDS and 1% deoxycholic acid sodium). The lysates were
quantified using the BCA protein assay kit (Thermo Fisher
Scientific). The lysates with equal amounts of protein were
separated on SDS-PAGE gels followed by electroblotting to
nitrocellulose membranes. Western blotting was then carried out
with appropriate primary and horseradish peroxidase-conjugated
secondary antibodies. Membranes were developed with enhanced
chemiluminescence (Bio-Rad, Richmond, CA, USA). Antibodies against
TMEM45A, TGFβ1, TGFβ2, RhoA and ROCK2 were purchased from Abcam
(Cambridge, MA, USA). GAPDH antibody was from Cell Signaling
Technology Inc. (Danvers, MA, USA).
Cell proliferation assay
A total of 3×103 cells/well were plated
in 96-well plates before viral infection and cultured for 24 h in
normal conditions. They were then infected with the TMEM45A-shRNA
virus or control virus (NC). Cell proliferation was detected at the
indicated times using CCK-8 (Dojindo Laboratories, Kumamoto, Japan)
according to the manufacturer’s instructions. Briefly, at the
indicated time-points, CCK-8 solution (10 μl in 100
μl DMEM) was added to each well and incubated for 1 h.
Optical density (OD) values at wavelength 450 nm were measured by a
microplate reader (Bio-Rad Laboratories Inc., Hercules, CA,
USA).
Cell cycle analysis
The cell cycle was evaluated by flow cytometry using
propidium iodide (PI; Sigma, St. Louis, MO, USA) staining on a flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Briefly, the
cells were plated in 6-well plates before viral infection and
cultured for 24 h under normal conditions. The cells were collected
and fixed in 70% ethanol at −20°C overnight 24 h after viral
infection. The cells were then washed in phosphate-buffered saline
(PBS) and resuspended in staining solution containing 20
μg/ml PI and 200 μg/ml RNase A. Experiments were
performed in triplicate and 3×104 cells were analyzed
per sample. G1, S, and G2/M fractions were quantified using
CellQuest software (BD Biosciences) and manual gating.
Cell adhesion assay
The adhesion assay was performed in 12-well plates.
The plates were pre-coated with 1 ml of fibronectin (5
μg/ml) for 2 h at room temperature. The cells were infected
with the indicated viral vectors 48 h before the assay was
performed. The cells were seeded into the coated plates at a
density of 105 cells/well and allowed to adhere at 37°C
for 1 h. Non-adherent cells were washed off with PBS and fixed in
4% paraformaldehyde and stained with Giemsa solution. The number of
adherent cells was determined as described previously (16).
In vitro invasion assay
The upper well of the Transwell (Corning Inc.,
Corning, NY, USA) was coated with Matrigel (BD Biosciences, San
Jose, CA, USA) at 37 °C in a 5% CO2 incubator for 1 h.
The indicated cells were serum-starved for 24 h, and then 500
μl of cell suspension containing 105 cells/ml
were placed in the upper compartment of the chamber. Culture medium
supplemented with 10% FBS (750 μl) was added into the lower
well of the chamber. The plates were incubated for 48 h. At the end
of the incubation, the cells on the upper surface of the filter
were completely removed by wiping with a cotton swab. The cells
that migrated into the lower well were washed with PBS, fixed in 4%
paraformaldehyde and stained by 0.2% crystal violet. The invading
cells were observed under a microscope (magnification, ×100). Cells
were counted in the central field of triplicate membranes.
Statistical analysis
The data were analyzed using the two-tailed
Student’s t-test to calculate the statistical significance of
difference between groups. The results are presented as the mean
value ± SD. Statistically significant differences were defined as
having a P-value <0.05.
