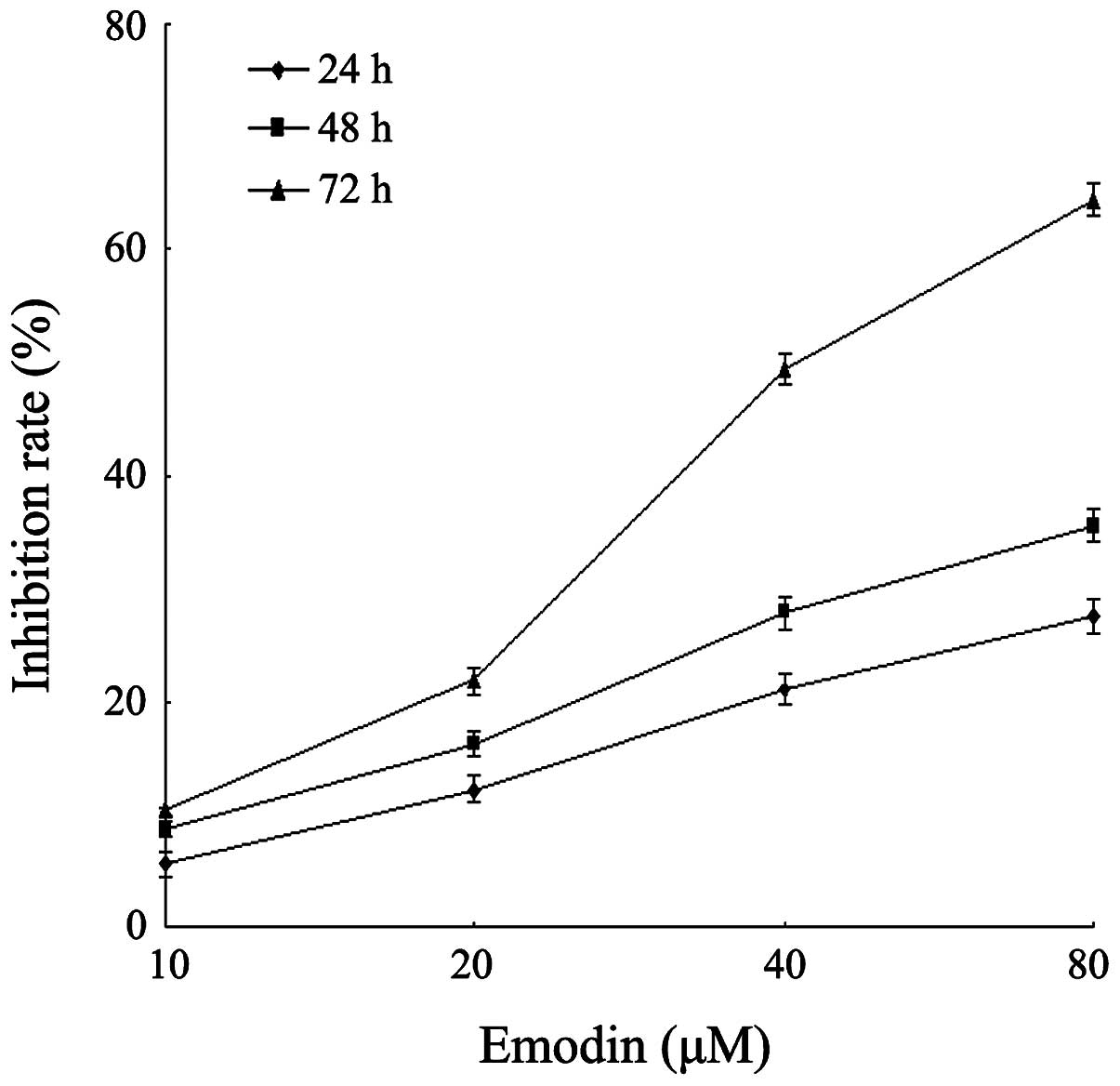
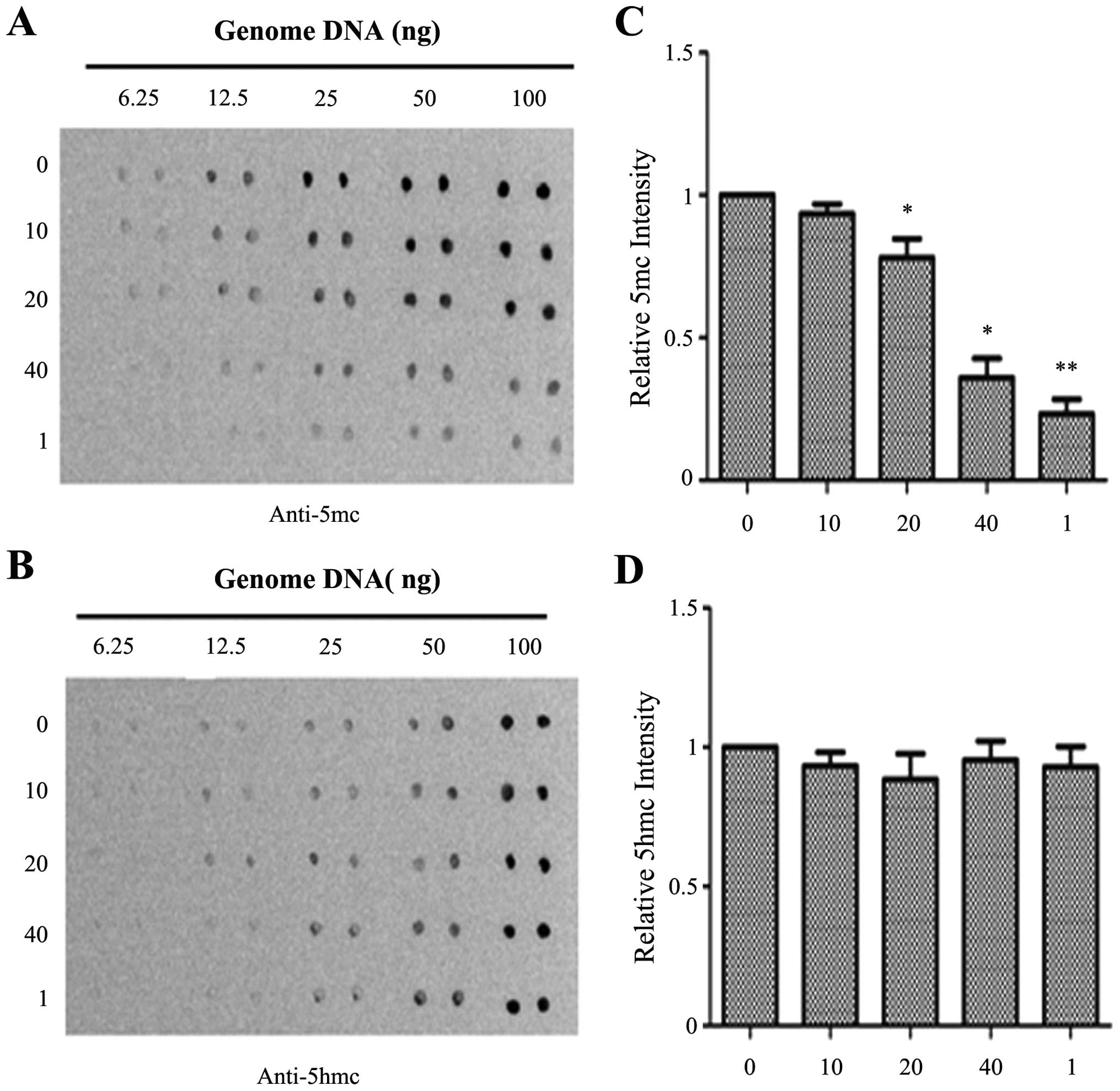
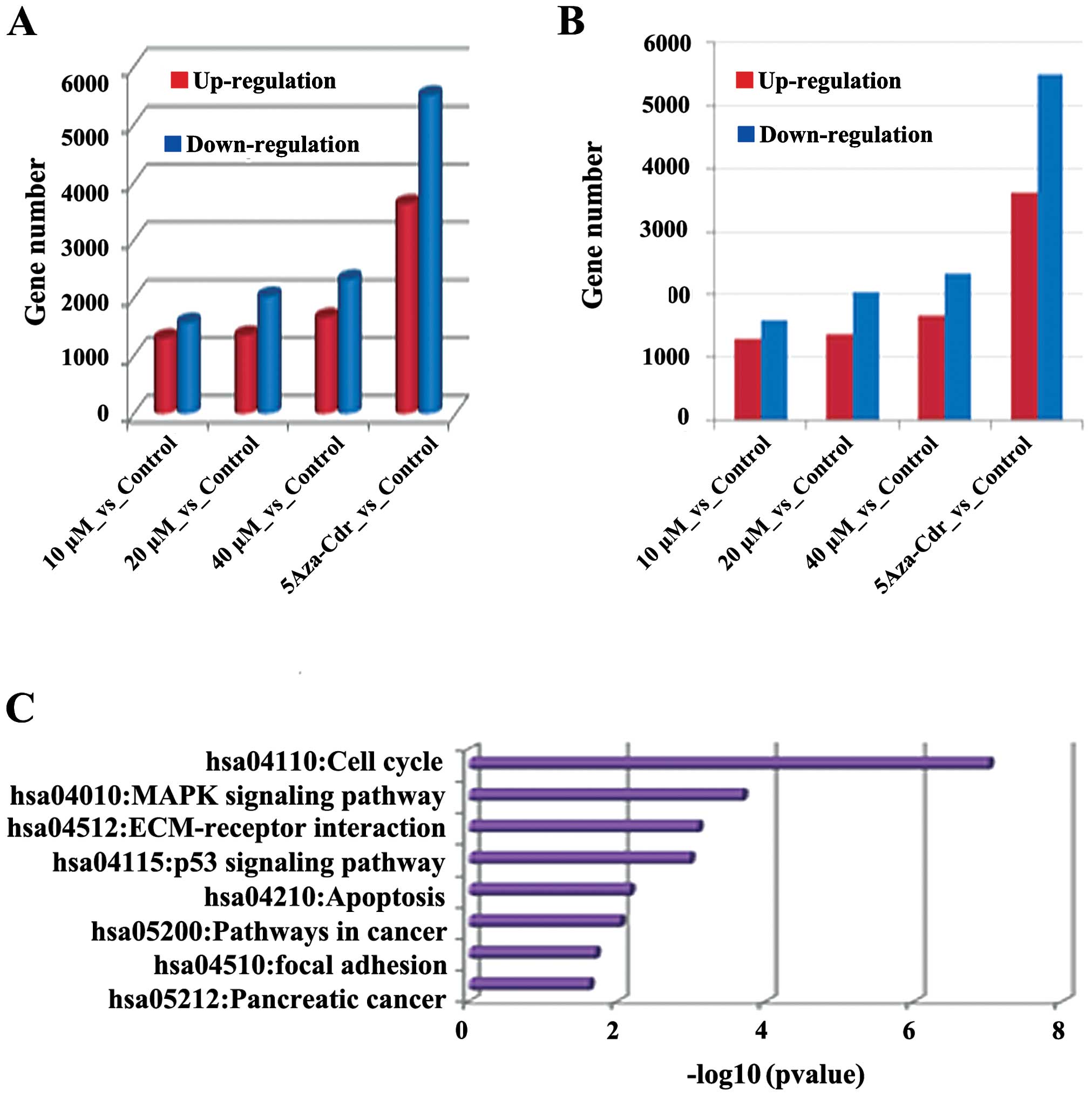
