Introduction
Gastric cancer is one of the most common cancers
worldwide (1). Approximately one
million new cases of stomach cancer were estimated to have been
diagnosed in 2008 (988,000 cases, 7.8% of the total), making it
currently the fourth most common malignancy in the world, behind
cancers of the lung, breast and colorectum. More than 70% of cases
(713,000 cases) occur in developing countries (467,000 in males and
246,000 in females) and half of the world total occurs in Eastern
Asia (mainly in China). The age of onset for developing gastric
cancer in the Chinese population is younger than that in the West.
Approximately 21,600 patients are diagnosed annually in the United
States, of whom 10,990 are expected to succumb to the disease
(2). Surgical treatment remains the
best treatment option for potential cure yet only for selected
patients suitable for surgery. Given the poor prognosis and the
fact that most gastric cancer cases are diagnosed at the advanced
or unresectable stage, new therapeutic strategies, treatment
options and novel therapeutic targets are desperately needed. Drug
development has been transformed following the identification and
ability to direct treatment at specific molecular targets.
Survivin, also called baculoviral inhibitor of
apoptosis repeat-containing 5 or BIRC5, is a protein that, in
humans, is encoded by the BIRC5 gene. Survivin is a member of the
inhibitor of apoptosis (IAP) family which encodes negative
regulatory proteins that prevent apoptotic cell death. IAP family
members usually contain multiple baculovirus IAP repeat (BIR)
domains, but this gene encodes proteins with only a single BIR
domain. The encoded proteins also lack a C-terminus RING finger
domain. Survivin has multiple functions including cytoprotection,
inhibition of cell death, and cell cycle regulation, particularly
at the mitotic stage of the cell cycle, all of which favor cancer
survival (3). Survivin plays
critical roles in cell division and cell survival (4,5). IAP
and BCL-2 family proteins are critically important for the
regulation of apoptosis (6).
Increased survivin mRNA or protein expression has previously been
reported to be a prognostic indicator of tumor progression in
different types of human cancer (7–12).
Cytoplasmic expression of survivin is common in breast cancer and
may be a useful clinical diagnostic and prognostic marker (13). Survivin, an apoptosis
inhibitor/cell-cycle regulator, is critically required for
suppression of apoptosis and ensuring normal cell division in the
G2/M phase of the cell cycle (14).
In cancer cells, elevated survivin is commonly associated with an
enhanced proliferative index, reduced level of apoptosis,
resistance to chemotherapy, and increased rate of tumor recurrence
(15–18). More recently, survivin expression
was also shown to enhance the metastatic potential of cancer cells
by promoting, together with XIAP, NF-κB-dependent transcription and
secretion of fibronectin (19).
Survivin expression has been reported to be low in normal
tissues.
RNA interference (RNAi) is one of the most important
technological breakthroughs in modern biology, allowing the direct
observation of the effects of the loss of function of specific
genes in mammalian systems. Recent studies advocate RNAi as a
promising therapeutic tool against infectious diseases and cancer
(20). Our recent research
demonstrated that lentiviral vector-mediated delivery of short
hairpin RNA (shRNA) resulted in the persistent knockdown of gene
expression (21,22). Α previous study reported that
knockdown of survivin expression in melanoma utilizing siRNA
inhibited cell proliferation and sensitized the cells to apoptosis
induced by chemotherapy (23).
However, stable suppression of survivin expression by RNA remains
to be investigated.
In the present study, we assessed the feasibility of
lentiviral vector-delivered shRNA against survivin for the
treatment of gastric cancer in vitro and in an experimental
animal model. Lentiviral-mediated survivin shRNA was utilized to
knock down survivin expression in gastric cancer cell lines
(SGC-7901, MGC-803 and MKN-28). The results showed that silencing
of survival expression inhibited cell proliferation and tumor
growth.
Materials and methods
Mice, cell lines, culture medium and
reagents
BALB/C-nu-nu male mice (weight 18–22 g, 6-weeks old)
and human gastric cancer cell lines SGC-7901, MGC-803, MKN-28 and
human embryonic kidney 293 cells (HEK93T) were obtained from the
Shanghai Tumor Institution. Classical Liquid Media/Dulbecco’s
modified Eagle’s medium (DMEM) and high glucose RPMI-1640 media
were purchased from HyClone (Thermo Fisher Scientific, Waltham, MA,
USA). Fetal bovine serum (FBS) was purchased from Gibco
(Invitrogen, Carlsbad, CA, USA). Dimethyl sulfoxide (DMSO) was
purchased from Sigma (St. Louis, MO, USA). Rabbit anti-survivin
polyclonal antibody and HRP-conjugated secondary antibodies were
purchased from Abcam (Cambridge, MA, USA).
All experimental procedures using animals in the
present study had received prior approval by the Institutional
Animal Care and Use Committee of Shandong University under contract
2011–0015.
Cell culture
The SGC-7901, MGC-803 and MKN-28 cell lines were
grown in RPMI-1640 medium supplemented with 10% FBS and 100 U/ml
penicillin and 100 mg/ml streptomycin (Gibco, Invitrogen). The
cells were grown at 37°C in a humidified atmosphere containing 5%
CO2. Stock cultures of each cell line were routinely
sub-cultured at least once a week, and the medium was changed every
2–3 days.
Immunocytochemistry
Stably transfected gastric cancer cell lines were
seeded into 4-chambered glass slides (Nunc Lab-Tek Chamber Slide
System). The cells were then incubated overnight. After 24 h, the
cells were rinsed with PBS, fixed with 3.7% w/v paraformaldehyde
(Sigma), rinsed with PBS and permeablized in 0.5% Triton X-100
(Sigma). Nonspecific immunoglobulin binding was blocked with 5%
normal goat serum and 0.5% NP-40 (Sigma). Primary antibodies
recognizing survivin (Abcam) were diluted 1:100 in blocking
solution. After incubation with the primary antibody, the cells
were rinsed with 0.05% Tween-20 (Bio-Rad, Hercules, CA, USA) in
PBS, and then incubated with a secondary antibody for 1 h at room
temperature. Staining with 3,3′-diaminobenzidine (DAB) was carried
out and observed under a light microscope.
Lentiviral shRNA vector constructs
pGIPZ-lentiviral shRNAmir vectors targeting the
human survivin gene and the non-silencing pGIPZ control vector were
purchased from Open Biosystems (Thermo Fisher Scientific). pGIPZ
non-silencing control vector was used as expression control to
generate non-silencing lentiviral stock to optimize expression
conditions in the mammalian cell line of interest. pGIPZ cloning
vector containing Turbo GFP reporter and also elements were
required to allow packaging of the expression construct into
virions (such as 5′ and 3′ LTRs and Ψ packaging signal). pGIPZ
vector also expresses a puromycin-resistant gene. The sequences of
survivin shRNA were: TCTTGAATGTAGAGATGCG, TTCCTAAGACATTGCTAAG and
AGCAGAAGAAACACTGGGC. 21-mer oligonucleotide TCTCGCTTGGGCGAGAGTAAG
that had no significant homology to any known human mRNA in the
databases was cloned in the same vector and used as the
control.