Results
Overexpression of TMEM45A in ovarian
cancer
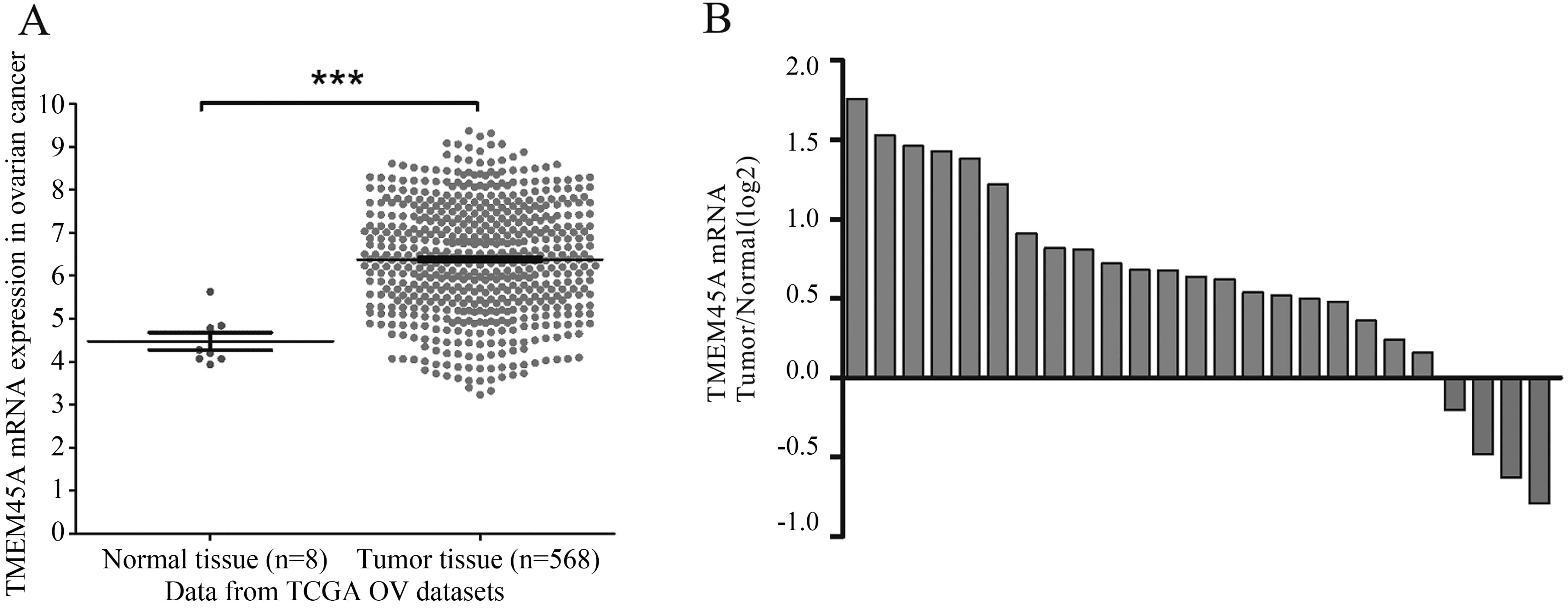
To explore the expression of TMEM45A in ovarian
cancer, we compared TMEM45A by analyzing high throughput
RNA-sequencing data of the ovarian cancer cohort from TCGA. TMEM45A
expression was significantly higher in the ovarian cancer tissues
than that in the adjacent tissues of the patients, which indicated
that TMEM45A may be an oncogene in ovarian cancer (Fig. 1A).
To further confirm the overexpression of TMEM45A in
ovarian cancer, we performed real-time PCR analysis on 25 pairs of
ovarian cancer and their matched non-cancerous tissue samples.
TMEM45A was found to be overexpressed in 84% (21/25) of the tested
ovarian cancer tissues (Fig. 1B).
Statistical analysis using the Student’s t-test indicated a
significant difference in the mRNA level of TMEM45A between the
ovarian tumor and normal tissues (P<0.001).
Downregulation of TMEM45A by RNA
interference (RNAi) in ovarian cancer cells
The protein and mRNA levels of TMEM45A in five
ovarian cancer cell lines were then detected. A high level of
TMEM45A was observed in the HO-8910 and A2780 cells (Fig. 2A). For that reason, these two cells
were selected for the following assays.