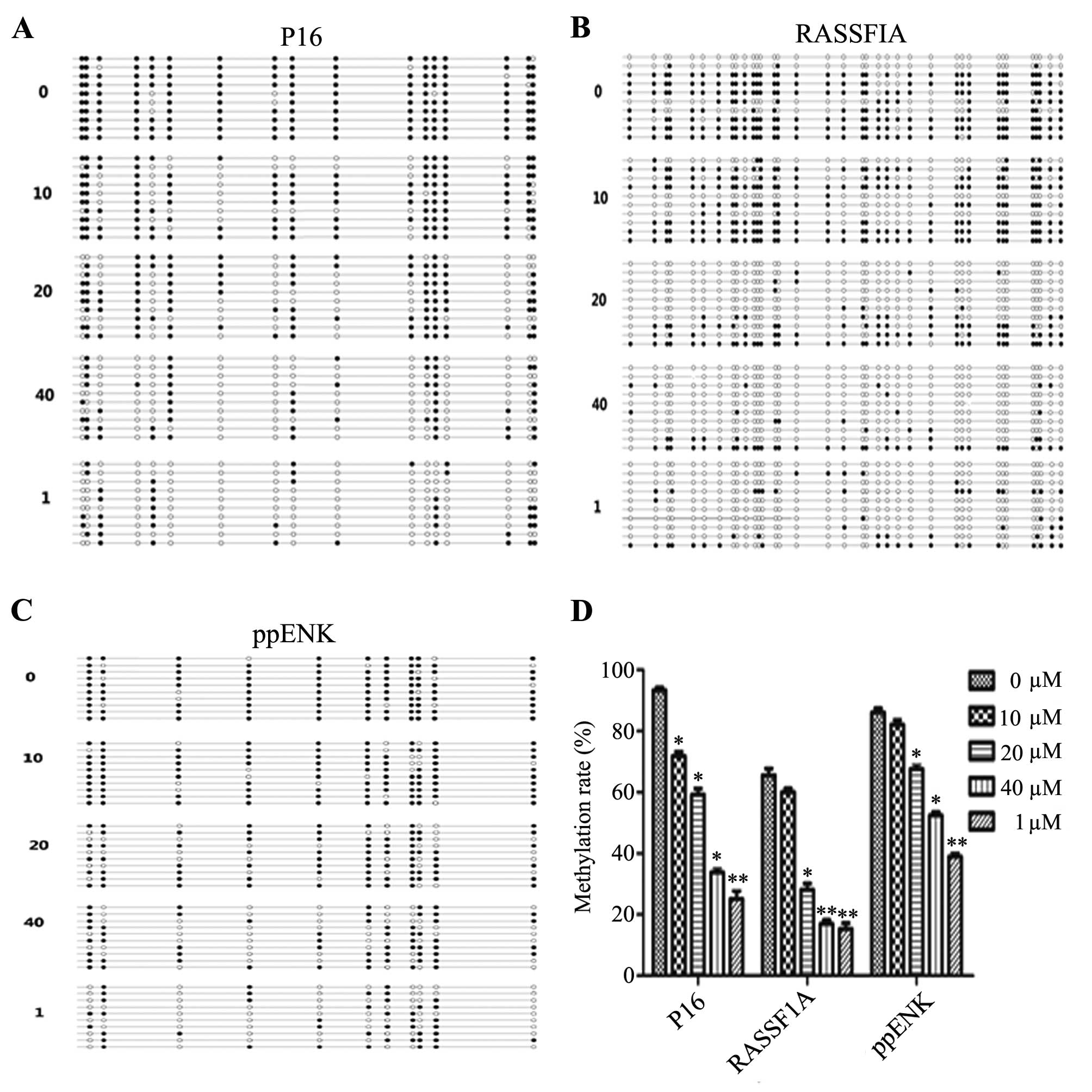
|
1
|
Reske SN: PET and PET-CT of malignant
tumors of the exocrine pancreas. Radiologe. 49:131–136. 2009.In
German. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hariharan D, Saied A and Kocher HM:
Analysis of mortality rates for pancreatic cancer across the world.