Virus production and transduction
Lentiviral shRNA was produced by co-transfection of
the Trans-Lentiviral packaging mix with a shRNA transfer vector
into HEK 293T packaging cells (Open Biosystems). The packaging mix
contains an optimized mixture of five packaging plasmids
(pTLA1-Pak, pTLA1-Enz, pTLA1-Env, pTLA1-Rev and pTLA1-TOFF) to
facilitate viral packaging of the transfer vector following
co-transfection into HEK293T producer cells. HEK293T cells stably
express the SV40 large T antigen (simian vacuolating viral
particles 40 TAg) which allows high levels of protein to been
expressed from vectors containing the SV40 origin of replication.
Following co-transfection, replication-incompetent virions were
released into the media which were collected after 48 and 72 h,
respectively. For cell infection, viral supernatants were
supplemented with 6 μg/ml polybrene and incubated with the
cells for 24 h. Gastric cancer cell lines were transduced by the
lentiviral particles followed by puromycin selection (1
μg/ml) for 10 days. The cell lines stably expressing shRNA
were maintained in puromycin (0.2 μg/ml).
RNA extraction and qRT-PCR
Total RNA extraction was performed using TRIzol
reagent (Invitrogen) according to the manufacturer’s instructions.
RNA concentration was measured by NanoDrop 1000 (Thermo Fisher
Scientific). One microgram of total RNA extracted from the cells
was subjected to reverse transcription (RT). The Verso cDNA kit
(Thermo Fisher Scientific) was used for cDNA synthesis. Real-time
RT-PCR was used to quantify the expression level of the survivin
gene in the gastric cancer cell lines SGC-7901, MGC-803 and MKN-28
using ABI 7300 real-time PCR thermal cycle instrument (ABI, USA),
according to the supplied protocol. Amplification conditions were
as follows: reverse-transcription reaction: 42°C, 30 min/cycle. PCR
cycling conditions were as follows: enzyme activation 95°C, 15
min/cycle; denaturation 95°C, 15 sec/40 cycles and
annealing/extension at 60°C for 60 sec.
A real-time PCR reaction was performed using the
Solaris qPCR Gene Expression Master Mix with Low ROX premixed and 1
μl of total cDNA in each well, Survivin-specific primers
were: forward, ACCGCATCTCTACATTCAAG and reverse,
CAAGTCTGGCTCGTTCTC. The relative expression levels were normalized
to the expression of endogenous β-actin. Primers were forward,
TCACCCACACTGTGCCCATCTACGA and reverse,
CAGCGGAACCGCTCATTGCCAATGG.
Protein extraction and western blot
analysis
For whole-cell protein extraction, the cells were
washed with cold PBS and subsequently lysed in cold RIPA lysis
buffer [50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM dithiothreitol
(DTT), 0.25% sodium deoxycholate, 0.1% NP-40] containing 1 mM
phenylmethysulfonyl fluoride (PMSF), 50 mM sodium pyrophosphate, 1
mM Na3VO4, 1 mM NaF, 5 mM EDTA, 5 mM EGTA and
a protease inhibitor cocktail (Roche Diagnostics, Mannheim,
Germany). Cell lysis was performed on ice for 30 min. Clear protein
extracts were obtained by centrifugation for 30 min at 4°C. Protein
concentrations were determined by the method of Bradford using the
Bio-Rad protein assay reagent (Bio-Rad) and 20–40 mg of protein
mixed with loading buffer was loaded per lane, and separated by 12%
SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were
transferred to PVDF membrane filters (Millipore, Billerrica, MA,
USA). Nonspecific binding was blocked by incubation in
phosphate-buffered saline (PBS) containing 0.1% Tween-20 (PBS-T)
and 5% skim milk. PVDF membranes were blocked with 5% dry milk for
1 h at 4°C. Membranes were incubated with the survivin primary
antibody (1:2,000) overnight at 4°C. The membranes were then
incubated with the corresponding secondary antibody (1:2,500,
horseradish peroxidase-conjugated anti-rabbit) in TBST-5% nonfat
milk for 1 h at room temperature and the immunoreactive bands were
visualized using the EZ ECL chemiluminescence detection kit for HRP
(Biological Industries Ltd., Kibbutz Beit Haemek, Israel). Images
were acquired using the LAS3000 Imager (Fujifilm, Tokyo, Japan).
Membranes were re-probed for β-actin as a loading control.
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8; Dojindo, Kunamoto,
Japan) was used for the cell proliferation assay. Five thousand
viable cells/well were placed into 96-well tissue culture plates in
a final volume of 100 μl. Every 24 h, a plate was subjected
to assay by adding 10 μl of CCK-8 solution to each well, and
the plate was further incubated for 4 h at 37°C. The absorbance at
450 nm was measured with a micro-plate reader. The experiment was
performed in 10 replicates.
Migration and invasion assays
For the Transwell migration assay, 30,000 cells were
added to the upper chamber in serum-free media and migration at
37°C towards 10% FBS containing growth media was determined either
after 24 or 48 h. The cells that migrated through the membrane were
fixed, stained with H&E (Sigma) and counted under a light
microscope. For the invasion assay, lower chambers of
Matrigel-coated invasion plates were coated with 10 mg/ml
fibronectin overnight at 4°C, and cells invading through the
Matrigel were fixed and stained after 48 h.
TUNEL assay
Terminal deoxynucleotidyl transferase-mediated dUTP
nick end labeling (In Situ Cell Death Detection kit, POD)
was used for confirming apoptosis of individual cells. The TUNEL
working procedure was carried out following the manufacturer’s
instructions (Roche). Endogenous peroxidase was blocked by
incubation in 1.3% H2O2 in PBS for 10 min at
room temperature before enzymatic labeling. During the TUNEL
procedure, the samples were washed in PBS. The fluorescent signal
conversion using anti-fluorescence antibody conjugated with
peroxidase and substrate color reaction applying chromogen DAB
(3,3′-diaminobenzidine tetrahydrochloride; Sigma-Aldrich,
Steinheim, Germany) were carried out after enzymatic labeling.
In vivo studies of gastric cancer
xenograft tumor models in nude mice
Six-week-old male BALB/c nude mice were housed in a
temperature-controlled, pathogen-free animal facility with a 12-h
light and dark cycle. The mice were injected subcutaneously into
bilateral flanks with untransfected cells, or cells transfected
with the non-silencing shRNA or survivin shRNA (2×106
cells in 200 μl PBS) to establish tumors. The tumor mass
(xenograft) volume was measured every week from week 3 to 7. After
week 7, the mice were sacrificed and tumors were harvested.