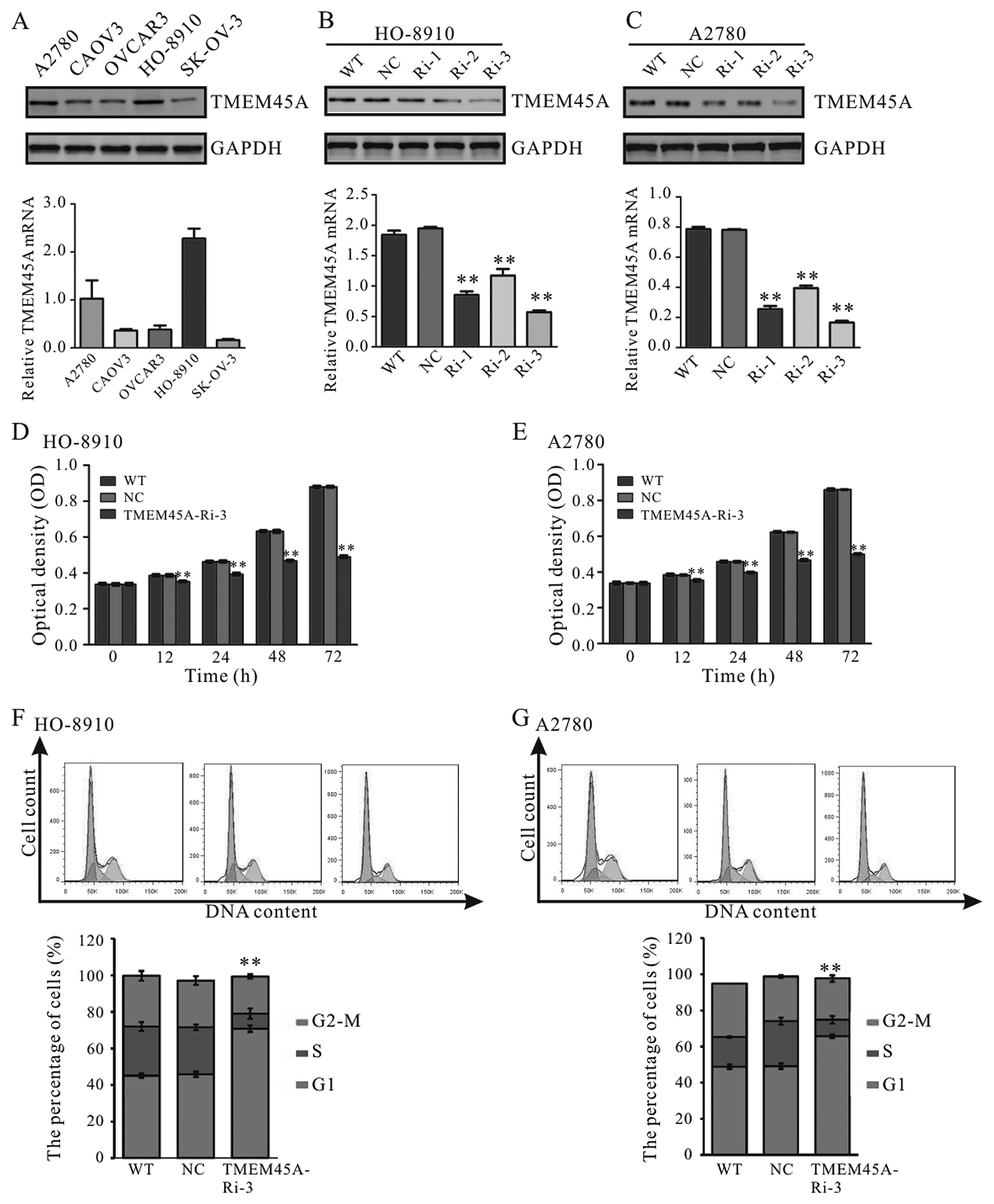 | Figure 2TMEM45A knockdown impairs cell
proliferation and cell cycle progression in the HO-8910 and A2780
cells. (A) TMEM45A expression in five ovarian cancer cell lines was
detected by western blotting and real-time PCR. GAPDH was used as
an internal control. High expression of TMEM45A was observed in the
A2780 and HO-8910 cells, which were selected for further analysis.
(B and C) Western blotting (upper panel) and real-time PCR (lower
panel) analyses showing the efficiency of TMEM45A knockdown in the
HO-8910 and A2780 cells. NC, scrambled shRNA viral-infected cells;
WT, wild-type cells; Ri-1, Ri-2 and Ri-3, TMEM45A-shRNA-1, -2 and
-3 viral-infected cells. (D and E) CCK-8 assay was performed to
evaluate the cell proliferation of the HO-8910 and A2780 cells. (F
and G) The percentages of cells in the G1, S and G2-M phases for
each sample at 48 h after viral infection are shown. NC, scrambled
shRNA viral-infected cells; WT, wild-type cells; TMEM45A-Ri-3,
TMEM45A-shRNA-3 viral-infected cells. Data are shown as the mean
value ± SD **P<0.01, vs. NC. |
To explore the functions of TMEM45A in ovarian
cancer, we suppressed TMEM45A levels in the HO-8910 and A2780 cells
by lentiviral-mediated delivery of shRNA. In the present study,
three pairs of shRNA sequences against the human TMEM45A gene were
designed and a non-specific scramble shRNA sequence was used as the
negative control (NC). The silencing effect of the TMEM45A-RNAi
virus was confirmed in the HO-8910 (Fig. 2B) and A2780 (Fig. 2C) cells by western blotting and
real-time PCR. Our results demonstrated that TMEM45A-Ri-3 was the
most efficient one and was selected for the following assays.