HPB Oxf. 10:58–62. 2008. View Article : Google Scholar
|
|
3
|
Guo X and Cui Z: Current diagnosis and
treatment of pancreatic cancer in China. Pancreas. 31:13–22. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cheng X and Blumenthal RM: Mammalian DNA
methyltransferases: a structural perspective. Structure.
16:341–350. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bird A: DNA methylation patterns and
epigenetic memory. Genes Dev. 16:6–21. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gambichler T, Sand M and Skrygan M: Loss
of 5-hydroxymeth-ylcytosine and ten-eleven translocation 2 protein
expression in malignant melanoma. Melanoma Res. 23:218–220. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Matsuoka S, Edwards MC, Bai C, Parker S,
Zhang P, Baldini A, Harper JW and Elledge SJ: p57KIP2, a
structurally distinct member of the p21CIP1 Cdk
inhibitor family, is a candidate tumor suppressor gene. Genes Dev.
9:650–662. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sato N, Fukushima N, Maehara N,
Matsubayashi H, Koopmann J, Su GH, Hruban RH and Goggins M:
SPARC/osteonectin is a frequent target for aberrant methylation in
pancreatic adenocarcinoma and a mediator of tumor-stromal
interactions. Oncogene. 22:5021–5030. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Attri J, Srinivasan R, Majumdar S, Radotra
BD and Wig J: Alterations of tumor suppressor gene P16INK4a in
pancreatic ductal carcinoma. BMC Gastroenterol. 5:222005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dammann R, Schagdarsurengin U, Liu L, Otto
N, Gimm O, Dralle H, Boehm BO, Pfeifer GP and Hoang-Vu C: Frequent
RASSF1A promoter hypermethylation and K-ras mutations in pancreatic
carcinoma. Oncogene. 22:3806–3812. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ueki T, Toyota M, Skinner H, Walter KM,
Yeo CJ, Issa JP, Hruban RH and Goggins M: Identification and
characterization of differentially methylated CpG islands in
pancreatic carcinoma. Cancer Res. 61:8540–8546. 2001.PubMed/NCBI
|
|
12
|
Fukushima N, Sato N, Ueki T, Rosty C,
Walter KM, Wilentz RE, Yeo CJ, Hruban RH and Goggins M: Aberrant
methylation of preproenkephalin and P16 genes in pancreatic
intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am
J Pathol. 160:1573–1581. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schutte M, Hruban RH, Geradts J, Maynard
R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff
I, Schmiegel W, et al: Abrogation of the Rb/P16 tumor-suppressive
pathway in virtually all pancreatic carcinomas. Cancer Res.
57:3126–3130. 1997.PubMed/NCBI
|
|
14
|
Moore PS, Sipos B, Orlandini S, Sorio C,
Real FX, Lemoine NR, Gress T, Bassi C, Klöppel G, Kalthoff H, et
al: Genetic profile of 22 pancreatic carcinoma cell lines. Analysis
of K-ras, p53, P16 and DPC4/Smad4. Virchows Arch. 439:798–802.
2001. View Article : Google Scholar
|
|
15
|
Brueckner B, Kuck D and Lyko F: DNA
methyltransferase inhibitors for cancer therapy. Cancer J.
13:17–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szyf M: DNA methylation and demethylation
as targets for anticancer therapy. Biochemistry (Mosc). 70:533–549.
2005. View Article : Google Scholar
|
|
17
|
Goffin J and Eisenhauer E: DNA
methyltransferase inhibitors - state of the art. Ann Oncol.
13:1699–1716. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lyko F and Brown R: DNA methyltransferase
inhibitors and the development of epigenetic cancer therapies. J
Natl Cancer Inst. 97:1498–1506. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiang N, Zhao R, Song G and Zhong W:
Selenite reactivates silenced genes by modifying DNA methylation
and histones in prostate cancer cells. Carcinogenesis.