Statistical analysis
For comparison of more than three groups, one-way
analysis of variance was used, followed by Tukey’s multiple
comparison. P-values <0.05 were considered to indicate
statistically significant results. One-way analysis of variance
(ANOVA), followed by the LSD post hoc test was used to
compare mean differences in two or more groups. All statistical
analysis was performed using IBM SPSS version 20.0.
Results
Survivin is highly upregulated in the
gastric cancer cell lines
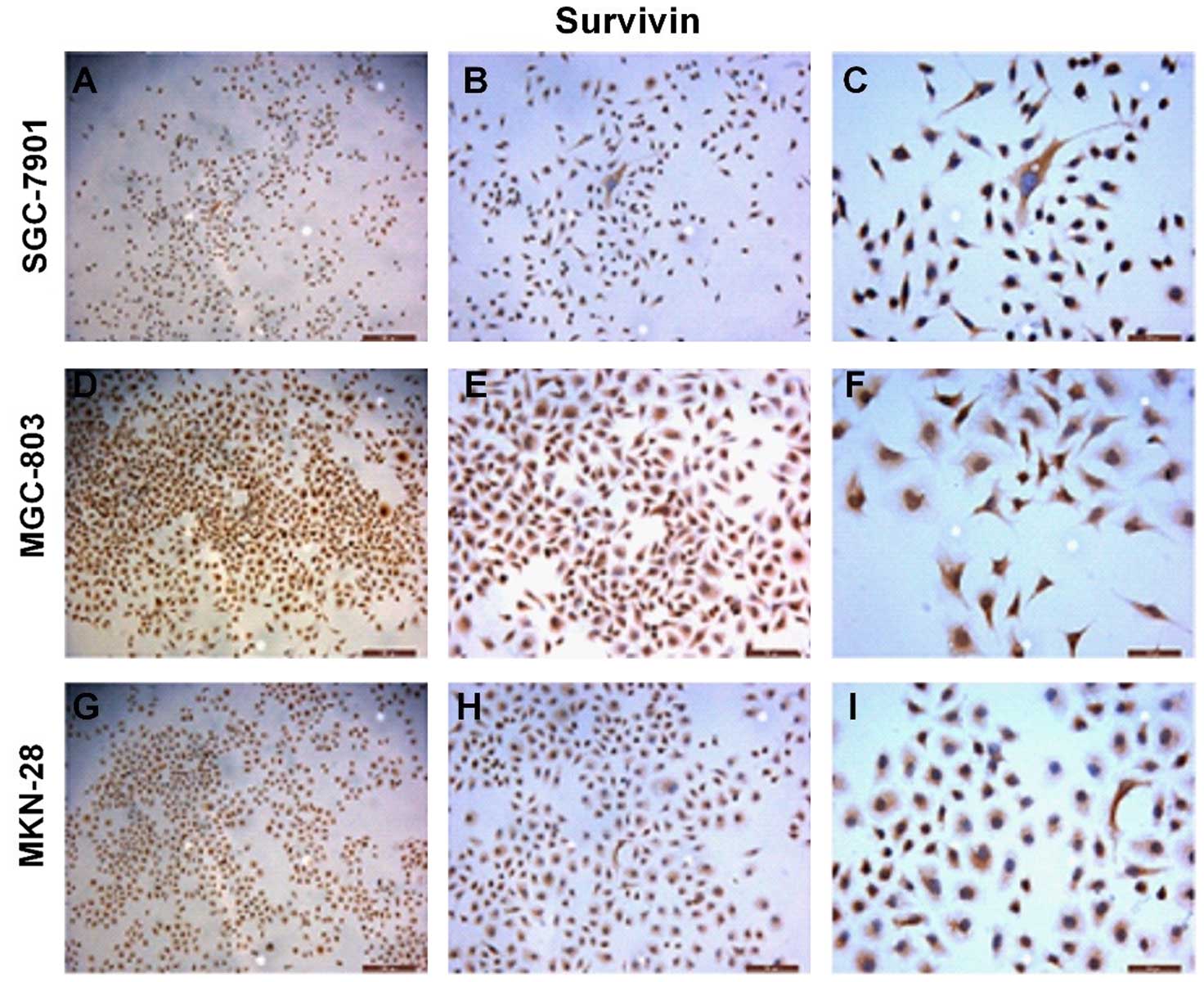
We assessed the survivin gene expression in the
gastric cancer-derived cell lines by immunocytochemistry (ICC)
(Fig. 1). Strong immunoreactivity
of survivin protein was detected in the cytoplasm of the gastric
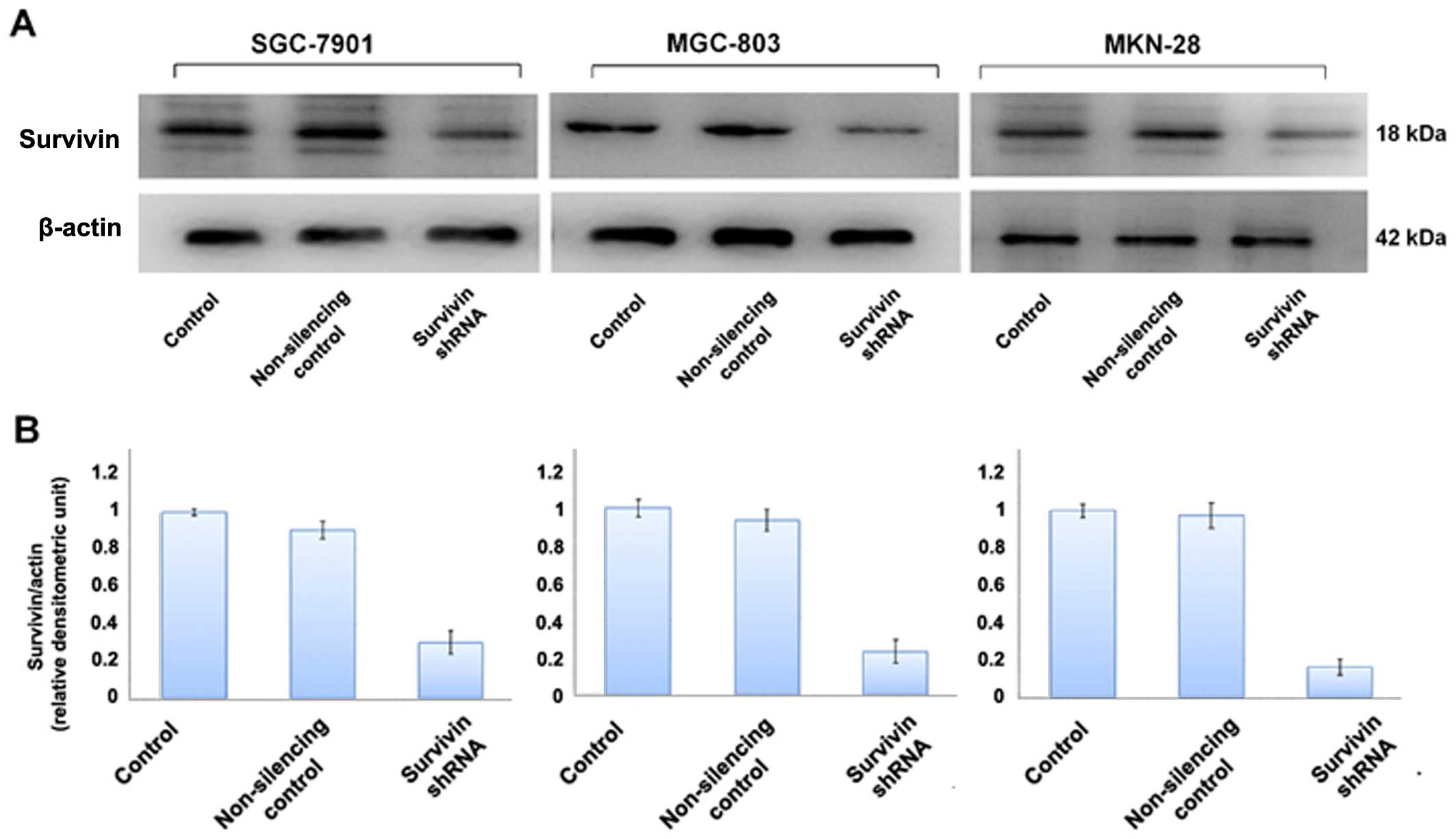
cancer cell lines. We also evaluated the level of survivin protein
expression in the cell lines by western blot analysis (Fig. 2) which was further confirmed by
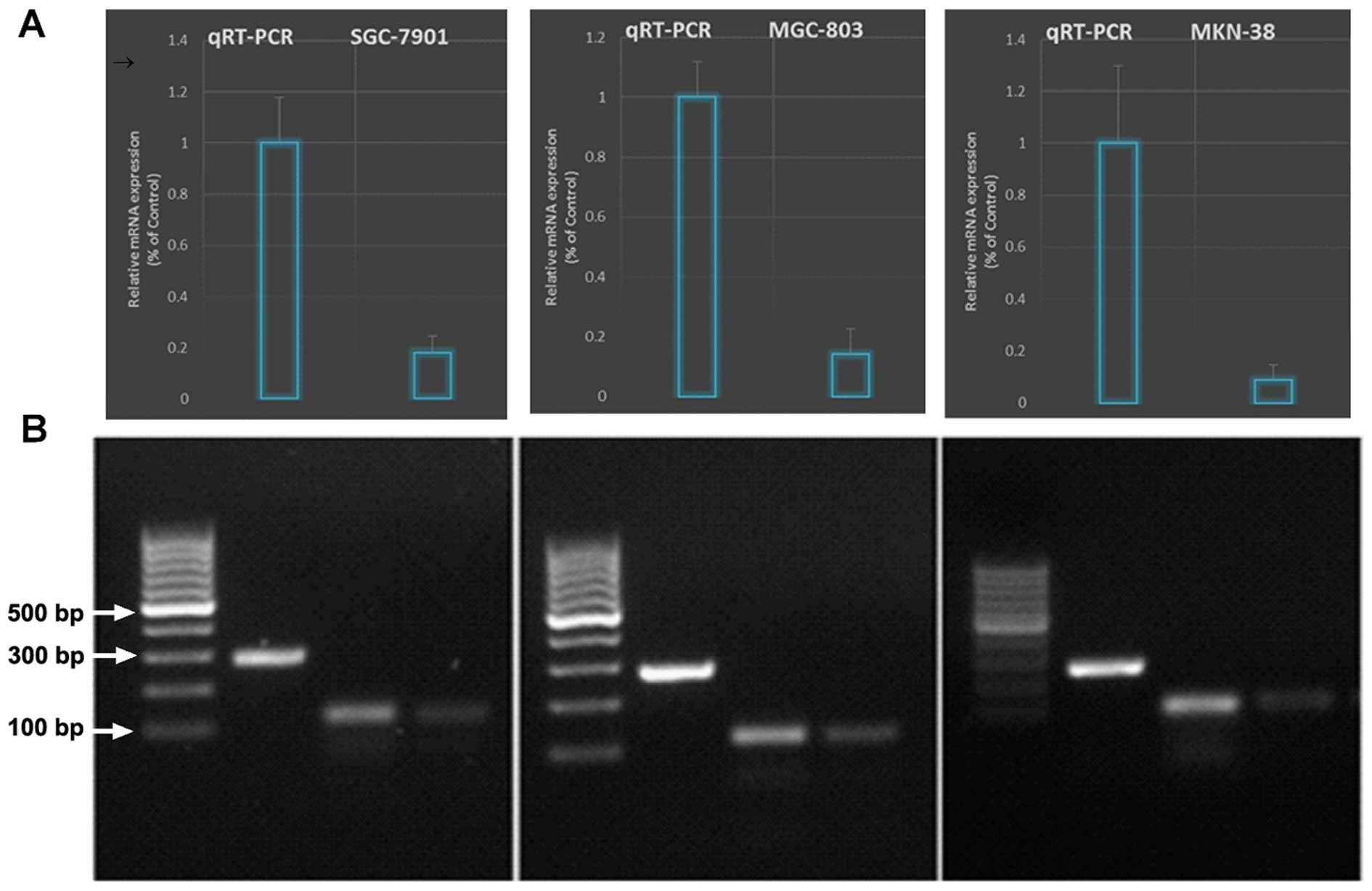
qRT-PCR (Fig. 3).
Lentiviral-mediated RNAi efficiently
suppresses survivin protein and mRNA expression in the gastric
cancer cell lines
Lentiviral-mediated survivin shRNAs specifically
knocked down the survivin expression and activity in the gastric
cancer cell lines. All gastric cancer cell lines used expressed
high levels of cytoplasmic survivin expression, and exhibited
aggressive growth and metastatic ability. To investigate the role
of survivin in gastric cancer cell growth and metastasis, we
constructed the lentiviral vector with survivin shRNA and infected
the cell lines. After viral infection, >95% of the cells were
GFP-positive, indicating a high efficiency of shRNA delivery.
Survivin shRNA more efficiently knocked down protein expression in
the cell lines as compared to survivin expression in the normal
control and non-silencing group (p<0.05) (Fig. 2). Survivin shRNA also efficiently
suppressed the survivin mRNA level as confirmed by qRT-PCR
(Fig. 3).
