Introduction
Hepatocellular carcinoma (HCC) is the most common
primary malignant tumor type observed in the human liver (1). HCC is also the third leading cause of
cancer-related mortality worldwide, and it is especially prevalent
in Southeast Asia and sub-Saharan Africa (2). Despite improvement in the treatment of
HCC over the last couple of decades, the mortality rate is still
high due to relapse and tumor metastasis (3,4).
Increased understanding of HCC progression at the molecular level
is crucial to develop novel targeted therapies which could be
promising therapeutic options for HCC patients.
Apurinic/apyrimidinic endonuclease-1 (APE1) is a
protein involved in DNA repair and transcriptional regulation of
gene expression (5). Several
studies have shown that APE1 is overexpressed in numerous types of
cancer, including prostate, non-small cell lung, colorectal,
ovarian and cervical cancer, and osteosarcoma (6–11).
APE1 overexpression was also found to be associated with a poor
prognosis and resistance to radiation and chemotherapeutic drugs
(12). Since APE1 is a vital enzyme
in the DNA repair pathway induced by irradiation damage, it may
have a critical role in the cancer cell response to chemotherapy
and radiotherapy. While the function of DNA repair has been
extensively investigated, the role of activating transcription
factors mediated by APE1 has not yet been widely studied (13). Previous studies indicated that APE1
stimulated the DNA binding activity of various transcription
factors including activator protein-1 (AP-1), NF-κB, HIF-1α and p53
(14,15). However, the link between gene
expression and related cell behavior is not well elucidated
hampering the understanding of the function of APE1 in cancer cell
biology.
Recently, APE1 was reported to be significantly
overexpressed in HCC tissues than that in the surrounding cirrhotic
tissues (16,17). In addition, we previously showed
that APE1 was highly expressed in a poorly differentiated group,
capsular invasion group and metastasis group (18). We also found that APE1 protein is
mainly expressed in the nuclei in normal liver tissues. While in
malignant liver tissues, APE1 protein is expressed in the nucleus
and cytoplasm. Therefore, we hypothesized that APE1 may be involved
in the metastatic progression of HCC. Based on previous results
(18), this study aimed to
investigate the knockdown effect of APE1 using shRNA in HCC and
demonstrate that silencing of APE1 in MHCC97-H cells can decrease
the oncogenic transforming potential in vitro and reduce the
growth of HCC tumor xenografts in vivo. It was demonstrated
that inhibition of APE1 may present a novel therapeutic approach
for the treatment of HCC.
Materials and methods
Cell line
The MHCC97-H cell line was obtained from the Liver
Cancer Research Institute of Fudan University (Shanghai, China).
The cells were cultured at 37°C in a humidified incubator under 5%
CO2 and grown in Dulbecco’s modified Eagle’s medium
(DMEM; Hyclone Company, Logan, UT, USA) supplemented with 10% fetal
bovine serum (FBS; PAA Laboratories GmbH, Pasching, Austria), 100
U/ml penicillin, and 100 µg/ml streptomycin.
Lentiviral vectors and infection
The APE1 RNA interference lentivirus vector named
LV-APE1-shRNA and LV-NC-shRNA (control) were constructed at
GeneChem Technology (Shanghai, China) as previously described
(19). The shRNA targeting
sequencing for APE1 was: 5′-GAGACCAAATGTTCAGAGAAC-3′. The MHCC97-H
cell line was infected with LV-APE1-shRNA or LV-NC-shRNA for 2 h
and subsequently placed in fresh medium, while the uninfected cells
were used as the blank control. The cells were cultured for the
next 48 h and then harvested for western blot analysis or prepared
for the following experiments.
Western blotting and RT-PCR
Western blot analysis was performed as previously
described (19) using the
antibodies as follows: mouse anti-APE1 (Novus Biologicals,
Littleton, CO, USA), anti-c-Fos polyclonal, anti-c-Jun,
anti-caspase-3 and anti-inducible matrix metalloproteinase (MMP)-2
and a monoclonal antibody against β-actin (Merck KGaA, Darmstadt,
Germany). The protein concentration was analyzed as APE1/β-actin.
Image J software (National Institutes of Health, Bethesda, MD, USA)
was used for densitometric analysis.
Total cell RNA was extracted from the infected cells
and control cells with TRIzol (Invitrogen, Carlsbad, CA, USA)
following the manufacturer’s instructions. Subsequently, reverse
transcription was performed using the PrimeScript RT reagent kit
(Takara Biotechnology, Dalian, China). Primers for APE1, c-Fos,
c-Jun, caspase-3 and MMP-2 were designed as listed in Table I. PCR reaction was performed with 30
cycles (Piko TCP9600; Thermo Scientific, Waltham, MA, USA). The
gene expression levels were analyzed as APE1/GAPDH. Densitometric
analysis was performed using Image J software.
 | Table IDetected genes and primer
sequences. |
Table I
Detected genes and primer
sequences.
| Gene | Forward primer | Reverse primer |
|---|
| APE1 |
5′-AGAGCCAGAGGCCAAGAAGAGTA-3′ |
5′-GAAGCCCATCCACATTCCAAGAG-3′ |
| c-Fos |
5′-GAGAAGCCAAGACTGAGCCG-3′ |
5′-CGTTGAAGCCCGAGAACATC-3′ |
| c-Jun |
5′-CTCAGACAGTGCCCGAGATG-3′ |
5′-GCTGCGTTAGCATGAGTTGG-3′ |
|
Caspase-3 |
5′-AGCCTGTTCCATGAAGGCAGA-3′ |
5′-CTGGCAGCATCATCCACACATAC-3′ |
| MMP-2 |
5′-GTGCCCAAGAATAGATGCTGAC-3′ |
5′-CGGTAGGGACATGCTAAGTAGAGT-3′ |
| GAPDH |
5′-CCTGCACCACCAACTGCTTAG-3′ |
5′-ACCACTGACACGTTGGCAG-3′ |
Cell proliferation
Cell proliferation of the infected and control cells
was assessed by
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay (Sigma, New York, NY, USA). Briefly, the cells were seeded at
3,000 cells/well and were cultured for 48 h prior to measurement.
