Introduction
Quercetin is a member of the flavonoids family and
one of the leading dietary antioxidants. It is present ubiquitously
in vegetables and fruits and is considered to exert beneficial
health effects (1,2). Quercetin is known to induce autophagy
in several cell types (3–5) and has attracted much attention in
recent years due to its anticancer effects in many types of cancer
(6,7).
Apoptosis is a morphologically obvious form of
programmed cell death that plays a critical role during
homeostasis, development, and diseases including cancer, acquired
immunodeficiency syndrome and neurodegenerative disorders (8). Apoptosis, also called programmed cell
death type I, can be induced in cancer cells by anticancer agents
(9,10). Tumor necrosis factor (TNF)-related
apoptosis-inducing ligand (TRAIL) was first confirmed as one of the
TNF super-family. It can induce apoptosis selectively in
tumorigenic or transformed cells, but not in normal cells,
highlighting its potential therapeutic application in cancer
treatment (11).
Autophagy is a conserved trafficking pathway that is
highly regulated by environmental conditions (12). The implication of autophagy as a
cell death mechanism in cancer with inactivated apoptosis is not
surprising. However, the role of autophagy in cancer and treatment
responsiveness is complicated (13). Autophagy is a general term for the
degradation of cytoplasmic ingredients within lysosomes (14). Macro-autophagy was first described
in mammals as the isolation of complete portions of cytosol,
including soluble proteins as well as complete organelles, in a
double membrane vesicle called autophagosome (15,16).
Once the 2 sides of the autophagosome fuse to each other, there is
a second fusion event between the autophagosome and the
lysosomes/vacuole that leads to the formation of the autolysosome
(17). The anti-malarial drug,
chloroquine (CQ) inhibits lysosomal acidification and prevents the
degradation of autophagosomes, thereby suppressing the autophagy
flux (18).
Microtubule-associated protein light chain 3 (LC3)
is localized and aggregated on the autophagosome and is, therefore,
considered as a marker of autophagy. LC3B undergoes lipidation and
is recruited to the phagophore where it is essential for membrane
elongation and closure (19). LC3B
transforms from LC3B-I to LC3B-II during autophagosome formation
(20). P62 is a multifunctional
signaling molecule, associated with a variety of cellular pathways.
It is one of the best-known autophagic substrates, and is,
therefore, extensively employed as an indicator of autophagic
degradation (21). P62 can deliver
ubiquitinylated cargos to the proteasome, though they are mainly
degraded by autophagy (21,22). P62 levels are generally inversely
related to autophagic degradation, since the loss of Atg genes or
factors required for the fusion of autophagosomes with lysosomes
all result in a marked increase of P62-positive aggregates
(23,24).
Apoptosis is blocked in various cancer cells, and
autophagy may be a major contributing mechanism of cancer cell
death; thus, the induction of autophagy may be employed as a
promising therapeutic strategy in cancer treatment (25). A549, human lung cancer cells, are
known to be TRAIL-resistant cells (26). We identified that treatment with
TRAIL or quercetin alone did not exert an influence on A549 human
cancer cells. We next investigated the effect of co-treatment of
TRAIL and quercetin on human cancer cells and quercetin-mediated
autophagy flux. We also examined the effect of quercetin-induced
autophagy on TRAIL-induced apoptosis in human lung cancer
cells.
Materials and methods
Cell culture
The human lung cancer cell line A549 was obtained
from the American Type Culture Collection (ATCC; Manassas, VA,
USA). Cells were cultured in RPMI-1640 (Invitrogen-Gibco, Carlsbad,
CA, USA) supplemented with 10% fetal bovine serum (FBS;
Invitrogen-Gibco), 100 U/ml penicillin, and 0.1 mg/ml gentamycin in
a humidified incubator maintained at 37°C and 5% CO2.
Cells were treated for 12 h with quercetin (Sigma-Aldrich, St.
Louis, MO, USA) and then exposed for 3 h to 200 ng/ml TRAIL, with
or without the autophagy inhibitor, chloroquine (10 µM)
(Sigma-Aldrich).
Crystal violet assay
Cell morphology was assessed microscopically
(inverted Microscope, Nikon Eclipse TS100; Nikon Corp., Tokyo,
Japan) and cell viability was determined by crystal violet staining
(C0775; Sigma-Aldrich), as previously described (27). Briefly, cells were stained for 10
min at RT with crystal violet solution (0.5% crystal violet in 30%
ethanol and 3% formaldehyde), washed 5 times with water, and then
dried. Subsequently, the cells were lysed with 1% SDS (sodium
dodecyl sulphate) and the absorbance was measured at 550 nm. Cell
viability was calculated from the relative dye intensity of the
samples compared to the controls.
Terminal deoxynucleotidyl transferase
dUTP nick end labeling (TUNEL) assay
TUNEL assay was carried out as previously described
(28). TUNEL analysis was performed
to measure the degree of cellular apoptosis using an in situ
ApoBrdu DNA fragmentation assay kit (BioVision, Mountain View, CA,
USA), following the manufacturer’s instructions. Cells were
counterstained with propidium iodide (PI) to show cell nuclei.