Effects of survivin shRNA on the
proliferation of SGC-7901, MGC-803 and MKN-28 cells
To determine the effects of the silencing of
survivin on the proliferation of SGC-7901, MGC-803 and MKN-28
cells, we treated these cells with different doses of survivin
shRNA for different periods. CCK-8 assay was then used to evaluate
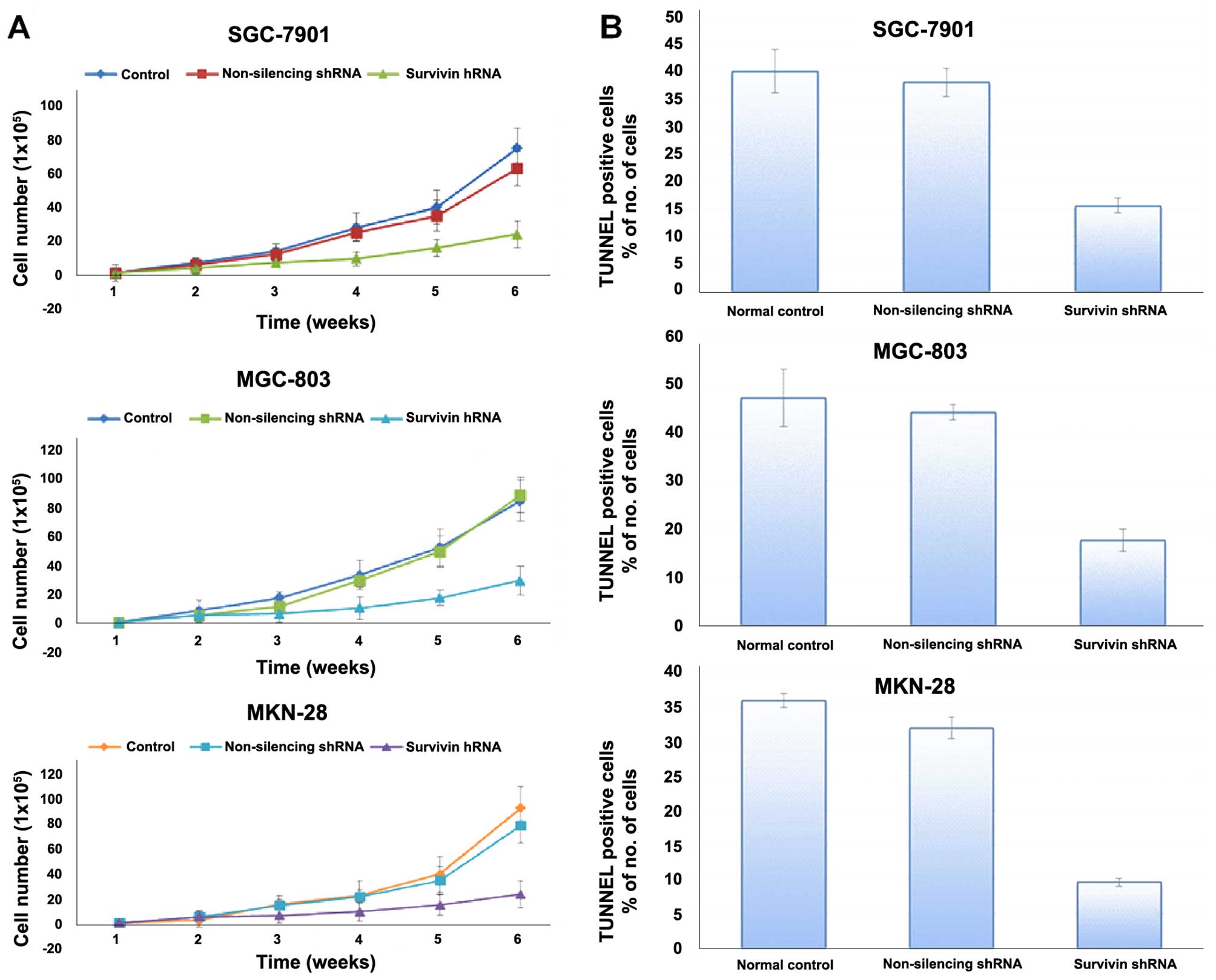
cell proliferation, and cell growth curves were generated. The
results revealed that the gastric cancer cell lines stably
transfected with survivin shRNA exhibited significantly reduced
cell proliferation relative to the normal control and non-silencing
group (Fig. 4A). Survivin
downregulation caused impairment of proliferation in the cells.
Survivin knockdown inhibited the proliferation of the gastric
cancer cell lines in vitro, indicating that the expression
of survivin significantly affects the growth of gastric cancer
cells.
Knockdown of survivin induces gastric
cancer cell apoptosis
As shown in Fig. 4B,
the percentage of apoptotic cells in the group infected with
survivin shRNA was much higher than that in the control shRNA group
(p<0.01). No significant difference in regards to apoptosis was
found between the control shRNA-infected cells and the non-infected
cells. These data indicated that knockdown of surviving expression
induced apoptosis in the gastric cancer cells.