TMEM45A knockdown inhibits cell
proliferation and induces G1-phase cell cycle arrest in ovarian
cancer cells
To investigate the role of TMEM45A downregulation on
cell proliferation, we assessed the proliferation of HO-8910 and
A2780 cells infected with TMEM45A-Ri-3 using the CCK-8 assay. As
shown in Fig. 2D and E, cell growth
was notably impaired in the TMEM45A-Ri-3 viral-infected cells
(TMEM45A-Ri-3) compared to the wild-type (WT) cells and the
scramble shRNA viral-infected (NC) cells. Our data indicated a role
of TMEM45A in the regulation of ovarian cancer cell
proliferation.
The possible effect of TMEM45A RNAi on cell cycle
progression was then determined. As shown in Fig. 2F, a higher number of HO-8910 cells
in the G1 phase (70.8±1.9%) was observed in cells infected with the
TMEM45A shRNA virus, compared with that in the WT cells (45.1±1.1%)
and NC cells (45.9±1.1%). Similar results were observed in the
A2780 cells (Fig. 2G). These
results indicated that the proliferation-promoting function of
TMEM45A was most likely mediated by promoting G1/S cell cycle
transition in the ovarian cancer cells.
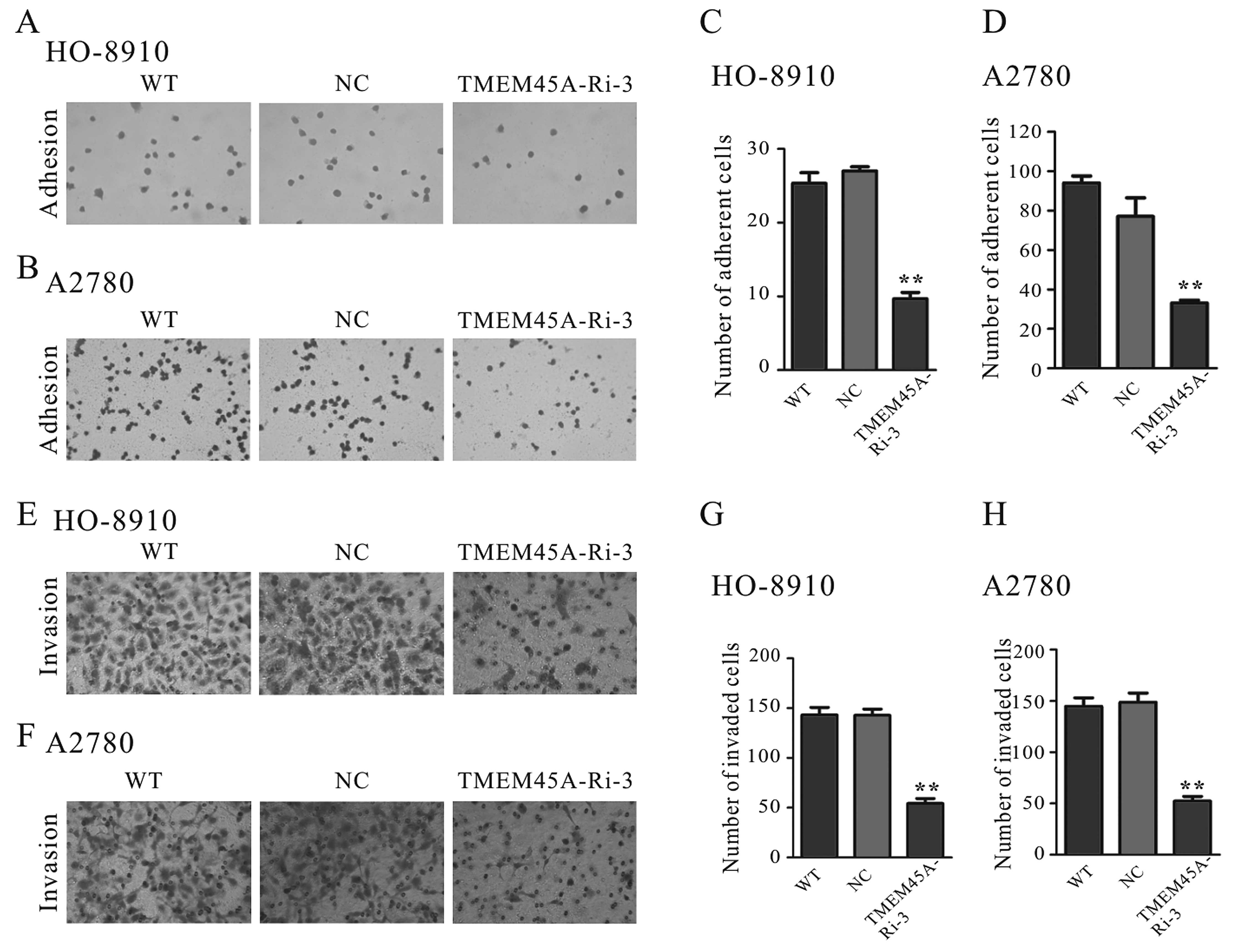
Suppression of TMEM45A expression
represses cell adhesion capacity
The effects of TMEM45A on cell adhesion capacity
were evaluated by a cell adhesion assay (Fig. 3). The adherent ability of ovarian
cells to fibronectin was significantly repressed by TMEM45A
knockdown. The number of adherent TMEM45A-Ri-3 cells was 37% of
that of the NC cells when the HO-8910 cells were used (Fig. 3A and C). Similar results were
obtained for the A2780 cells (Fig. 3B
and D). These data suggest that TMEM45A may regulate ovarian
cancer cell adhesion.