Trypan blue exclusion assay
The number of viable cells was determined by trypan
blue dye exclusion (Sigma-Aldrich) using a hemocytometer. The
result was expressed as a percentage relative to vehicle-treated
controls.
BacMam transduction
Wild-type or mutant GFP-tagged LC3B was expressed in
cells by adding the appropriate concentrations of appropriate virus
from the Premo Autophagy Sensor LC3B-GFP (BacMam 2.0) kit (P36235;
Life Technologies) to the growth medium as indicated in the figure
legends.
Western blot analysis
A549 cells were lysed in lysis buffer [25 mM HEPES
(4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), pH 7.4, 100
mM NaCl, 1 mM EDTA (ethylene diamine tetra acetic acid), 5 mM
MgCl2, 0.1 mM DTT (dithiothreitol), and a protease
inhibitor mixture]. Whole cell proteins were electrophoretically
resolved on a 10–15% sodium dodecyl sulfate polyacrylamide gel and
transferred to a nitrocellulose membrane. Immunoreactivity was
detected through sequential incubation with primary antibodies,
horseradish peroxidase-conjugated secondary antibodies, and
enhanced chemiluminescence reagents i.e. West Save Gold detection
kit (AbFrontier Co., Ltd., Seoul, Korea). The primary antibodies
used for immunoblotting were anti-LC3B (#4108; Cell Signaling
Technology, Danvers, MA, USA), anti-P62 (#MABC32; Millipore,
Billerica, MA, USA), anti-phospho-AKT (#2118-1; Epitomics,
Burlingame, CA, USA), anti-caspase-3 (#9665; Cell Signaling
Technology), anti-cleaved caspase-3 (#9661; Cell Signaling
Technology) and anti-β-actin (A5441; Sigma Aldrich). Images were
examined using a Fusion FX7 imaging system (Vilber Lourmat, Marne
La Vallee, France). Densitometry of the signal bands was analyzed
using Bio-1D software (Vilber Lourmat).
Statistical analysis
The unpaired t-test or Welch’s correction was used