The cells were then washed with phosphate-buffered saline (PBS;
Biovision, San Francisco Bay, CA, USA) and then incubated with 0.45
mg/ml MTT solution made up in serum-free media at 37°C. Once the
formation of blue formazan crystals was apparent, the MTT solution
was removed. Subsequently, 100 µl of DMSO (Sigma) was added
to solubilize the blue formazan crystal, and the absorbance was
read at 570 nm.
Cell adhesion
The adhesion assay was performed as previously
described (20). Briefly, a 96-well
plate was pre-coated with Matrigel and fibronectin (FN) (both from
BD Biosciences, New York, NY, USA), dissolved in 50 µl of
PBS, incubated for 2 h at 37°C and then blocked for non-specific
sites by the addition of 1% bovine serum albumin (BSA) in 100
µl of PBS/well for 30 min. Cells with 90% confluency were
harvested with a brief treatment of 0.25% trypsin (Invitrogen). The
cells were plated in 96-well plates at 1×105 density and
cultured for 1 h. The medium was aspirated and the cells were
washed twice with PBS to remove unattached cells. The viability of
the attached cells was determined by MTT assay. All experiments
were performed in triplicate with 4–8 replicates/experiment.
Cell migration and invasion
Cell migration and invasion assays were performed in
BioCoat Transwell chambers (Corning Costar, Tewksbury, MA, USA).
While cell migration was determined with uncoated porous inserts,
cell invasion was measured using a filter precoated with Matrigel.
Cells were starved under a serum-free condition 24 h prior to
experimentation. Subsequently, the cells were plated on the upper
chamber with 0.2% FBS and cultured for 24 h. Migrated and invaded
cells on the inserts were fixed using 90% ethanol followed by
staining with 0.1% crystal violet (Sigma) for counting. The numbers
of migrated and invaded cells were counted over three fields per
one filter for triplicate experiments.
Xenograft tumor growth
BALB/c nude mice (Shanghai Slaccas Co., Shanghai,
China) (4-5 weeks old) were used in the xenograft growth study. The
animal study was performed according to procedures approved by the
Animal Care and Ethics Committee of Fujian Medical University.
Tumor growth was measured as follows. Briefly, a volume of 0.2 ml
of 2×106 viable LV-APE1-shRNA or LV-NC-shRNA cells was
subsequently injected into the right axilla of BALB/c nude mice
(n=6 per group). Tumor volumes were analyzed every two days by
measuring the major axis (a) and the minor axis (b) by Vernier
calipers. The total volume (V) was calculated by the equation: V =
1/6πab2. After 3 weeks, the nude mice were euthanized,
and the xenografts were resected for weight measurement. The
inhibitory rate of tumor growth was calculated using the following
equation: Inhibitory rate = 1 – (average weight of experimental
group – average weight of control group) × 100%. Subsequently,
tissue samples were collected for histological and RT-PCR
analyses.
Histological analysis
Histological analysis was performed as described
previously (21). In brief,
5-µm sections were stained with hemotoxylin and 1% eosin Y
solution separately. After that, the sections were rehydrated in a
graded series of ethanol solutions, and then the ethanol was
extracted with xylene. Mounting medium was added and the slide was
covered with a coverslip prior to observation using a microscope.
Apoptosis was measured using the terminal-deoxynucleotidyl
transferase dUTP nick end labeling (TUNEL) method carried out using
the TUNEL apoptosis assay kit (C1091; Beyotime, Shanghai, China)
according to the manufacturer’s instructions. Briefly, the sections
were incubated with 0.3% H2O2 for 30 min.
After washing with PBS, the sections were treated with
biotin-16-dUTP in reaction buffer at 37°C for 60 min. Subsequently,
the specimens were incubated with streptavidin-peroxidase complex
and stained with DAB after twice with PBS (both from Maixin,
Fuzhou, China). Finally, cell morphology was observed under a light
microscope (Olympus BX43; Olympus Co., Tokyo, Japan).
Statistics
Data are expressed as mean ± SEM from a
representative experiment conducted in triplicate. Results were
analyzed using unpaired two-tailed Student’s t-test and P-values
<0.05 were applied to determine statistically significance by
comparing data sets.
Results
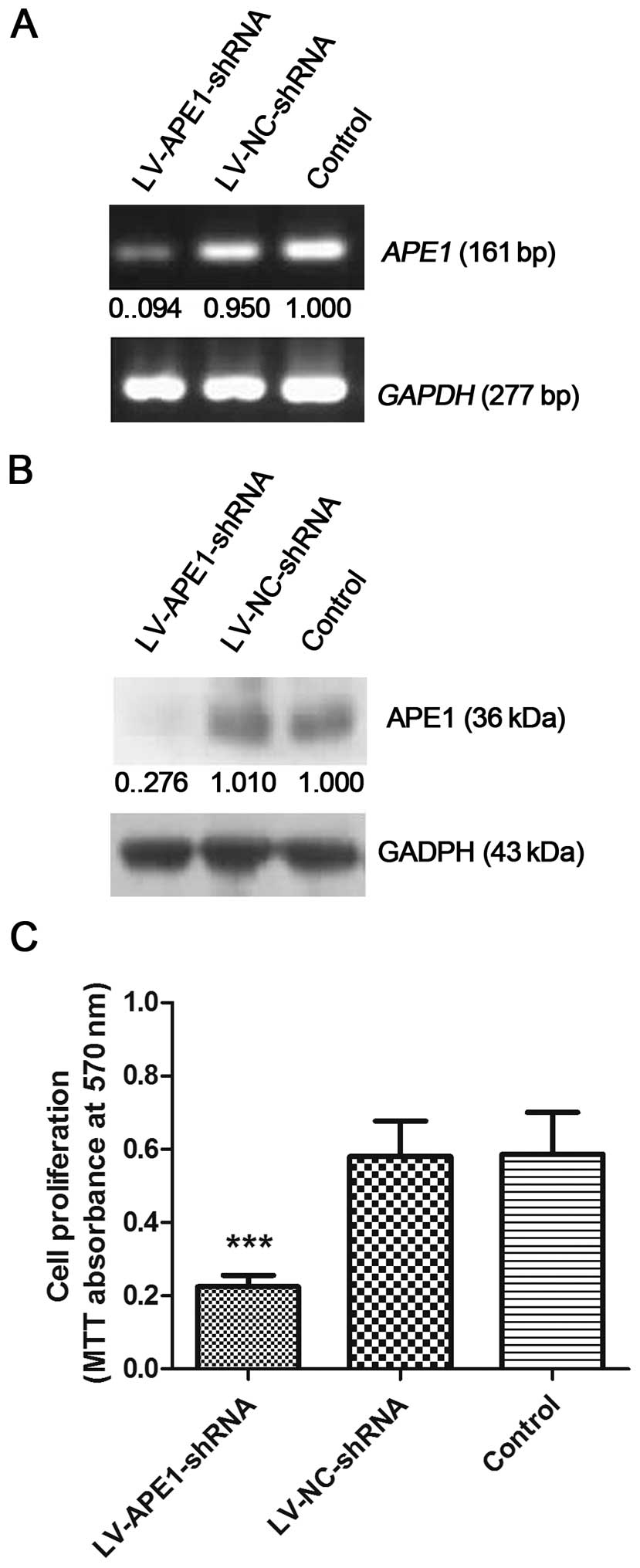
shRNA silencing of APE1 expression
effectively reduces APE1 mRNA and protein expression in the
MHCC97-H cells
MHCC97-H is a highly metastatic HCC cell line, which
expresses high endogenous APE1 according to our previous results.