29:2175–2181. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Brueckner B, Garcia Boy R, Siedlecki P,
Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M and Lyko F:
Epigenetic reactivation of tumor suppressor genes by a novel
small-molecule inhibitor of human DNA methyltransferases. Cancer
Res. 65:6305–6311. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cui X, Wakai T, Shirai Y, Yokoyama N,
Hatakeyama K and Hirano S: Arsenic trioxide inhibits DNA
methyltransferase and restores methylation-silenced genes in human
liver cancer cells. Hum Pathol. 37:298–311. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin SZ, Chen KJ, Tong HF, Jing H, Li H and
Zheng SS: Emodin attenuates acute rejection of liver allografts by
inhibiting hepatocellular apoptosis and modulating the Th1/Th2
balance in rats. Clin Exp Pharmacol Physiol. 37:790–794.
2010.PubMed/NCBI
|
|
23
|
Li HL, Chen HL, Li H, Zhang KL, Chen XY,
Wang XW, Kong QY and Liu J: Regulatory effects of emodin on NF-κB
activation and inflammatory cytokine expression in RAW 264.7
macrophages. Int J Mol Med. 16:41–47. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li-Weber M: Targeting apoptosis pathways
in cancer by Chinese medicine. Cancer Lett. 332:304–312. 2013.
View Article : Google Scholar
|
|
25
|
Liu A, Chen H, Tong H, Ye S, Qiu M, Wang
Z, Tan W, Liu J and Lin S: Emodin potentiates the antitumor effects
of gemcitabine in pancreatic cancer cells via inhibition of nuclear
factor-κB. Mol Med Rep. 4:221–227. 2011.PubMed/NCBI
|
|
26
|
Liu Z, Xie Z, Jones W, Pavlovicz RE, Liu
S, Yu J, Li PK, Lin J, Fuchs JR, Marcucci G, et al: Curcumin is a
potent DNA hypomethylation agent. Bioorg Med Chem Lett. 19:706–709.
2009. View Article : Google Scholar
|
|
27
|
Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H,
Welsh W and Yang CS: Tea polyphenol (−)-epigallocatechin-3-gallate
inhibits DNA methyltransferase and reactivates methylation-silenced
genes in cancer cell lines. Cancer Res. 63:7563–7570.
2003.PubMed/NCBI
|
|
28
|
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim
SH, Ito S, Yang C, Wang P, Xiao MT, et al: Oncometabolite
2-hydroxyglutarate is a competitive inhibitor of
α-ketoglutarate-dependent dioxy-genases. Cancer Cell. 19:17–30.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin SZ, Wei WT, Chen H, Chen KJ, Tong HF,
Wang ZH, Ni ZL, Liu HB, Guo HC and Liu DL: Antitumor activity of
emodin against pancreatic cancer depends on its dual role:
promotion of apoptosis and suppression of angiogenesis. PLoS One.
7:e421462012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu A, Luo J and Zhang JH: Emodin combined
gemcitabine inhibited the growth of pancreatic cancer in vitro and
in vivo and its mechanisms study. Zhongguo Zhong Xi Yi Jie He Za
Zhi. 32:652–656. 2012.In Chinese. PubMed/NCBI
|
|
31
|
Chen H, Wei W, Guo Y, Liu A, Tong H, Wang
Z, Tan W, Liu J and Lin S: Enhanced effect of gemcitabine by emodin
against pancreatic cancer in vivo via cytochrome C-regulated
apoptosis. Oncol Rep. 25:1253–1261. 2011.PubMed/NCBI
|
|
32
|
Tahiliani M, Koh KP, Shen Y, Pastor WA,
Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et
al: Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in
mammalian DNA by MLL partner TET1. Science. 324:930–935. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu SC and Zhang Y: Active DNA
demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol.
11:607–620. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu
J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al: Tumor development is
associated with decrease of TET gene expression and
5-methylcytosine hydroxylation. Oncogene. 32:663–669. 2013.
View Article : Google Scholar
|