TMEM45A knockdown inhibits the
invasiveness of ovarian cancer cells
To discover whether TMEM45A affects the invasive
ability of ovarian cancer cells, we performed a Matrigel-coated
membrane chamber invasion assay. As shown in Fig. 3, a radical reduction in the invasive
ability was observed in the TMEM45A-knockdown cells compared to the
control cells. The number of invaded TMEM45A-Ri-3 cells decreased
to 38% of that of the NC cells when the HO-8910 cells were used
(Fig. 3E and G). Similar results
were observed for the A2780 cells (Fig.
3F and H).
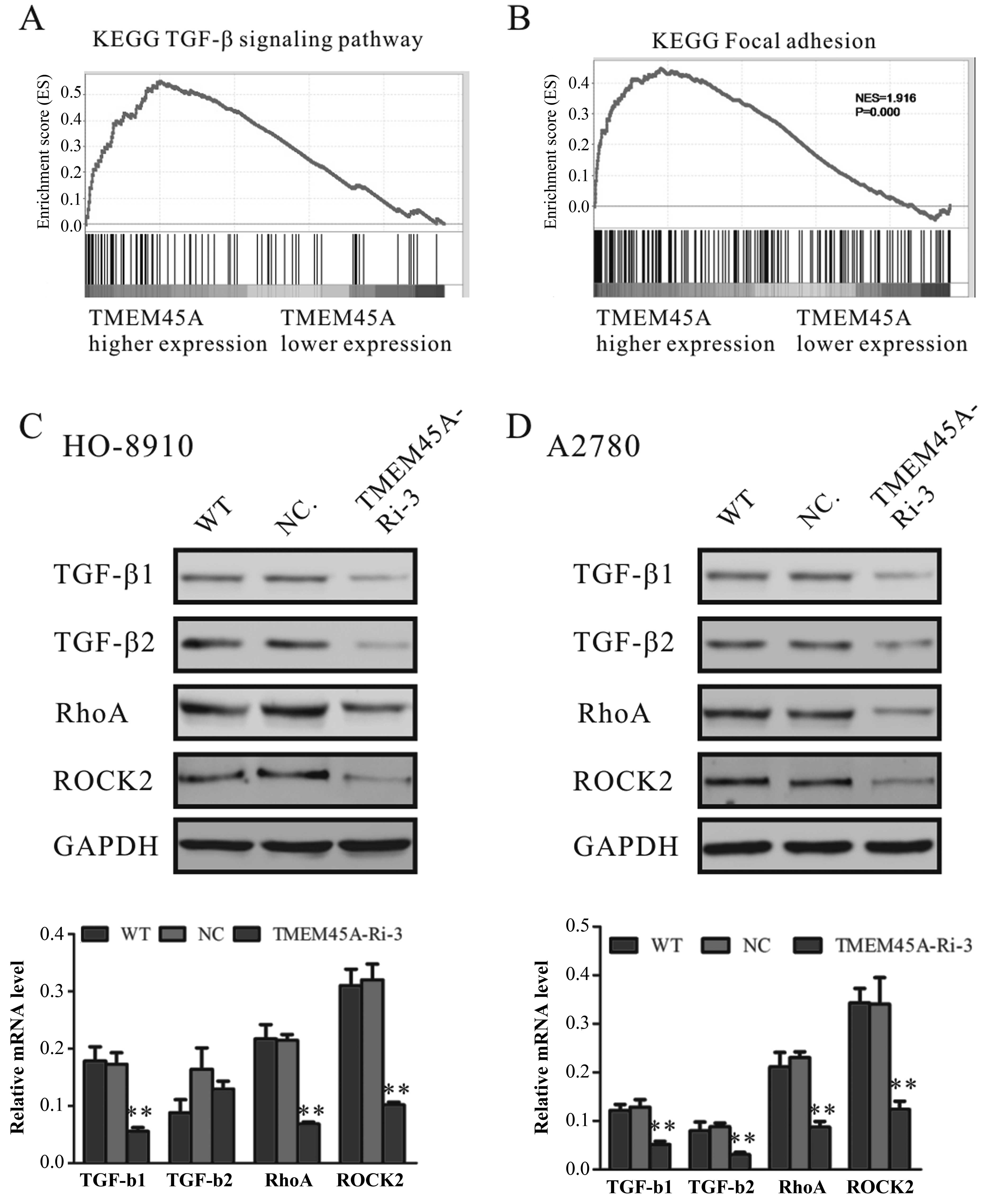
Identification of genes and
signaling-associated biological pathways and processes by GSEA
The exact pathway that TMEM45 may regulate in
ovarian cancers remains unclear. To probe the TMEM45A-associated
pathways on an unbiased basis, we performed GSEA using high
throughput RNA-sequencing data of the ovarian cancer cohort from
TCGA. GSEA is designed to detect coordinated differences in
expression of predefined sets of functionally related genes. Among
all the 188 predefined ‘KEGG pathway’ gene sets, the TGF-β
signaling pathway and focal adhesion pathway were identified as
having a significant association with higher expression of TMEM45A
(Fig. 4A and B).
TMEM45A knockdown downregulates the mRNA
and protein levels of TGF-β1, TGF-β2, RhoA and ROCK2
To further confirm the association of TMEM45A
expression and the TGF-β signaling and focal adhesion pathways, we
detected the expression of important regulators of the TGF-β
signaling pathway and cell adhesion molecules in the TMEM45A-Ri-3
and NC cells. The mRNA and protein levels of TGF-β1, TGF-β2, RhoA
and ROCK2 were significantly decreased in both the HO-8910
(Fig. 4C) and A2780 (Fig. 4D) cells following downregulation of
TMEM45A.
Discussion
Recently, the expression and functions of TMEM45A in
cancers have been reported (10–13).