for comparison between the 2 groups. The one-way ANOVA followed by
the Tukey-Kramer test was used for multiple comparison. All
statistical analyses were performed with GraphPad Prism software.
Results were considered significant for values
**P<0.01 or ***P< 0.001.
Results
Quercetin enhanced TRAIL-mediated cell
death in human lung cancer cells
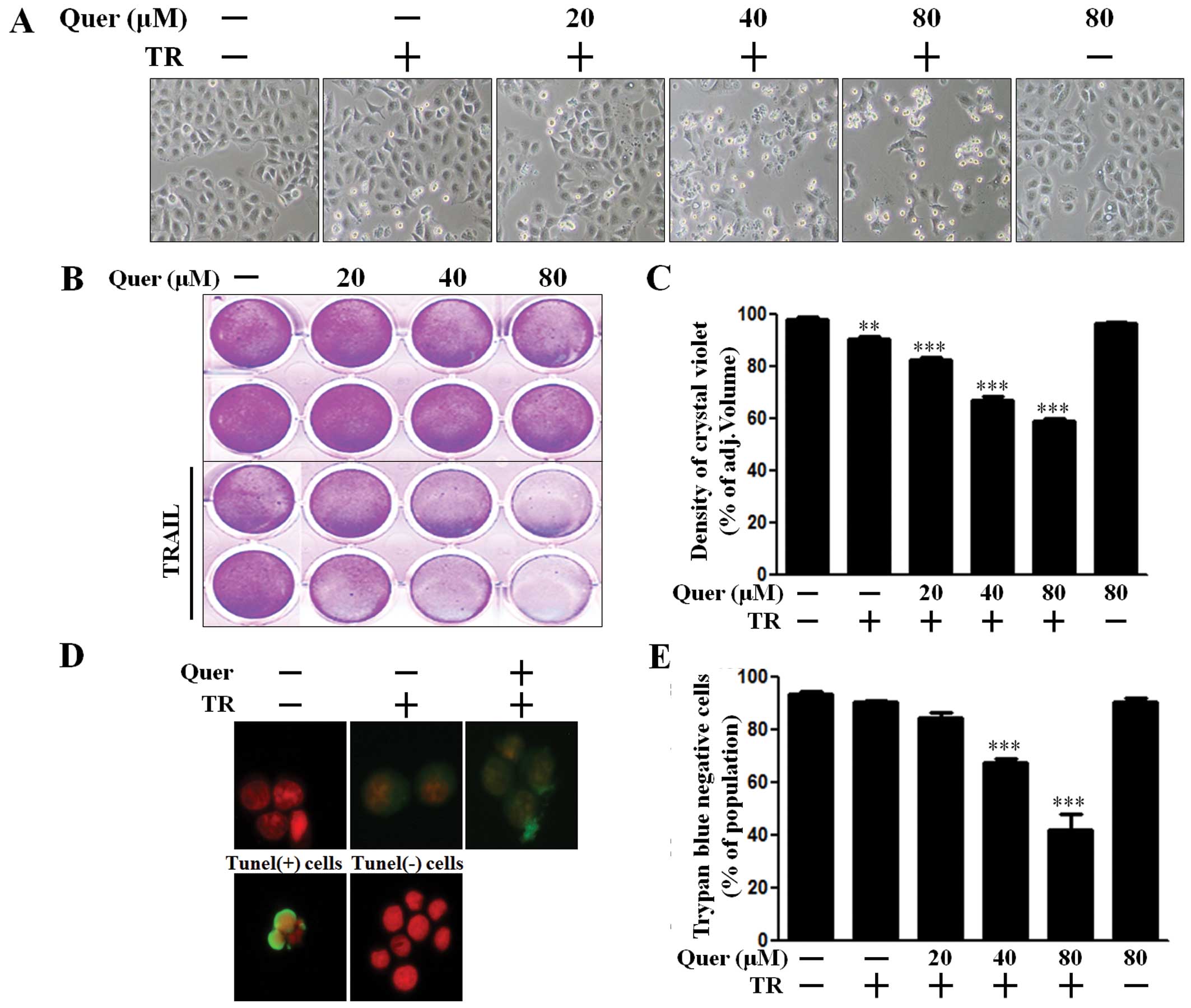
We conducted several types of cell viability assays
to investigate the effect of co-treatment with quercetin and TRAIL
on A549 human lung cancer cells. First, we examined the
photographed image of cell amounts using a light microscope and the
crystal violet assay. The cell viability of cells treated with
quercetin only was comparable to that of untreated controls. Cell
death resulted in 3–5% of TRAIL-treated A549 cells. Importantly,
quercetin treatment enhanced cell death to ~70% on photographed
images (Fig. 1A) and augmented cell
death to ~50% in the crystal violet assay (Fig. 1B and C). We additionally performed a
TUNEL assay (Fig. 1D) and trypan
blue exclusion assay (Fig. 1E). As
shown in Fig. 1D, apoptosis in
quercetin and TRAIL treated cells emitted green fluorescence
indicative of DNA strand breakage. In Fig. 1E, quercetin dose-dependently
increased TRAIL-mediated cell death. These results indicated that
quercetin was effective in promoting TRAIL-induced cell death in
A549 human lung cancer cells.
Quercetin treatment induces autophagy
flux and apoptosis
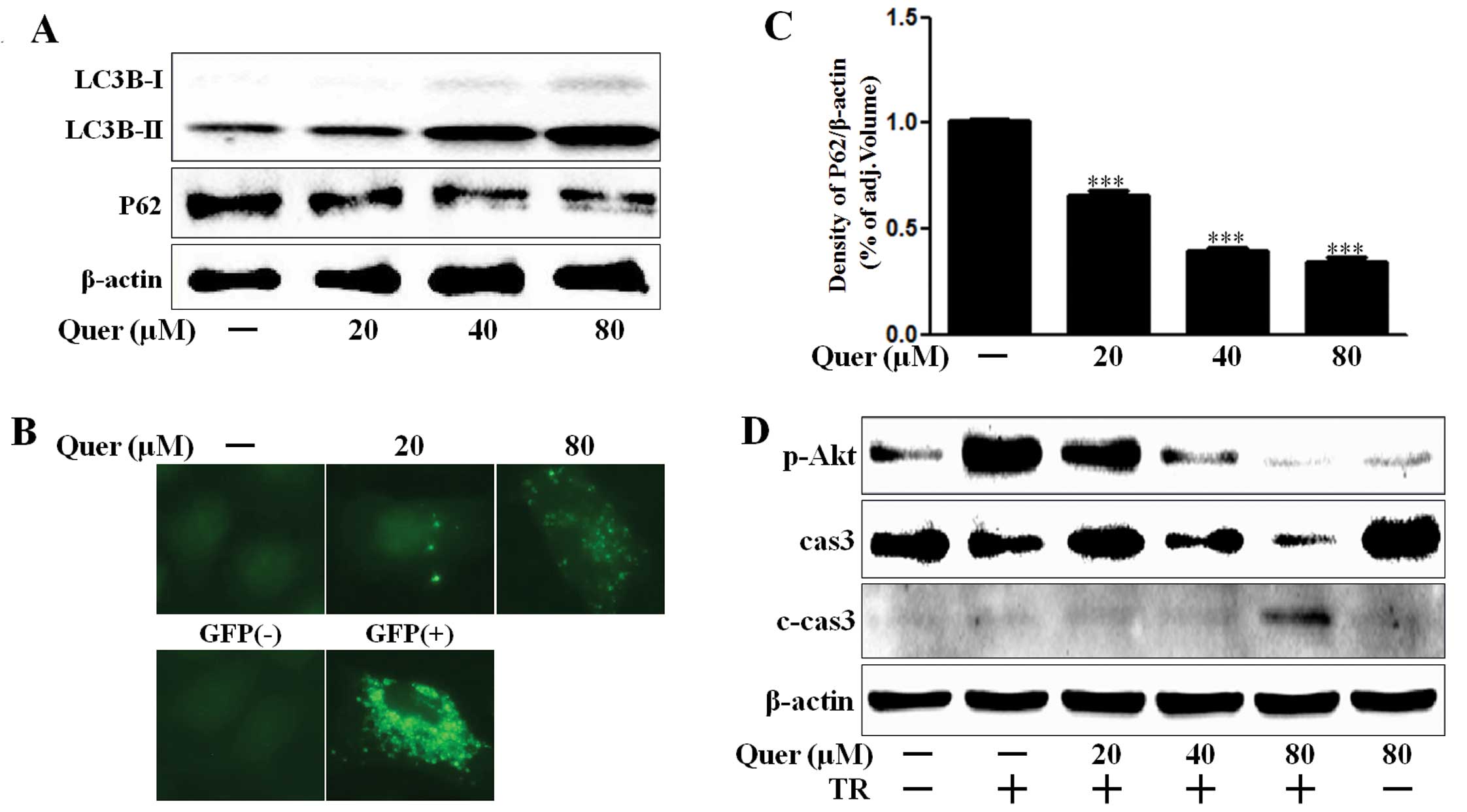
We evaluated quercetin mediated autophagy flux by
estimating LC3B transformation and P62 expression. Levels of the
late autophagosome marker LC3-II increased in the quercetin-treated
group in a dose-dependent manner as compared with the control group
on western blot analysis (Fig. 2A).