In this study, MHCC97-H cells were infected by a lentivirus which
contained APE1 shRNA (LV-APE1-shRNA) or an empty vector
(LV-NC-shRNA). Following infection, the cells were collected for
mRNA and protein expression analysis by RT-PCR and western
blotting. Uninfected MHCC97-H cells were included as the negative
control. Compared with the LV-NC-shRNA cells and uninfected
negative control cells, the mRNA and protein expression levels of
APE1 in the LV-APE1-shRNA cells was decreased. Based on the RT-PCR
results, the mRNA level of APE1 was reduced by 90.6% in the
LV-APE1-shRNA cells, while the protein level of APE1 was
significantly reduced by 72.4% in the LV-APE1-shRNA cells vs. that
in the LV-NC-shRNA and control cells as detected by western
blotting (Fig. 1A and B).
APE1 silencing significantly reduces
MHCC97-H cell proliferation
After infection, MHCC97-H cell proliferation rates
were evaluated in the LV-APE1-shRNA, LV-NC-shRNA and negative
control cells. As shown in Fig. 1C,
cell proliferation was markedly decreased by knockdown of APE1 in
the LV-APE1-shRNA cells (0.2871±0.1118) compared to the LV-NC-shRNA
(0.5213±0.2551) and negative control (0.5515±0.2659) cells.
However, the proliferation rate was almost equal in the LV-NC-shRNA
and blank control groups.
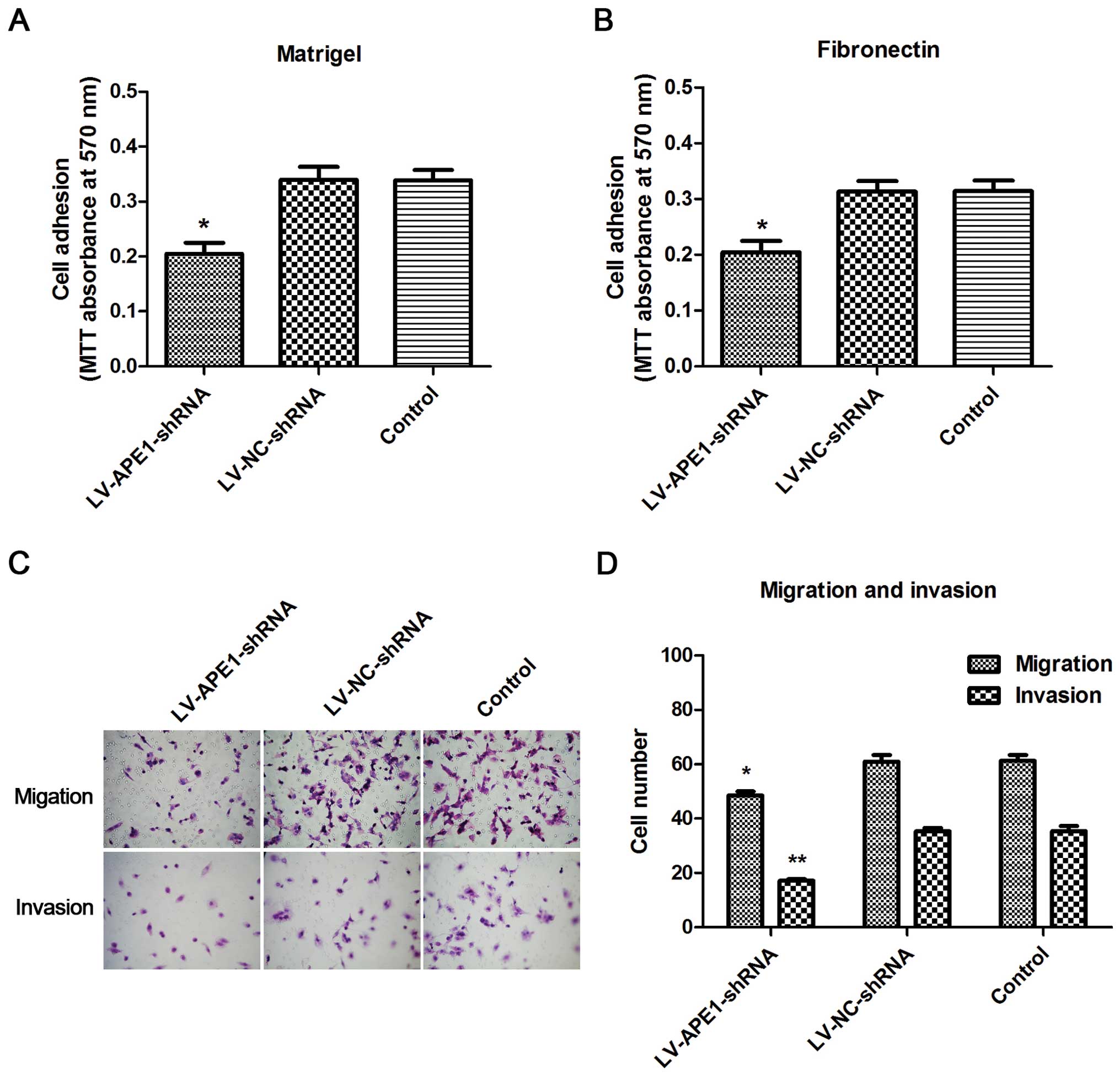
APE1 silencing inhibits HCC cell adhesion
ability
Cell adhesion was evaluated by cell-matrix adhesion,
which was measured by MTT assay to reflect the cell adhesion
ability. As shown in Fig. 2A, cell
adhesion was significantly inhibited in the LV-APE1-shRNA cells
compared to that in the LV-NC-shRNA and negative control cells in
the Matrigel-coated plates. The data were similar with the results
using FN-coated plates (Fig. 2B).