In the present study, bioinformatic analysis using high throughput
RNA-sequencing data from TCGA demonstrated a higher expression of
TMEM45A in ovarian cancer compared to that in normal tissues
(Fig. 1A). We then evaluated the
mRNA levels of TMEM45A in 25 pairs of ovarian tumor and normal
tissues by real-time PCR. We found that TMEM45A was overexpressed
in 84% (21/25) of the tested ovarian cancer tissues (Fig. 1B).
To further investigate the functions of TMEM45A in
ovarian cancer, we suppressed the expression of TMEM45A in two
ovarian cell lines, HO-8910 and A2780, by RNA interference
(Fig. 2B and C). Our data showed
that suppression of TMEM45A expression markedly repressed the cell
proliferation (Fig. 2D and E).
Abnormal regulation of the cell cycle is frequently observed in
most common malignancies, resulting in aberrant cell proliferation
(17,18). In the present study, silencing of
TMEM45A by RNAi significantly stimulated cell cycle arrest in the
G1-phase (Fig. 2F and G), which
indicated that TMEM45A promoted cell proliferation by promoting
G1/S cell cycle transition in ovarian cancer cells. Moreover, cell
adhesion and invasion of ovarian cells were also inhibited by
TMEM45A downregulation (Fig. 3).
These data suggest a role of TMEM45A in ovarian cell
carcinogenesis.
The exact pathway that TMEM45 may regulate in
ovarian cancers remains unclear. Our GSEA data indicated that
TMEM45A overexpression was positively correlated with the TGF-β
signaling pathway (Fig. 4A). TGF-β
signaling participates in a variety of cellular processes,
including cell differentiation, proliferation, apoptosis, and
determination of developmental fate (19). The TGF-β signaling pathway has also
been considered to promote tumor progression and invasion. In
response to elevated TGF-β levels, the tumor cell becomes more
migratory and invasive (20,21).