The activation of the autophagy through the formation of
autophagosomes in A549 lung cancer cells was visualized by the
Premo Autophagy Sensor (LC3B-FP) BacMam 2.0 system. LC3B-FP and
LC3B (G120A)-FP viral vectors (MOI=30) were transduced in A549
cells, enabling the expression of fluorescent LC3B protein. We
consequently monitored autophagosomes dynamics using inverted
fluorescent microscopy. The mutant chimera LC3B (G120A)-FP was used
as a negative control. According to the results shown in Fig. 2B, BacMam LC3B (G120A)-FP transduced
cells showed a marked diffuse cytosolic expression pattern. A549
cells treated with quercetin, exhibited an extensive punctate
fluorescent distribution pattern, suggesting LC3B-FP protein
accumulation in autophagosome. P62 was also decreased
dose-dependently by quercetin (Fig. 2A
and C). These results suggested that quercetin dose-dependently
mediated autophagy flux. Akt is a key signaling molecule that
associates oncogenic receptors to many essential pro-survival
cellular functions in human cancer (29). We determined that quercetin
treatment interrupted Akt activation and enhanced caspase-3
cleavage (Fig. 2D). Caspase-3 plays
a key role in regulating programmed cell death or apoptosis, a
normal process required for regulatory maintenance of physiological
functions (30). Thus, these
results suggested that quercetin mediated autophagy flux and
enhanced TRAIL-induced apoptosis in A549 human lung cancer
cells.
Quercetin regulates autophagy flux
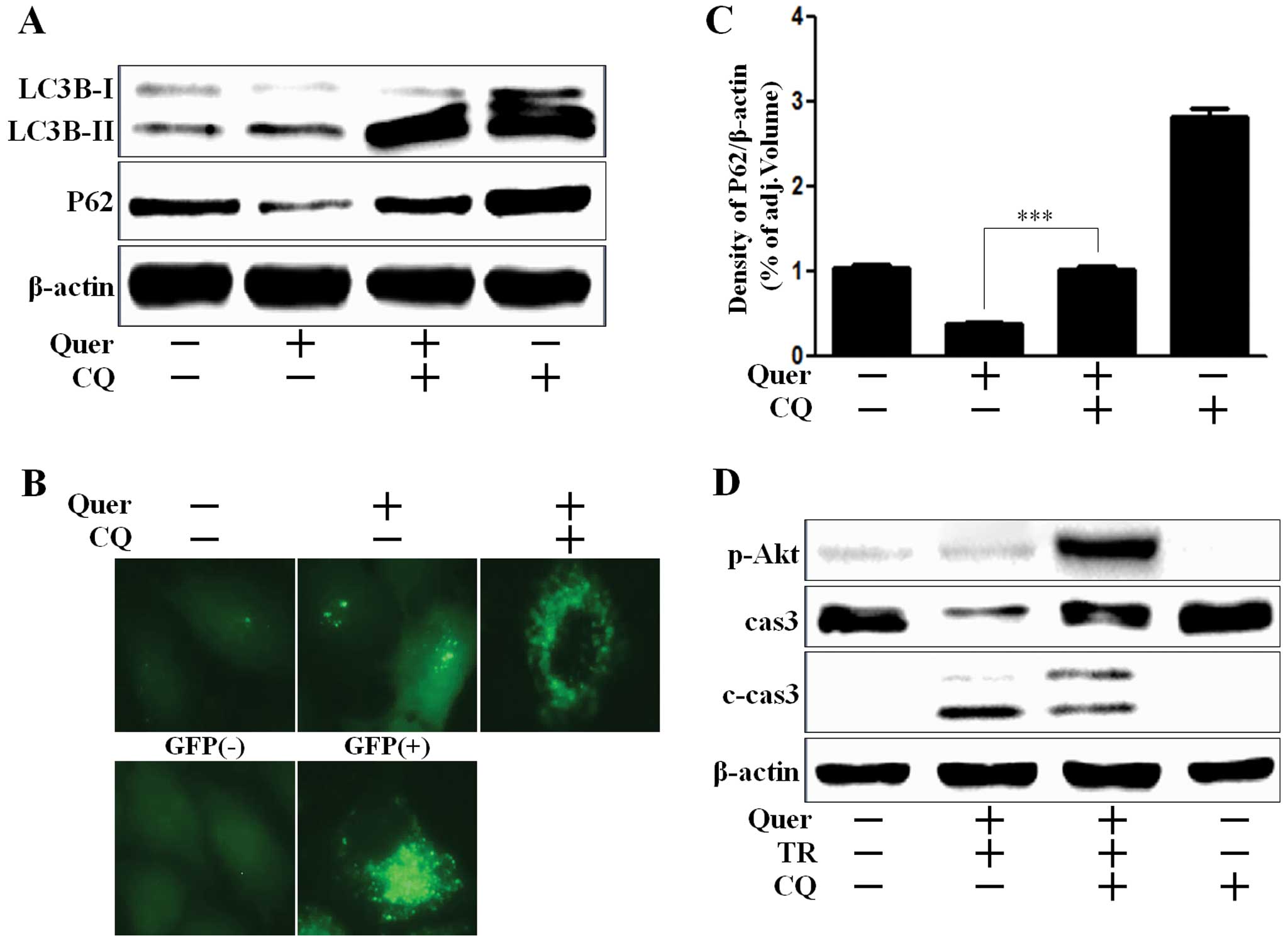
We investigated whether quercetin induced autophagy
flux or regulated chloroquine (CQ)-mediated autophagy flux. CQ is
widely used to inhibit the maturation of autophagosome into
degradative autolysosome (31,32).
We showed that upregulation of LC3B-II by quercetin was potentiated
by CQ, since CQ inhibited the fusion of autophagosome and
autolysosome (Fig. 3A). A549 cells
treated with quercetin, presented an extensive punctate fluorescent
distribution pattern which was also augmented by CQ (Fig. 3B). P62 is a crucial mediator to
target protein to the autophagy system in the removal of aggregated