Thus, silencing of APE1 significantly inhibited HCC cell adhesion
ability.
APE1 silencing reduces cell invasive
ability
Cell migration and invasion were investigated by
Transwell assays. Cell migration was slightly decreased following
the silencing of APE1 in vitro. However, cell invasion was
markedly reduced in the LV-APE1-shRNA cells compared to that in the
LV-NC-shRNA and negative control cells (Fig. 2C). Cell counting results showed that
the cell migration in the LV-APE1-shRNA cells was decreased ~26%
compared to the LV-NC-shRNA and negative control cells (Fig. 2D).
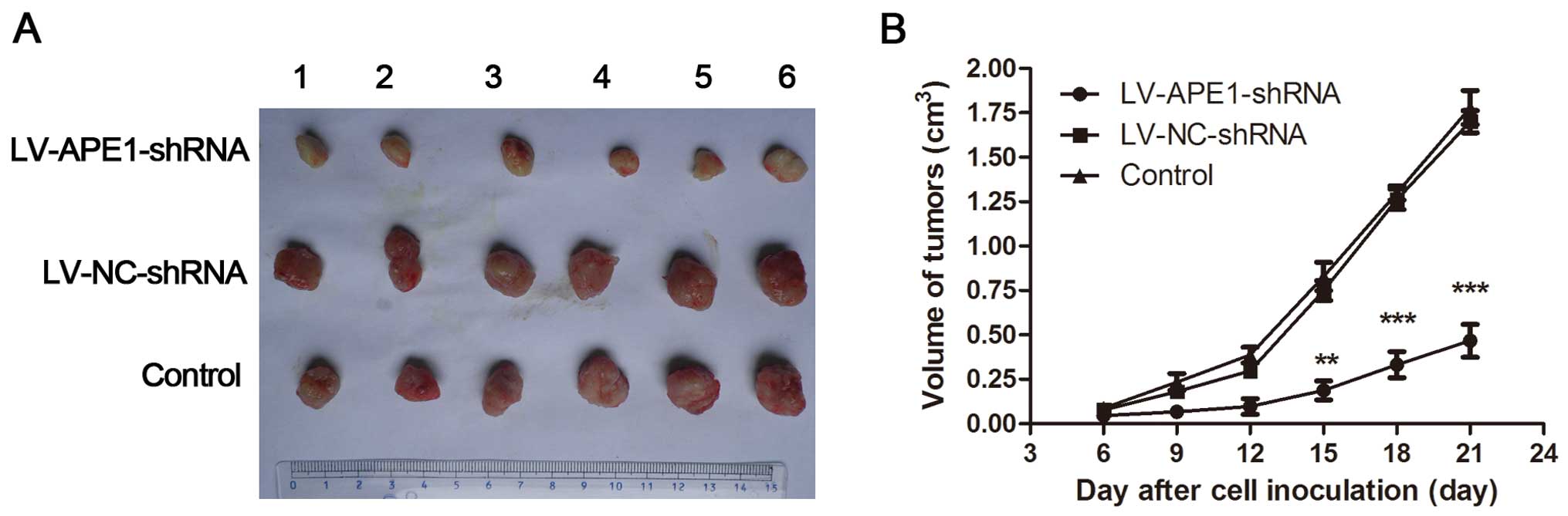
APE1 knockdown inhibits HCC tumor
xenograft growth
APE1-knockdown MHCC97-H cells were subcutaneously
injected into nude mice to investigate tumor growth in vivo.
Consistent with the in vitro data presented above indicating
that proliferation was significantly enhanced in cells infected
with LV-NC-shRNA and markedly reduced in APE1-depleted cells, tumor
growth was observed in the nude mice injected with LV-NC-shRNA
MHCC97-H cells and compared to those injected with LV-APE1-shRNA
MHCC97-H cells. The volume of tumors was also measured. Firstly,
the volume of the tumor xenografts from the LV-APE1-shRNA group
showed a marked reduction in growth rate compared to tumors
obtained from the LV-NC-shRNA and negative control groups after 9
days. Furthermore, the final weight of the tumor xenografts from
the LV-APE1-shRNA group was ~315% less than the final weight of the
tumor xenografts from the LV-NC-shRNA and negative control groups
(Fig. 3). The inhibition rate was
75.9% in the LV-APE1-shRNA group compared to the control groups in
the final stage by measuring the weight of the xenografts (Table II).
 | Table IISilencing of APE1 inhibits tumor
formation of the MHCC97-H cell-derived tumors in vivo. |
Table II
Silencing of APE1 inhibits tumor
formation of the MHCC97-H cell-derived tumors in vivo.