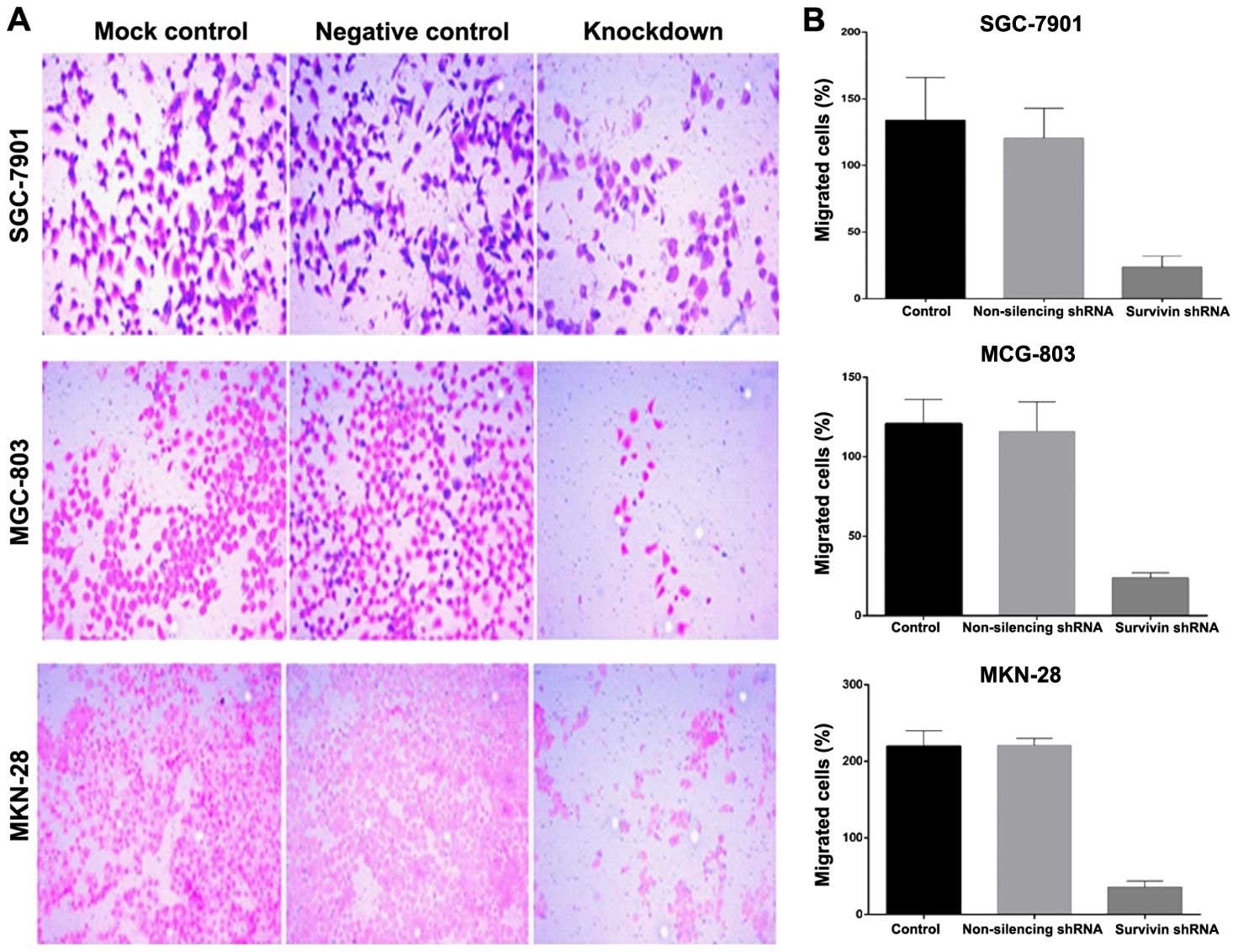
Lentiviral-mediated survivin shRNA
significantly impairs migration and invasion of gastric cancer
cells
Cell migration was evaluated in the Boyden migration
assay 2 days after the gastric cancer cell lines were stably
transfected with survivin shRNA or transfected with the control
shRNA. Cells with motile capacity were able to migrate through the
pores of the Transwell filters due to attraction to 10% FBS in the
lower chamber. Cells transfected with survivin shRNA displayed
lower migratory ability compared with this ability in the
non-transfected control cells (Fig.
5). The introduction of survivin shRNA into the cells, however,
markedly decreased the cell migration, in comparison to the
non-transfected cells. To evaluate the function of survivin shRNA
on gastric cancer cell invasion, Matrigel invasion chambers were
utilized. Silencing of survivin expression led to a significant
decrease in the invasive ability of the gastric cancer cells.
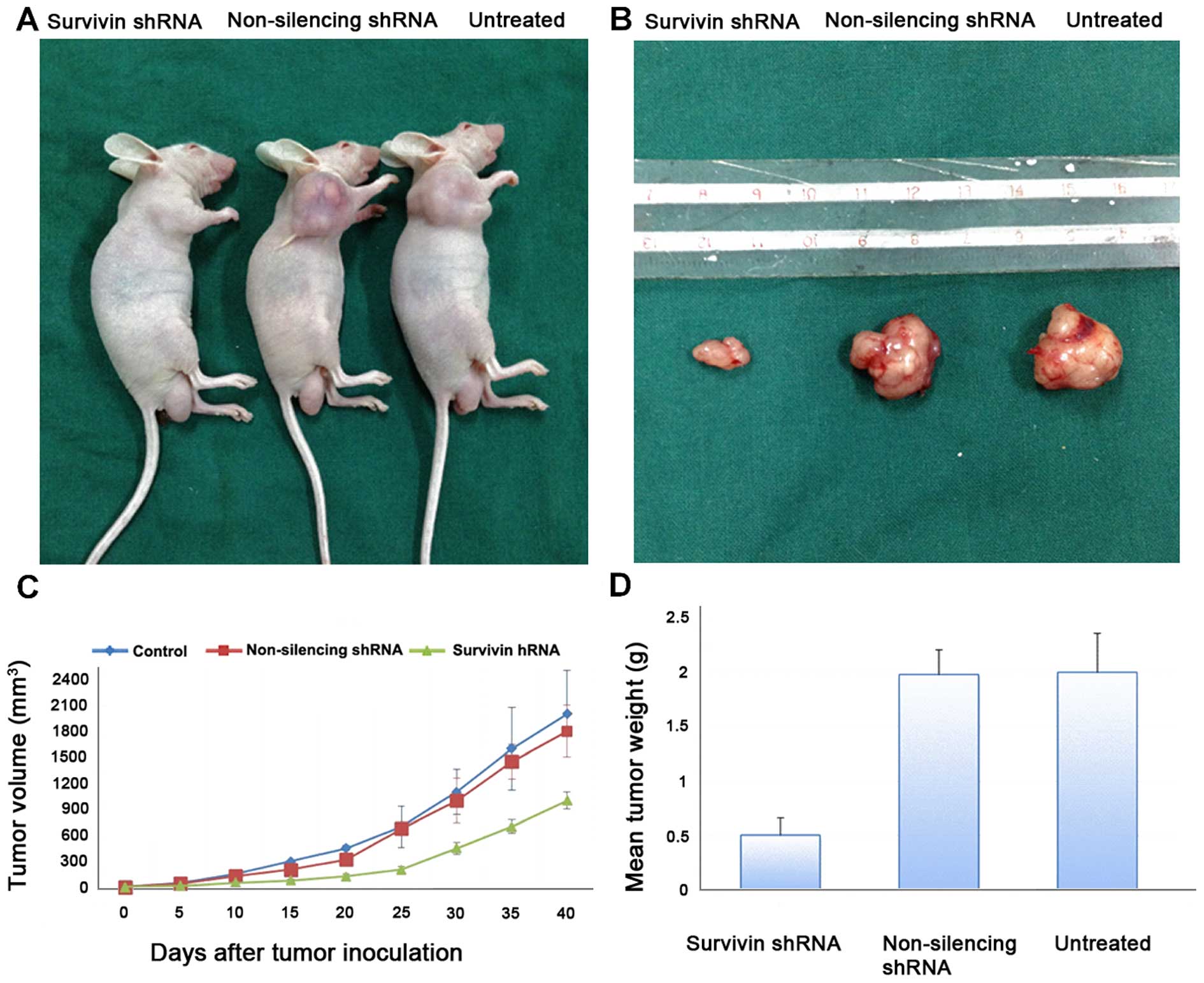
In vivo studies of gastric cancer
xenograft tumor models in nude mice
To further evaluate the effects of reduced survivin
expression on the tumorigenic phenotype and in particular its
contribution to in vivo tumor growth, gastric cancer cell
lines stably transfected with the non-silencing shRNA or survivin
shRNA or the untreated cells were injected into mice
(2×106 cells in 200 μl PBS) to develop a
xenograft model of human gastric cancer. Tumors were measured using
Vernier calipers for calculation of tumor size. Tumors derived from
the survivin shRNA-transfected cells grew less rapidly as compared
to the negative control (p<0.05). Photographic image of nude
mice bearing xenograft tumors (Fig.