proteins. P62 is itself degraded during autophagy (33). Reduction of P62 level by quercetin
treatment indicated autophagy flux activation, and also the
activation of autophagy flux was inhibited by CQ treatment, given
the increase of P62 levels (Fig. 3A and
C). According to the results shown in Fig. 3D, A549 cells co-treated with CQ were
augmented Akt activation and decreased caspase-3 cleavage, whereas
caspase-3 activation were induced in cells treated with quercetin
and TRAIL treatment, which indicated that CQ played a protective
role against apoptotic cell death in A549 cells.
Autophagy regulates quercetin-induced
TRAIL sensitivity
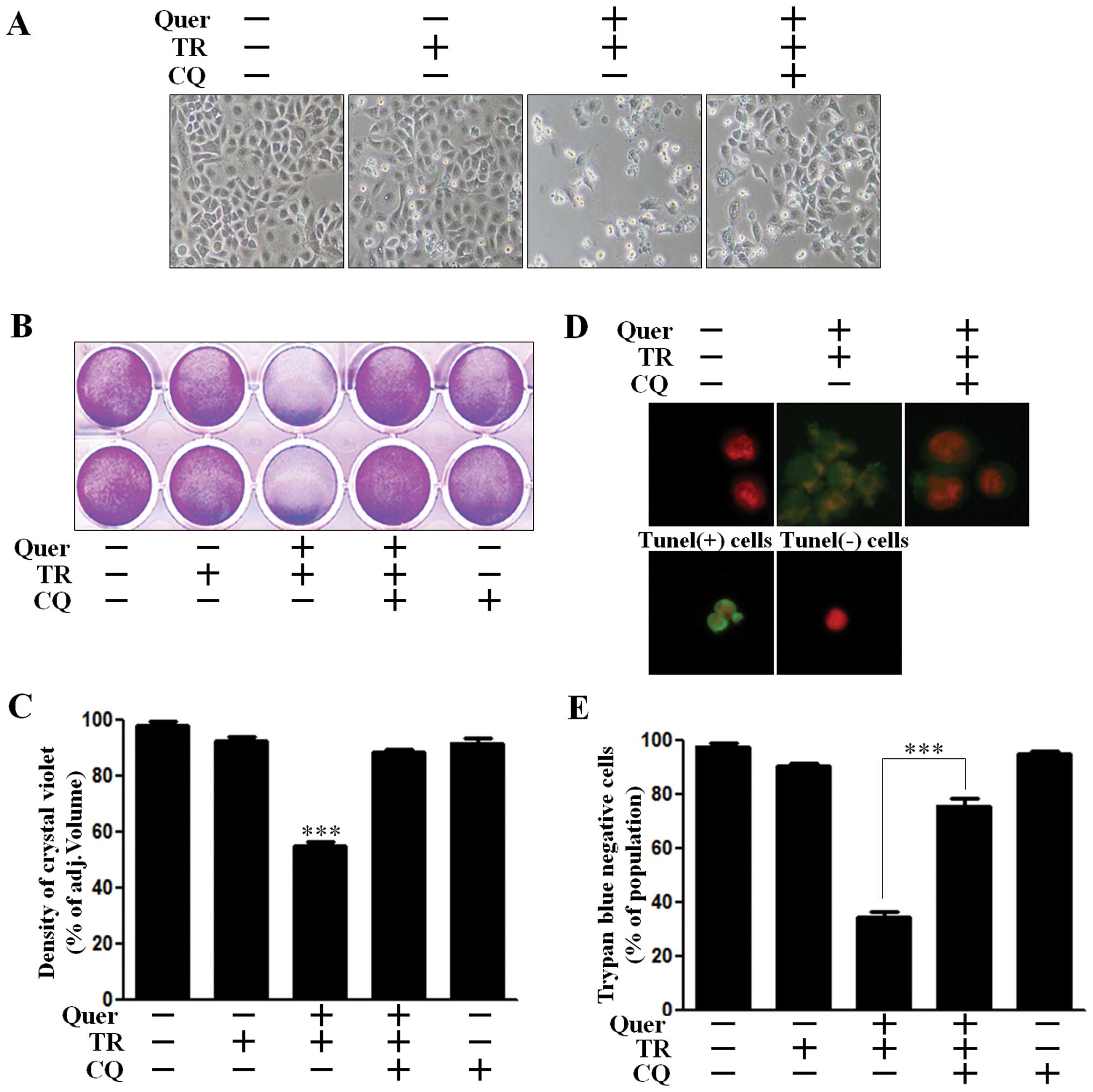
We analyzed cell viability using CQ, to investigate
the effect of quercetin-mediated autophagy on TRAIL-sensitivity.
A549 cells treated with CQ recovered Akt activation and were
protected against cleavage of caspase-3 (Fig. 3B). We accordingly investigated
whether autophagy inhibition using CQ exerted an influence on cell
viability. We examined the photographed image of cell amounts using
light microscopy and performed the crystal violet assay. Quercetin
treatment in TRAIL treated cells enhanced cell death to ~70% in the
photographed image (Fig. 4A), and
to 50% in the crystal violet assay (Fig. 4B and C). However, cell death
recovered to ~60% cell survival in the photographed image (Fig. 4A) and 85% cell survival in the
crystal violet assay (Fig. 4B and
C). We performed a TUNEL assay (Fig. 1D) and trypan blue exclusion assay
(Fig. 1E). As shown in Fig. 4D, the apoptotic process in quercetin
and TRAIL treated cells emitted green fluorescence, indicative of
DNA strand breakage, however, green fluorescence was weak in
CQ-treated cells. In Fig. 1E, CQ
treatment alleviated cell death by inhibiting quercetin-mediated
TRAIL sensitivity. These results indicated that CQ was effective in
inhibiting quercetin-induced TRAIL sensitivity by regulating
autophagy flux in A549 human lung cancer cells. Collectively our
data suggested that quercetin-induced autophagy flux had a harmful
effect on TRAIL sensitivity and inhibition of autophagy flux played