| Group | n | Weight of tumors
(g) | Inhibition ratio
(%) |
|---|
| LV-APE1-shRNA | 6 | 0.267±0.082a | 73.772 |
| LV-NC-shRNA | 6 | 1.018±0.160 | – |
| Control | 6 | 1.107±0.178 | – |
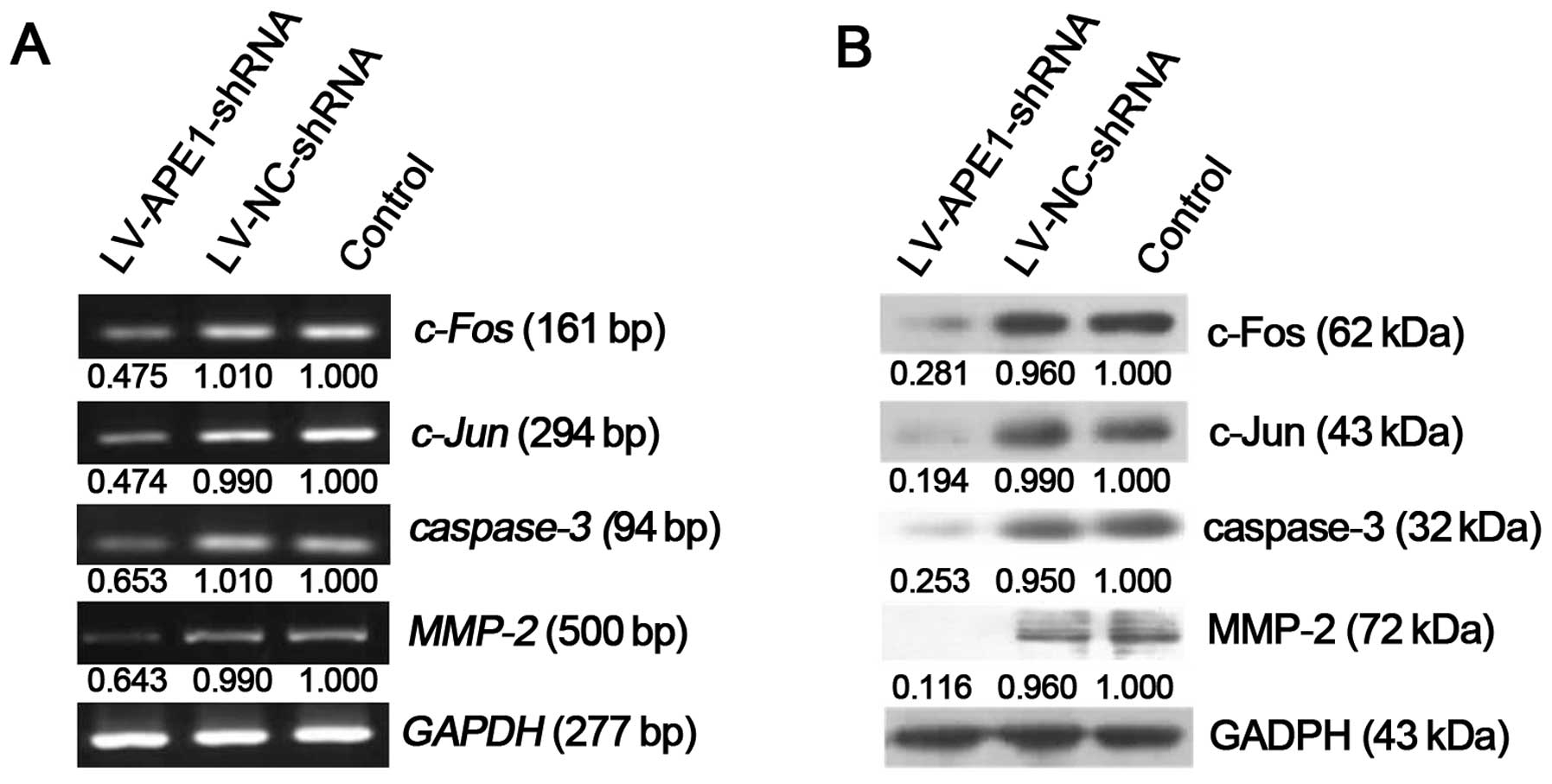
Gene and protein expression profile
analysis
RNA and protein were extracted from the xenografts,
and expression levels of various genes were assessed by RT-PCR and
western blotting. Compared with negative control, the LV-APE1-shRNA
group showed a significantly decrease in mRNA expression of
c-Fos (47.580%), c-Jun (47.402%), caspase-3
(65.355%) and MMP-2 (64.369%) (Fig. 4A). These observations were
consistent at the protein level, where the LV-APE1-shRNA group
exhibited a significant decrease in c-Fos, c-Jun, caspase-3 and
MMP-2 expression by 71.872, 80.399, 75.616 and 88.393%,
respectively (Fig. 4B).