In the present study, TMEM45A knockdown significantly decreased the
expression of TGF-β1 and TGF-β2 (Fig.
4), which indicates an association between TMEM45A function and
the regulation of TGF-β signaling in ovarian cancer cells.
Moreover, our GSEA results indicated a positive
correlation between TMEM45A overexpression and focal adhesion genes
(Fig. 4B). Rho is required for the
formation and maintenance of focal adhesion. It has been shown that
RhoA, a member of the Rho subfamily, plays a central role in the
regulation of cell survival, motility, apoptosis and invasion
(22,23). RhoA expression was reported to be
upregulated in lung, breast, colon and ovarian cancer. The
expression level of RhoA may be positively correlated with the
progression of these carcinomas, suggesting that RhoA may play an
important role in tumorigenesis and tumor progression (24–28).
The malignant phenotype in gastric cancer (29) and breast cancer (30) cells can be reversed by the
inhibition of RhoA expression. Suppression of ROCK2 expression was
reported to impair anchorage-independent growth and invasion of
non-small cell lung cancer (31).
In the present study, TMEM45A knockdown markedly decreased the mRNA
and protein levels of RhoA and ROCK2. The dowregulation of RhoA and
ROCK2 may be associated with the impaired adhesion and invasive
ability (Fig. 3) of the TMEM45A
RNAi cells. To our knowledge, our data are the first to associate
the functions of TMEM45A with the RhoA/ROCK2 signaling pathway.
In conclusion, our study demonstrated the
overexpression of TMEM45A in ovarian cancer, which was related with
cancerous transformation. Our study is the first to link TMEM45A
with the regulation of TGF-β signaling and the RhoA/ROCK signaling
pathway, and thus provides useful information for targeted
therapy.
References
|
1
|
Zeinoun Z, Teugels E, De Bleser PJ, Neyns
B, Geerts A and De Greve J: Insufficient TGF-beta 1 production
inactivates the autocrine growth suppressive circuit in human
ovarian cancer cell lines. Anticancer Res. 19:413–420.
1999.PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cannistra SA: Cancer of the ovary. N Engl
J Med. 351:2519–2529. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Spentzos D, Levine DA, Ramoni MF, Joseph
M, Gu X, Boyd J, Libermann TA and Cannistra SA: Gene expression
signature with independent prognostic significance in epithelial
ovarian cancer. J Clin Oncol. 22:4700–4710. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Berchuck A, Iversen ES, Lancaster JM,
Pittman J, Luo J, Lee P, Murphy S, Dressman HK, Febbo PG, West M,
et al: Patterns of gene expression that characterize long-term
survival in advanced stage serous ovarian cancers. Clin Cancer Res.
11:3686–3696. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hartmann LC, Lu KH, Linette GP, Cliby WA,
Kalli KR, Gershenson D, Bast RC, Stec J, Iartchouk N, Smith DI, et
al: Gene expression profiles predict early relapse in ovarian
cancer after platinum-paclitaxel chemotherapy. Clin Cancer Res.
11:2149–2155. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spentzos D, Levine DA, Kolia S, Otu H,
Boyd J, Libermann TA and Cannistra SA: Unique gene expression
profile based on pathologic response in epithelial ovarian cancer.
J Clin Oncol. 23:7911–7918. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Helleman J, Jansen MP, Span PN, van
Staveren IL, Massuger LF, Meijer-van Gelder ME, Sweep FC, Ewing PC,
van der Burg ME, Stoter G, et al: Molecular profiling of platinum
resistant ovarian cancer. Int J Cancer. 118:1963–1971. 2006.
View Article : Google Scholar
|
|
9
|
Dressman HK, Berchuck A, Chan G, Zhai J,
Bild A, Sayer R, Cragun J, Clarke J, Whitaker RS, Li L, et al: An
integrated genomic-based approach to individualized treatment of
patients with advanced-stage ovarian cancer. J Clin Oncol.