a protective role against quercetin-mediated TRAIL sensitivity in
A549 human lung cancer cells.
Discussion
The purpose of the present study was to investigate
the role of quercetin-induced autophagy flux and the regulation of
TRAIL-mediated sensitivity by quercetin treatment in A549 human
lung cancer cells. The results suggested that quercetin-induced
autophagy and the resultant enhancement in quercetin-induced TRAIL
sensitivity might be a key underlying mechanism of autophagy
flux.
Quercetin belongs to an extensive class of
polyphenolic flavonoid compounds and is practically ubiquitous in
plants and plant food sources. It has been studied as a promising
chemoprevention component in a variety of cancer models (34). Consistent with previous reports, the
present study showed that quercetin promoted TRAIL-mediated
apoptosis in lung cancer cells.
Autophagy is an evolutionary conserved, dynamic
lysosome-mediated process that entails the sequestration and
delivery of cytoplasmic material to the lysosome where it is
degraded and recycled (35,36). Some studies have insisted that
autophagy is a double-edged sword, with both useful and harmful
potential in cancer (37). Wang
et al (38) suggested that
quercetin induces protective autophagy in gastric cancer cells.
Their study indicated that quercetin-induced apoptosis and
autophagy played a protective role during apoptosis. The present
study, on the other hand, showed that quercetin-induced autophagy
induced TRAIL-sensitivity, and then mediated apoptosis. We
demonstrated that quercetin-induced autophagy did not play a
protective function by using an autophagy inhibitor in A549 lung
cancer cells. However, this experimental evidence was insufficient
to elucidate function of autophagy flux.
Some reports confirmed that TRAIL induces autophagy
in several types of cancer cells (39,40).
However, our results indicated that TRAIL treatment did not mediate
transformation from LC3B-I to LC3B-II (data not shown). On the
contrary, TRAIL-treated LC3-II transformation decreased slightly
than control. We concluded that TRAIL was possibly not associated
with autophagy in A549 lung cancer cells.