6A) and image of the xenografts dissected from the nude mice
are shown (Fig. 6B). These results
demonstrate that the in vivo tumor growth was inhibited by
shRNA-mediated knockdown of survivin expression in the gastric
cancer cell lines.
Tumor growth and weight and evaluation of
the antitumoral effect
Tumor volume was evaluated twice a week by measuring
two perpendicular diameters with calipers (Fig. 6C). Tumor volume (V) was calculated
using the following equation: V = (a2 × b)/2 where a is
the width of the tumor (small diameter) and b the length (large
diameter), both in millimeters. The mean tumor weight of the
resected tumors is shown in Fig.
6D. Tumor weight was significantly reduced in the survivin
shRNA knockdown xenograft tumors.
Discussion
The major aim of the present study was to
investigate the effect of lentiviral-mediated knockdown of survivin
on cell proliferation in vitro and tumor growth in
vivo. Survivin is a recently discovered IAP that is unique for
its expression in a wide range of embryonic and fetal tissues but
is lowly expressed in terminally differentiated normal adult
tissues. Survivin is widely implicated in processes related to
tumor development and progression due to its ability to inhibit
apoptosis, promote cell cycle progression, accelerate metastasis
and enhance angiogenesis (24). The
IAP and BCL-2 family proteins are critically important for the
regulation of apoptosis (6).
Survivin is also required for the maintenance of the spindle
assembly checkpoint to allow proper microtubule alignment to ensure
cell propagation (25). Lack of
survivin during cell division causes polyploidy as well as
apoptosis (26). The robust
expression of survivin in gastric cancer vs. normal cells, its
correlation with poor prognosis and resistance to therapy suggest
that survivin is an inducible resistance factor in cancer cells and
is involved in the emergence of a refractory phenotype to
anticancer therapies. Similar to the conserved mechanism in the
developmental process, survivin inhibition has been shown to
enhance cell death (27). Various
approaches have been attempted to downregulate or block survivin in
cancer cells to inhibit cell survival and at the same time enhance
cell death.
RNAi is a relatively new technology and holds
promise for the development of therapeutic gene silencing. However,
the knockdown effect of regular synthesized siRNA only lasts for a
short time and does not allow the stable inhibition of target gene
function. Lentiviral vectors allow efficient delivery and stable
transfection of a gene of interest. In previous studies, we tested
the efficacies of lentiviral vectors for shRNA delivery in cancer
cells (21,22). An approach using short shRNA
successfully reduced survivin expression, and induced apoptosis and
growth inhibition in a lymphoma cell line (28). In the present research, we used
lentivirus-based vectors to silence survivin expression in gastric
cancer cell lines (SGC-7901, MGC-803 and MKN-28). We found
lentiviral-mediated shRNA was highly capable of stable knockdown of
survivin expression in the cell lines.
Increased survivin mRNA or protein expression has
previously been reported to be a prognostic indicator of tumor
progression in different types of human cancer (7–12).
Cytoplasmic expression of survivin is common in breast cancer and
may be a useful clinical diagnostic and prognostic marker (13). Survivin expression has previously
been observed in gastric cancer, yet no in vitro or in
vivo data are available to date regarding the role of
lentiviral-mediated knockdown of survivin expression in this cancer
type. Therefore, we quantified the expression levels of survivin in
three different gastric cancer cell lines and downregulated the
survivin expression using shRNA. The results of ICC, western blot
analysis and qRT-PCR revealed that survivin expression was
upregulated in all three gastric cancer cell lines. We constructed
a lentiviral vector against survivin and stably transduced gastric
cell lines with survivin shRNA. The results showed that expression
of survivin protein in all three cell lines transfected with
survivin shRNA was reduced compared with that in the non-silencing
control cells. Furthermore, the proteins extracted from the cell
lines were analyzed with western blot analysis. The transfection
results were stable, persistence and consistent with the mRNA
levels. The duration of gene downregulation was better than the
transient cell transfection. The present study provides in
vitro and in vivo evidence for a role of survivin shRNA
in the pathogenesis and progression of gastric cancer.
In the present study, we investigated the function
of survivin in gastric cancer and demonstrated that silencing of
survivin by RNAi led to reduced proliferation and migration in
gastric cancer SGC-7901, MGC-803 and MKN-28 cell lines. The
findings indicate that survivin may take part in the development,
progression and metastasis of gastric cancer or prognosis of the
patients by regulating proliferation and migration of gastric
cancer cells. Researcher has demonstrated that simultaneous
inhibition of survivin and XIAP by siRNA significantly reduced cell
proliferation and increased caspase-3/7 activity in pancreatic
carcinoma cells (29). Inhibition
of apoptosis is believed to be through direct binding to caspase-3
and -7, preventing their activation (30). The effect of survivin knockdown by
siRNA on the proliferation of two human cancer cell lines has also
been reported (31). At least one
mechanism of survivin involves the suppression of default apoptosis
in the G2/M phase (14).
Furthermore, in the xenograft model, survivin shRNA
potently inhibited the in vivo growth of gastric cancer
xenografts, resulting in tumor regression, while the mouse tumors
derived from the untreated cells grew rapidly and aggressively.
Tumor growth was significantly decreased in the survivin shRNA
transfection groups as compared with the control groups. Various
therapeutics are associated with toxic side effects such as weight
loss. These systemic toxicities may be related with higher
morbidity and a lower response rate, resulting in poorer survival.
However, in our animal model, weight loss and toxic effects of
treatment were not obvious (data not shown). In the present study,
the measured body weights of mice had no differences (data not
shown) and there were no significant pathologic findings from
tissue obtained from the shRNA-treated group. For these reasons,
shRNA targeting survivin can be used in gastric cancer therapy
without systemic toxic effects in vivo. In the present
study, despite an effective reduction in tumor size, we observed
marked tumor regression, with tumors displaying extensive areas of
necrotic tissue and a significant decrease in the number of blood
vessels (data not shown).