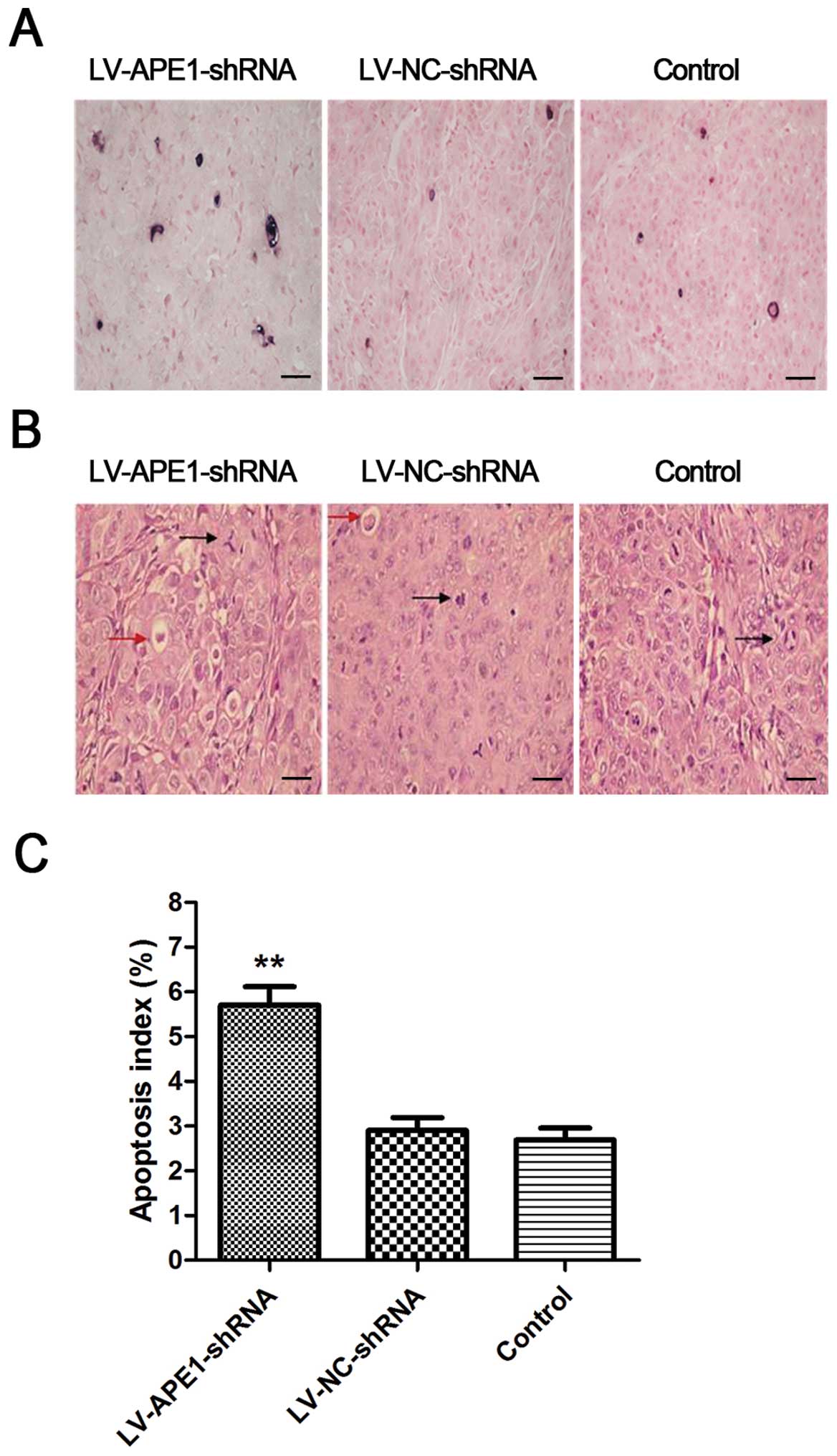
APE1 silencing accelerates the apoptosis
of HCC cells and reduces the number of irregular mitoses
When comparing the different samples from the
LV-APE1-shRNA, LV-NC-shRNA and negative control groups, we
identified a positive qualitative difference caused by silencing of
APE1. From the H&E staining results, less tissue necrosis and
more small cancer cell apoptotic bodies (indicating as red dye and
small round body) were observed in the LV-APE1-shRNA tumors
compared to the LV-NC-shRNA and negative control tumors (Fig. 5A). Furthermore, according to the
pathological classification, the irregular mitosis was grouped into
different types, such as tri-pole, asymmetrical and popcorn-like.
By counting the number of irregular mitoses, we found that
irregular mitotic tumor cells numbered <5/HPF in the
LV-APE1-shRNA group, while these numbers in the LV-NC-shRNA and
negative control groups were >5/HPF.
Apoptosis index can be calculated by counting the
apoptotic cell number using the TUNEL method. As shown in Fig. 5B, a higher number of apoptotic cells
was observed as black dots in the LV-APE1-shRNA group when compared
to the LV-NC-shRNA and negative control groups in the TUNEL-stained
tissue samples. The apoptosis index was significantly increased in
the LV-APE1-shRNA group which was almost double than that in the
LV-NC-shRNA and negative control groups (Fig. 5C).
Discussion
The poor prognosis of HCC patients is mainly due to
the high frequency of late-stage cancer and metastasis at
diagnosis. Despite benefits from surgical resection and a low risk
of complications, only a limited proportion of HCC patients are
eligible to undergo optional resection at diagnosis. Chemotherapy
represents the main therapeutic option for these HCC patients, but
neither single drug nor multiple drug treatments prolong the
survival of late-stage HCC patients (22). In order to improve therapeutic
efficacy, targeted drugs such as sorafenib are currently used in
late-stage HCC patient. Sorafenib is a multikinase inhibitor that
has already been approved for the treatment of advanced HCC
(23). Combination of chemotherapy
drugs with a targeted drug offers a novel therapeutic approach for
HCC treatment. However, the adverse effects and acquisition of drug
resistance of patients to sorafenib remain a problem in the clinic
(24). It is necessary to explore
more promising and specific targeted drugs for HCC particularly for
late-stage HCC patients with metastasis.
APE1 is a multifunctional gene which regulates DNA
repair and related gene expression during cell damage and cancer
development. Based on our previous study, APE1 was shown to be
highly expressed in poorly differentiated, capsular invasion and
metastasis groups (18). In
addition, we also found that APE1 protein is mainly expressed in
the nuclei in normal liver tissues, while in malignant liver
tissues, APE1 protein is expressed in the nucleus and cytoplasm.
Such a differentiated expression pattern of APE1 indicates that it
can be a potential marker for cancer diagnosis and treatment.
Previously, the function of APE1 in HCC was found to be associated
with cell apoptosis and resistance to radiotherapy and chemotherapy
(10). Cun et al found that
knockdown of APE1 increased the sensitivity of human HCC cells to
radiotherapy in vitro and in vivo (12) and Zou et al demonstrated that
inhibition of APE1 by a small-molecule inhibitor suppresses
pancreatic cancer cell proliferation and migration (25). Moreover, inhibition of APE1 by
small-molecular agents also inhibited the growth of tumor
endothelium and endothelial progenitor cells (26). These results suggested an important
role of APE1 in cancer and that the function of APE1 can be
investigated by silencing of APE1. Lentiviral-mediated silencing of
genes through RNA interference is an ideal approach that can
achieve long-lasting transgene expression (27). In the present study, we selected
lentiviral-mediated shRNA silencing of the APE1 gene, which can
provide more stable expression compared to atopic transfection
using liposome transfection.
Previous studies have shown that APE1 is a redox
activator in the cell nucleus, which can activate the DNA binding
activity of transcription factors associated with cancer
development such as AP-1, NF-κB and P53 (28,29).