According to the results reported in Figs. 2D and 3D, Akt signaling was activated by TRAIL
treatment and decreased activation of Akt was observed on treatment
with quercetin. Akt signal is activated by TRAIL-resistance in
breast cancer cells (41) and TRAIL
phosphorylated PI3k and Akt in leukemic T Jurkat cells (42). Akt or protein kinase B (PKB) is one
of the most critical kinases in the regulation of cell survival.
Enhanced activity of the PI3K/Akt pathway is found in many
malignancies and is associated with the stimulation of cell growth
and cell survival (43).
Cross-talk between autophagy and apoptosis is
complicated and sometimes contradictory; however, it is a critical
determinant of the overall fate of the cell. This study determined
that quercetin-induced autophagy flux might play a crucial role in
TRAIL sensitivity in A549 human lung cancer cells. Quercetin may
thus be a useful regulator for TRAIL-mediated cancer therapy in
lung cancer.
Acknowledgments
The present study was supported by a grant from the
National Research Foundation of Korea (NRF), funded by the Korean
government (MISP) (2013R1A4A1069486).
References
|
1
|
Baowen Q, Yulin Z, Xin W, Wenjing X, Hao
Z, Zhizhi C, Xingmei D, Xia Z, Yuquan W and Lijuan C: A further
investigation concerning correlation between anti-fibrotic effect
of liposomal quercetin and inflammatory cytokines in pulmonary
fibrosis. Eur J Pharmacol. 642:134–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boots AW, Haenen GR and Bast A: Health
effects of quercetin: From antioxidant to nutraceutical. Eur J
Pharmacol. 585:325–337. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qu L, Liang X, Gu B and Liu W: Quercetin
alleviates high glucose-induced Schwann cell damage by autophagy.
Neural Regen Res. 9:1195–1203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim H, Moon JY, Ahn KS and Cho SK:
Quercetin induces mitochondrial mediated apoptosis and protective
autophagy in human glioblastoma U373MG cells. Oxid Med Cell Longev.
2013:5964962013.
|
|
5
|
Wei L, Liu JJ, Cao J, Du NC, Ji LN and
Yang XL: Role of autophagy in quercetin-induced apoptosis in human
bladder carcinoma BIU-87 cells. Zhonghua Zhong Liu Za Zhi.
34:414–418. 2012.In Chinese. PubMed/NCBI
|
|
6
|
Wei YQ, Zhao X, Kariya Y, Fukata H,
Teshigawara K and Uchida A: Induction of apoptosis by quercetin:
Involvement of heat shock protein. Cancer Res. 54:4952–4957.
1994.PubMed/NCBI
|
|
7
|
Murakami A, Ashida H and Terao J:
Multitargeted cancer prevention by quercetin. Cancer Lett.
269:315–325. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Steller H: Mechanisms and genes of
cellular suicide. Science. 267:1445–1449. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hickman JA: Apoptosis induced by
anticancer drugs. Cancer Metastasis Rev. 11:121–139. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang JY: Apoptosis-based anticancer
drugs. Nat Rev Drug Discov. 1:101–102. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
MacFarlane M: TRAIL-induced signalling and
apoptosis. Toxicol Lett. 139:89–97. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang CW and Klionsky DJ: The molecular
mechanism of autophagy. Mol Med. 9:65–76. 2003.PubMed/NCBI
|
|
13
|
Chen N and Karantza V: Autophagy as a
therapeutic target in cancer. Cancer Biol Ther. 11:157–168. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cuervo AM: Autophagy: In sickness and in
health. Trends Cell Biol. 14:70–77. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Seglen PO, Berg TO, Blankson H, Fengsrud
M, Holen I and Strømhaug PE: Structural aspects of autophagy. Adv
Exp Med Biol. 389:103–111. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mortimore GE, Miotto G, Venerando R and
Kadowaki M: Autophagy. Subcell Biochem. 27:93–135. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cuervo AM: Autophagy: Many paths to the
same end. Mol Cell Biochem. 263:55–72. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu B, Bao JK, Yang JM and Cheng Y:
Targeting autophagic pathways for cancer drug discovery. Chin J
Cancer. 32:113–120. 2013. View Article : Google Scholar :
|
|
19
|
Klionsky DJ: Autophagy: From phenomenology
to molecular understanding in less than a decade. Nat Rev Mol Cell
Biol. 8:931–937. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rubinsztein DC, Cuervo AM, Ravikumar B,
Sarkar S, Korolchuk V, Kaushik S and Klionsky DJ: In search of an
‘autophagomometer’. Autophagy. 5:585–589. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sahani MH, Itakura E and Mizushima N:
Expression of the autophagy substrate SQSTM1/p62 is restored during
prolonged starvation depending on transcriptional upregulation and
autophagy-derived amino acids. Autophagy. 10:431–441. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Myeku N and Figueiredo-Pereira ME:
Dynamics of the degradation of ubiquitinated proteins by
proteasomes and autophagy: Association with sequestosome 1/p62. J
Biol Chem. 286:22426–22440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bartlett BJ, Isakson P, Lewerenz J,
Sanchez H, Kotzebue RW, Cumming RC, Harris GL, Nezis IP, Schubert
DR, Simonsen A, et al: p62, Ref(2)P and ubiquitinated proteins are
conserved markers of neuronal aging, aggregate formation and
progressive autophagic defects. Autophagy. 7:572–583. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Klionsky DJ, Abdalla FC, Abeliovich H,
Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M,
Agostinis P, Aguirre-Ghiso JA, et al: Guidelines for the use and
interpretation of assays for monitoring autophagy. Autophagy.