25:517–525. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Crijns AP, Fehrmann RS, de Jong S, Gerbens
F, Meersma GJ, Klip HG, Hollema H, Hofstra RM, te Meerman GJ, de
Vries EG, et al: Survival-related profile, pathways, and
transcription factors in ovarian cancer. PLoS Med. 6:e242009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Flamant L, Roegiers E, Pierre M, Hayez A,
Sterpin C, De Backer O, Arnould T, Poumay Y and Michiels C: TMEM45A
is essential for hypoxia-induced chemoresistance in breast and
liver cancer cells. BMC Cancer. 12:3912012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Urquidi V, Goodison S, Cai Y, Sun Y and
Rosser CJ: A candidate molecular biomarker panel for the detection
of bladder cancer. Cancer Epidemiol Biomarkers Prev. 21:2149–2158.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jinawath N, Chamgramol Y, Furukawa Y,
Obama K, Tsunoda T, Sripa B, Pairojkul C and Nakamura Y: Comparison
of gene expression profiles between Opisthorchis viverrini and
non-Opisthorchis viverrini associated human intrahepatic
chol-angiocarcinoma. Hepatology. 44:1025–1038. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hsieh TC and Wu JM: Differential control
of growth, cell cycle progression, and gene expression in human
estrogen receptor positive MCF-7 breast cancer cells by extracts
derived from polysaccharopeptide I’m-Yunity and Danshen and their
combination. Int J Oncol. 29:1215–1222. 2006.PubMed/NCBI
|
|
15
|
Subramanian A, Kuehn H, Gould J, Tamayo P
and Mesirov JP: GSEA-P: a desktop application for gene set
enrichment analysis. Bioinformatics. 23:3251–3253. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Silletti S, Paku S and Raz A: Autocrine
motility factor and the extracellular matrix. I coordinate
regulation of melanoma cell adhesion, spreading and migration
involves focal contact reorganization. Int J Cancer. 76:120–128.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Molinari M: Cell cycle checkpoints and
their inactivation in human cancer. Cell Prolif. 33:261–274. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi Y and Massagué J: Mechanisms of
TGF-beta signaling from cell membrane to the nucleus. Cell.
113:685–700. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wakefield LM and Roberts AB: TGF-beta
signaling: Positive and negative effects on tumorigenesis. Curr
Opin Genet Dev. 12:22–29. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vergara D, Merlot B, Lucot JP, Collinet P,
Vinatier D, Fournier I and Salzet M: Epithelial-mesenchymal
transition in ovarian cancer. Cancer Lett. 291:59–66. 2010.
View Article : Google Scholar
|
|
22
|
Schmitz AA, Govek EE, Böttner B and Van
Aelst L: Rho GTPases: signaling, migration, and invasion. Exp Cell
Res. 261:1–12. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Aznar S and Lacal JC: Rho signals to cell
growth and apoptosis. Cancer Lett. 165:1–10. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fritz G, Just I and Kaina B: Rho GTPases
are over-expressed in human tumors. Int J Cancer. 81:682–687. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Abraham MT, Kuriakose MA, Sacks PG, Yee H,
Chiriboga L, Bearer EL and Delacure MD: Motility-related proteins
as markers for head and neck squamous cell cancer. Laryngoscope.
111:1285–1289. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kamai T, Arai K, Tsujii T, Honda M and
Yoshida K: Over-expression of RhoA mRNA is associated with advanced
stage in testicular germ cell tumour. BJU Int. 87:227–231. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Horiuchi A, Imai T, Wang C, Ohira S, Feng
Y, Nikaido T and Konishi I: Up-regulation of small GTPases, RhoA
and RhoC, is associated with tumor progression in ovarian
carcinoma. Lab Inves. 83:861–870. 2003. View Article : Google Scholar
|
|
28
|
Kamai T, Tsujii T, Arai K, Takagi K, Asami
H, Ito Y and Oshima H: Significant association of Rho/ROCK pathway
with invasion and metastasis of bladder cancer. Clin Cancer Res.
9:2632–2641. 2003.PubMed/NCBI
|
|
29
|
Liu N, Bi F, Pan Y, Sun L, Xue Y, Shi Y,
Yao X, Zheng Y and Fan D: Reversal of the malignant phenotype of
gastric cancer cells by inhibition of RhoA expression and activity.
Clin Cancer Res. 10:6239–6247. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pille JY, Denoyelle C, Varet J, Bertrand
JR, Soria J, Opolon P, Lu H, Pritchard LL, Vannier JP, Malvy C, et
al: Anti-RhoA and anti-RhoC siRNAs inhibit the proliferation and
invasiveness of MDA-MB-231 breast cancer cells in vitro and in
vivo. Mol Ther. 11:267–274. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vigil D, Kim TY, Plachco A, Garton AJ,
Castaldo L, Pachter JA, Dong H, Chen X, Tokar B, Campbell SL, et
al: ROCK1 and ROCK2 are required for non-small cell lung cancer
anchorage-independent growth and invasion. Cancer Res.
72:5338–5347. 2012. View Article : Google Scholar : PubMed/NCBI
|