The AP-1 complex has been implicated in the transformation and
progression of cancer (30). It was
reported that activation of AP-1 transcription factors is
frequently found in the early event of human HCC development
(31). Firstly, AP-1 is comprised
of heterodimers of Fos and Jun family proteins, which bind to a
consensus DNA sequence that are often located in the target
promoter region of genes. c-fos and c-jun are
proto-genes that are important in cell cycle progression as well as
proliferation (32). Secondly,
altering the expression of AP-1 component proteins also affects
cellular invasion (33).
AP-1-regulated genes play a role in the invasive process. For
example, inducible matrix metalloproteinases (MMPs) share a
consensus AP-1 binding site in their promoter, which is found to be
expressed in various invasive tumors (34). In addition, Fishel et al
found that c-Fos can induce expression of MMP-1, MMP-2 and MMP-3
and initiate tumor progression (35). On the contrary, invasiveness of
cells was significantly reduced when expression of the AP-1
component and AP-1 protein activity were inhibited (32). In the present study, we found that
silencing of APE1 decreased c-Fos and MMP-2 expression, which
consequently inhibited cell-matrix adhesion, cell migration and
invasion. These results indicate that AP-1 is one of the downstream
effectors of APE1, confirming APE1 as regulator of cancer cell
proliferation and migration.
Targeted APE1 therapy was assessed in a previous
study, suggesting that APE1 expression level and its deregulation
in cells can be used as a therapeutic target for tumor radiotherapy
and chemotherapy sensitivity prediction (35). Our data validated the possibility of
using APE1 as a target for inhibiting tumor growth in vivo.
In the LV-APE1-shRNA group, the final weight of the tumor
xenografts was ~315% less than that in the control group. The
inhibition rate was 75.9% in the LV-APE1-shRNA group compared to
the control groups in the final stage by measuring the weight of
xenografts. This result is in line with previous results which
found that inhibition of APE1 reduced the expression of c-Fos,
resulting in a decrease in cell migration and invasion (28). Moreover, silencing of APE1 enhanced
the sensitivity of HCC cells to radiotherapy (12). Therefore, APE1 as a tumor suppressor
in HCC could be used in targeted therapy combined with other
treatment strategies such as surgery, chemotherapy, radiotherapy
and immunotherapy.
In conclusion, the present study demonstrated that
APE1 silencing by shRNA suppressed the proliferation and mobility
of HCC cells. Firstly, silencing of APE1 expression in HCC cells
decreased cell viability and survival compared with the control
cells in vitro while the cell proliferation rate in the
APE1-silenced group was significantly higher than that in the
control cells. In addition, APE1 was important in cell-matrix
adhesion, cell migration and invasion. Silencing of APE1 expression
markedly decreased cell adhesion in Matrigel or FN-coated plates.
Moreover, cell migration and invasion were also decreased when APE1
expression was depleted. Finally, xenograft growth was inhibited by
silencing of APE1 expression compared with that in the control
groups in nude mice. Activation of protein expression levels were
found to be associated with APE1 expression by western blot and
RT-PCR analyses. Expression of c-Fos and c-Jun were downregulated
by silencing of APE1, while expression of caspase-3 and MMP-2 were
also decreased. These results suggest that APE1 is an important
gene in the regulation of HCC cell proliferation and mobility by
regulating related gene expression. These findings suggest that
APE1 may be useful in cancer targeted drug development for
inhibition of cancer metastasis.
Acknowledgments
This study was supported by the United Health and
Education Research Project of Fujian Province (grant no. WKJFJ-04),
the Scienctific Research Project of Education Department of Fujian
Province (grant no. 2013B009), as well as the Research Fund of the
Health System for Young Talents of Fujian Province (grant no.
2014-ZQN-JC-24).
References
|
1
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tang ZY: Hepatocellular carcinoma – cause,
treatment and metastasis. World J Gastroenterol. 7:445–454.
2001.
|
|
4
|
Bruix J and Sherman M: American
association for the study of liver diseases: management of
hepatocellular carcinoma: an update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abbotts R and Madhusudan S: Human AP
endonuclease 1 (APE1): from mechanistic insights to druggable
target in cancer. Cancer Treat Rev. 36:425–435. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kelley MR, Cheng L, Foster R, Tritt R,
Jiang J, Broshears J and Koch M: Elevated and altered expression of
the multifunctional DNA base excision repair and redox enzyme
Ape1/ref-1 in prostate cancer. Clin Cancer Res. 7:824–830.
2001.PubMed/NCBI
|
|
7
|
Yoo DG, Song YJ, Cho EJ, Lee SK, Park JB,
Yu JH, Lim SP, Kim JM and Jeon BH: Alteration of APE1/ref-1
expression in non-small cell lung cancer: the implications of
impaired extracellular superoxide dismutase and catalase
antioxidant systems. Lung Cancer. 60:277–284. 2008. View Article : Google Scholar
|
|
8
|
Xiang DB, Chen ZT, Wang D, Li MX, Xie JY,
Zhang YS, Qing Y, Li ZP and Xie J: Chimeric adenoviral vector
Ad5/F35-mediated APE1 siRNA enhances sensitivity of human
colorectal cancer cells to radiotherapy in vitro and in vivo.
Cancer Gene Ther. 15:625–635. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tanner B, Grimme S, Schiffer I,
Heimerdinger C, Schmidt M, Dutkowski P, Neubert S, Oesch F, Franzen
A, Kölbl H, et al: Nuclear expression of apurinic/apyrimidinic
endonuclease increases with progression of ovarian carcinomas.
Gynecol Oncol. 92:568–577. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Herring CJ, West CM, Wilks DP, Davidson
SE, Hunter RD, Berry P, Forster G, MacKinnon J, Rafferty JA, Elder
RH, et al: Levels of the DNA repair enzyme human
apurinic/apyrimidinic endonuclease (APE1, APEX, Ref-1) are
associated with the intrinsic radiosensitivity of cervical cancers.