In conclusion, these findings indicate for the first
time that targeting cytoplasmic survivin expression utilizing
lentiviral survivin shRNA in gastric cancer cell lines reduced the
progression of cancer. We also showed that the lentiviral vector
targeting survivin expression blocked the growth and induced cell
death in the gastric carcinoma cell lines in vitro and in
vivo.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamamoto H, Ngan CY and Monden M: Cancer
cells survive with survivin. Cancer Sci. 99:1709–1714. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ambrosini G, Adida C and Altieri DC: A
novel anti-apoptosis gene, survivin, expressed in cancer and
lymphoma. Nat Med. 3:917–921. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li F, Ambrosini G, Chu EY, Plescia J,
Tognin S, Marchisio PC and Altieri DC: Control of apoptosis and
mitotic spindle checkpoint by survivin. 396:580–584. 1998.
|
|
6
|
Jäättelä M: Escaping cell death: Survival
proteins in cancer. Exp Cell Res. 248:30–43. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Adida C, Berrebi D, Peuchmaur M,
Reyes-Mugica M and Altieri DC: Anti-apoptosis gene, survivin, and
prognosis of neuroblastoma. Lancet. 351:882–883. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kawasaki H, Altieri DC, Lu C-D, Toyoda M,
Tenjo T and Tanigawa N: Inhibition of apoptosis by survivin
predicts shorter survival rates in colorectal cancer. Cancer Res.
58:5071–5074. 1998.PubMed/NCBI
|
|
9
|
Monzó M, Rosell R, Felip E, Astudillo J,
Sánchez JJ, Maestre J, Martín C, Font A, Barnadas A and Abad A: A
novel anti-apoptosis gene: Re-expression of survivin messenger RNA
as a prognosis marker in non-small-cell lung cancers. J Clin Oncol.
17:2100–2104. 1999.PubMed/NCBI
|
|
10
|
Swana HS, Grossman D, Anthony JN, Weiss RM
and Altieri DC: Tumor content of the antiapoptosis molecule
survivin and recurrence of bladder cancer. N Engl J Med.
341:452–453. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sarela AI, Macadam RC, Farmery SM, Markham
AF and Guillou PJ: Expression of the antiapoptosis gene, survivin,
predicts death from recurrent colorectal carcinoma. Gut.
46:645–650. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kato J, Kuwabara Y, Mitani M, Shinoda N,
Sato A, Toyama T, Mitsui A, Nishiwaki T, Moriyama S, Kudo J, et al:
Expression of survivin in esophageal cancer: Correlation with the
prognosis and response to chemotherapy. Int J Cancer. 95:92–95.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sohn DM, Kim SY, Baek MJ, Lim CW, Lee MH,
Cho MS and Kim TY: Expression of survivin and clinical correlation
in patients with breast cancer. Biomed Pharmacother. 60:289–292.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shin S, Sung B-J, Cho Y-S, Kim HJ, Ha NC,
Hwang JI, Chung CW, Jung YK and Oh BH: An anti-apoptotic protein
human survivin is a direct inhibitor of caspase-3 and -7.
Biochemistry. 40:1117–1123. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Adamkov M, Kajo K, Vybohova D, Krajcovic
J, Stuller F and Rajcani J: Correlations of survivin expression
with clinicomor-phological parameters and hormonal receptor status
in breast ductal carcinoma. Neoplasma. 59:30–37. 2012. View Article : Google Scholar
|
|
16
|
Sui L, Dong Y, Ohno M, Watanabe Y,
Sugimoto K and Tokuda M: Survivin expression and its correlation
with cell proliferation and prognosis in epithelial ovarian tumors.
Int J Oncol. 21:315–320. 2002.PubMed/NCBI
|
|
17
|
Tanaka K, Iwamoto S, Gon G, Nohara T,
Iwamoto M and Tanigawa N: Expression of survivin and its
relationship to loss of apoptosis in breast carcinomas. Clin Cancer
Res. 6:127–134. 2000.PubMed/NCBI
|
|
18
|
Zaffaroni N, Pennati M, Colella G, Perego
P, Supino R, Gatti L, Pilotti S, Zunino F and Daidone MG:
Expression of the anti-apoptotic gene survivin correlates with
taxol resistance in human ovarian cancer. Cell Mol Life Sci.
59:1406–1412. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mehrotra S, Languino LR, Raskett CM,
Mercurio AM, Dohi T and Altieri DC: IAP regulation of metastasis.
Cancer Cell. 17:53–64. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yano J, Hirabayashi K, Nakagawa S,
Yamaguchi T, Nogawa M, Kashimori I, Naito H, Kitagawa H, Ishiyama
K, Ohgi T, et al: Antitumor activity of small interfering
RNA/cationic liposome complex in mouse models of cancer. Clin
Cancer Res. 10:7721–7726. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Akhtar J, Wang Z, Yu C and Zhang ZP:
Effectiveness of local injection of lentivirus-delivered stathmin1
and stathmin1 shRNA in human gastric cancer xenograft mouse. J
Gastroenterol Hepatol. 29:1685–1691. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Akhtar J, Wang Z, Zhang ZP and Bi MM:
Lentiviral-mediated RNA interference targeting stathmin1 gene in
human gastric cancer cells inhibits proliferation in vitro and
tumor growth in vivo. J Transl Med. 11:2122013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chawla-Sarkar M, Bae SI, Reu FJ, Jacobs
BS, Lindner DJ and Borden EC: Downregulation of Bcl-2, FLIP or IAPs
(XIAP and survivin) by siRNAs sensitizes resistant melanoma cells
to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 11:915–923.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Altieri DC: Survivin, cancer networks and
pathway-directed drug discovery. Nat Rev Cancer. 8:61–70. 2008.
View Article : Google Scholar
|
|
25
|
Lens SMA, Wolthuis RMF, Klompmaker R, Kauw
J, Agami R, Brummelkamp T, Kops G and Medema RH: Survivin is
required for a sustained spindle checkpoint arrest in response to
lack of tension. EMBO J. 22:2934–2947. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang D, Welm A and Bishop JM: Cell
division and cell survival in the absence of survivin. Proc Natl
Acad Sci USA. 101:15100–15105. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Altieri DC: Validating survivin as a
cancer therapeutic target. Nat Rev Cancer. 3:46–54. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gu CM, Zhu YK, Ma YH, Zhang M, Liao B, Wu
HY and Lin HL: Knockdown of survivin gene by vector-based short
hairpin RNA technique induces apoptosis and growth inhibition in
Burkitt’s lymphoma Raji cell line. Neoplasma. 53:206–212. 2006.
|
|
29
|
Yang J, Ouyang J, Ouyang L, Ouyang L and
Chen Y: Inhibition of cell proliferation and increase of
chemosensitivity by simultaneous knockdown of XIAP and survivin in
pancreatic carcinoma cells. Oncol Res. 21:43–50. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tamm I, Wang Y, Sausville E, Scudiero DA,
Vigna N, Oltersdorf T and Reed JC: IAP-family protein survivin
inhibits caspase activity and apoptosis induced by Fas (CD95), Bax,
caspases, and anticancer drugs. Cancer Res. 58:5315–5320.
1998.PubMed/NCBI
|
|
31
|
Guan HT, Xue XH, Wang XJ, Li A and Qin ZY:
Knockdown of survivin expression by small interfering RNA
suppresses proliferation of two human cancer cell lines. Chin Med
Sci J. 21:115–119. 2006.PubMed/NCBI
|