8:445–544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu B, Cheng Y, Liu Q, Bao JK and Yang JM:
Autophagic pathways as new targets for cancer drug development.
Acta Pharmacol Sin. 31:1154–1164. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang L, Hao JL, Jin ML, Zhang YG and Wei
P: Effect of embelin on TRAIL receptor 2 mAb-induced apoptosis of
TRAIL-resistant A549 non-small cell lung cancer cells. Asian Pac J
Cancer Prev. 14:6115–6120. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chaudhari AA, Seol JW, Lee YJ, Seol DW and
Park SY: Hypoxia protects articular chondrocytes from
thapsigargin-induced apoptosis. Biochem Biophys Res Commun.
381:513–517. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Seo JS, Moon MH, Jeong JK, Seol JW, Lee
YJ, Park BH and Park SY: SIRT1, a histone deacetylase, regulates
prion protein-induced neuronal cell death. Neurobiol Aging.
33:1110–1120. 2012. View Article : Google Scholar
|
|
29
|
Agarwal E, Brattain MG and Chowdhury S:
Cell survival and metastasis regulation by Akt signaling in
colorectal cancer. Cell Signal. 25:1711–1719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Degterev A, Boyce M and Yuan J: A decade
of caspases. Oncogene. 22:8543–8567. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kroemer G and Jäättelä M: Lysosomes and
autophagy in cell death control. Nat Rev Cancer. 5:886–897. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Boya P, González-Polo RA, Casares N,
Perfettini JL, Dessen P, Larochette N, Métivier D, Meley D,
Souquere S, Yoshimori T, et al: Inhibition of macroautophagy
triggers apoptosis. Mol Cell Biol. 25:1025–1040. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Matsumoto G, Wada K, Okuno M, Kurosawa M
and Nukina N: Serine 403 phosphorylation of p62/SQSTM1 regulates
selective autophagic clearance of ubiquitinated proteins. Mol Cell.
44:279–289. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jeong JH, An JY, Kwon YT, Rhee JG and Lee
YJ: Effects of low dose quercetin: Cancer cell-specific inhibition
of cell cycle progression. J Cell Biochem. 106:73–82. 2009.
View Article : Google Scholar :
|
|
35
|
Pattingre S, Tassa A, Qu X, Garuti R,
Liang XH, Mizushima N, Packer M, Schneider MD and Levine B: Bcl-2
antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell.
122:927–939. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang Z and Klionsky DJ: An overview of the
molecular mechanism of autophagy. Curr Top Microbiol Immunol.
335:1–32. 2009.PubMed/NCBI
|
|
37
|
White E and DiPaola RS: The double-edged
sword of autophagy modulation in cancer. Clin Cancer Res.
15:5308–5316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang K, Liu R, Li J, Mao J, Lei Y, Wu J,
Zeng J, Zhang T, Wu H, Chen L, et al: Quercetin induces protective
autophagy in gastric cancer cells: Involvement of Akt-mTOR- and
hypoxia-induced factor 1α-mediated signaling. Autophagy. 7:966–978.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Singh K, Sharma A, Mir MC, Drazba JA,
Heston WD, Magi-Galluzzi C, Hansel D, Rubin BP, Klein EA and
Almasan A: Autophagic flux determines cell death and survival in
response to Apo2L/TRAIL (dulanermin). Mol Cancer. 13:702014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Park KJ, Lee SH, Kim TI, Lee HW, Lee CH,
Kim EH, Jang JY, Choi KS, Kwon MH and Kim YS: A human scFv antibody
against TRAIL receptor 2 induces autophagic cell death in both
TRAIL-sensitive and TRAIL-resistant cancer cells. Cancer Res.
67:7327–7334. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shankar E, Sivaprasad U and Basu A:
Protein kinase C epsilon confers resistance of MCF-7 cells to TRAIL
by Akt-dependent activation of Hdm2 and downregulation of p53.
Oncogene. 27:3957–3966. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zauli G, Sancilio S, Cataldi A, Sabatini
N, Bosco D and Di Pietro R: PI-3K/Akt and NF-kappaB/IkappaBalpha
pathways are activated in Jurkat T cells in response to TRAIL
treatment. J Cell Physiol. 202:900–911. 2005. View Article : Google Scholar
|
|
43
|
Fresno Vara JA, Casado E, de Castro J,
Cejas P, Belda-Iniesta C and González-Barón M: PI3K/Akt signalling
pathway and cancer. Cancer Treat Rev. 30:193–204. 2004. View Article : Google Scholar : PubMed/NCBI
|