Br J Cancer. 78:1128–1133. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang D, Luo M and Kelley MR: Human
apurinic endonuclease 1 (APE1) expression and prognostic
significance in osteosarcoma: Enhanced sensitivity of osteosarcoma
to DNA damaging agents using silencing RNA APE1 expression
inhibition. Mol Cancer Ther. 3:679–686. 2004.PubMed/NCBI
|
|
12
|
Cun Y, Dai N, Xiong C, Li M, Sui J, Qian
C, Li Z and Wang D: Silencing of APE1 enhances sensitivity of human
hepatocellular carcinoma cells to radiotherapy in vitro and in a
xenograft model. PLoS One. 8:e553132013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tell G, Quadrifoglio F, Tiribelli C and
Kelley MR: The many functions of APE1/Ref-1: not only a DNA repair
enzyme. Antioxid Redox Signal. 11:601–620. 2009. View Article : Google Scholar
|
|
14
|
Bhakat KK, Mantha AK and Mitra S:
Transcriptional regulatory functions of mammalian AP-endonuclease
(APE1/Ref-1), an essential multifunctional protein. Antioxid Redox
Signal. 11:621–638. 2009. View Article : Google Scholar
|
|
15
|
Li M and Wilson DM III: Human
apurinic/apyrimidinic endo-nuclease 1. Antioxid Redox Signal.
20:678–707. 2014. View Article : Google Scholar :
|
|
16
|
Di Maso V, Avellini C, Crocè LS, Rosso N,
Quadrifoglio F, Cesaratto L, Codarin E, Bedogni G, Beltrami CA,
Tell G, et al: Subcellular localization of APE1/Ref-1 in human
hepatocellular carcinoma: possible prognostic significance. Mol
Med. 13:89–96. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang QH, Xiang DB, Li MX, Liao PL, Li ZP
and Wang D: Expression of DNA repair gene apurinic/apyrimidinic
endonuclease 1 and its correlation with the expression of mutant
p53 in hepatocellular carcinoma. Chin J Dig Surg. 8:453–456.
2009.
|
|
18
|
Huang AM, Zheng ZH, Liu JF, Zang SB, Gao
LY and Chen SP: The expression of APE1 gene and its clinical
implication in hepatocellular carcinoma: a study using tissue chip
assay. Zhonghua Gan Zang Bing Za Zhi. 16:542–543. 2008.In Chinese.
PubMed/NCBI
|
|
19
|
Li YJ, Zheng ZH, Liu JF, Gao MQ and Huang
AM: Construction and identification of a lentiviral vector for RNA
interference of APE1 gene. J Fujian Med Univ. 44:86–90. 2010.
|
|
20
|
Casey RC, Oegema TR Jr, Skubitz KM,
Pambuccian SE, Grindle SM and Skubitz AP: Cell membrane
glycosylation mediates the adhesion, migration, and invasion of
ovarian carcinoma cells. Clin Exp Metastasis. 20:143–152. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fischer AH, Jacobson KA, Rose J and Zeller
R: Hematoxylin and eosin staining of tissue and cell sections. CSH
Prot. 2008:pdb.prot4986. 2008.
|
|
22
|
Di Maio M, De Maio E, Perrone F, Pignata S
and Daniele B: Hepatocellular carcinoma: systemic treatments. J
Clin Gastroenterol. 35(Suppl 2): S109–S114. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Llovet JM, Ricci S, Mazzaferro V, Hilgard
P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A,
et al: SHARP investigators study group: sorafenib in advanced
hepatocellular carcinoma. N Engl J Med. 359:378–390. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Llovet JM and Bruix J: Molecular targeted
therapies in hepato-cellular carcinoma. Hepatology. 48:1312–1327.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zou GM and Maitra A: Small-molecule
inhibitor of the AP endo-nuclease 1/REF-1 E3330 inhibits pancreatic
cancer cell growth and migration. Mol Cancer Ther. 7:2012–2021.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zou GM, Karikari C, Kabe Y, Handa H,
Anders RA and Maitra A: The APE-1/Ref-1 redox antagonist E3330
inhibits the growth of tumor endothelium and endothelial progenitor
cells: therapeutic implications in tumor angiogenesis. J Cell
Physiol. 219:209–218. 2009. View Article : Google Scholar
|
|
27
|
Morris KV and Rossi JJ:
Lentiviral-mediated delivery of siRNAs for antiviral therapy. Gene
Ther. 13:553–558. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ando K, Hirao S, Kabe Y, Ogura Y, Sato I,
Yamaguchi Y, Wada T and Handa H: A new APE1/Ref-1-dependent pathway
leading to reduction of NF-kappaB and AP-1, and activation of their
DNA-binding activity. Nucleic Acids Res. 36:4327–4336. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eferl R and Wagner EF: AP-1: a
double-edged sword in tumorigenesis. Nat Rev Cancer. 3:859–868.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu P, Kimmoun E, Legrand A, Sauvanet A,
Degott C, Lardeux B and Bernuau D: Activation of NF-kappa B, AP-1
and STAT transcription factors is a frequent and early event in
human hepa-tocellular carcinomas. J Hepatol. 37:63–71. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yuen MF, Wu PC, Lai VC, Lau JY and Lai CL:
Expression of c-Myc, c-Fos, and c-jun in hepatocellular carcinoma.
Cancer. 91:106–112. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ozanne BW, McGarry L, Spence HJ, Johnston
I, Winnie J, Meagher L and Stapleton G: Transcriptional regulation
of cell invasion: AP-1 regulation of a multigenic invasion
programme. Eur J Cancer. 36:1640–1648. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Westermarck J and Kähäri VM: Regulation of
matrix metallo-proteinase expression in tumor invasion. FASEB J.
13:781–792. 1999.PubMed/NCBI
|
|
34
|
Lam E, Kilani RT, Li Y, Tredget EE and
Ghahary A: Stratifin-induced matrix metalloproteinase-1 in
fibroblast is mediated by c-fos and p38 mitogen-activated protein
kinase activation. J Invest Dermatol. 125:230–238. 2005.PubMed/NCBI
|
|
35
|
Fishel ML and Kelley MR: The DNA base
excision repair protein Ape1/Ref-1 as a therapeutic and
chemopreventive target. Mol Aspects Med. 28:375–395. 2007.
View Article : Google Scholar : PubMed/NCBI
|