Introduction
Renal cell carcinoma (RCC) is the most common
carcinoma of the adult kidney and accounts for ~3% of all cancer
cases in adults with the highest mortality rate of over 40%
(1). Clear cell RCC (ccRCC) is the
major subtype of RCC accounting for more than 80% of all RCC types
(2). If ccRCC is limited to the
kidneys and has no systemic metastatic spread, it is a potentially
curable disease, particularly when diagnosed at an early stage
(3). Metastasis significantly
affects the clinical outcome of RCC patients. Despite the approval
of several targeted therapies that have led to an improvement in
the progression-free survival rate of these patients, advanced and
metastatic RCC remains difficult to treat (4). Understanding the molecular mechanisms
involved in metastatic RCC may improve the outcome of RCC patients.
Thus, more efforts are urgently needed to demonstrate the potential
mechanisms underlying the metastasis of RCC. Recently, microRNAs, a
class of small non-coding RNAs, were found to play critical roles
in the metastatic dissemination of tumor cells in different types
of cancer, including ccRCC (5).
miR-19a has been identified as an oncogenic miRNA in
gastric (6) and bladder cancer
(7) and glioma (8). In addition, miR-19a appears to be a
tumor suppressor in some types of cancer, such as multiple myeloma
(9). Thus, miR-19a is a
bifunctional miRNA depending on various cancer types. However, the
role of miR-19a in renal cancer remains unknown.
It has been reported that modulation of the
miR-19/PTEN/AKT/p53 axis by curcumin inhibited bisphenol
A-associated breast cancer progression (10). The PIK3/AKT signaling pathway has
been implicated in the metastasis of urothelial carcinoma, such as
renal carcinoma (11).
Phosphoinositide-3-kinase catalytic α subunit (PIK3CA) and PTEN,
integral parts of this pathway, play a critical role in controlling
malignant growth, cell cycle progression and proliferation
(11). The mutations identified in
PIK3CA and PTEN are present in renal metastasis of adenoid cystic
carcinoma of the breast, coinciding with a decrease in their
expression levels, suggesting that inactivation of the PI3K/AKT
pathway may be responsible for the unusually aggressive course of
adenoid cystic carcinoma (12).
Thus, we hypothesized that miR-19a/PIK3CA signaling may play a role
in metastatic renal carcinoma.
In the present study, we found that miR-19a
expression was significantly upregulated in metastatic ccRCC when
compared with that in adjacent and primary carcinoma tissues using
qPCR and in situ hybridization experiments. In addition,
these results were confirmed in renal carcinoma cell lines. miR-19a
expression in the cell lines derived from a metastatic site was
higher than that of cell lines derived from a primary site. By
gain- and loss-of-function experiments, we found that miR-19a acted
as an oncogenic miRNA regulating renal cancer cell proliferation,
migration and invasion by directly targeting PIK3CA. Furthermore,
we also explored the downstream molecules of miR-19a/PIK3CA
signaling. Recent studies have indicated that the Notch signaling
pathway plays an important role in renal function (13). A previous study demonstrated that
high expression of Notch signaling molecules, Notch1 and its ligand
Jagged1, increased the risk of metastasis in T1 stage ccRCC
(14). In the present study, we
found that Notch signaling was induced by upregulation of miR-19a,
and inactivation of Notch signaling attenuated cell proliferation,
migration and invasion promoted by miR-19a. Thus, we provide
evidence to demonstrate that downregulation of miR-19a may be
therapeutically beneficial for metastatic renal carcinoma.
Materials and methods
Sample collection
A total of 60 clear cell renal cell carcinoma tissue
samples, including 20 adjacent, 20 primary and 20 metastatic tumor
tissues were obtained from the Hunan Provincial People’s Hospital
according to the Legislation and Ethics Boards of Hunan Provincial
People’s hospital. Informed consent was obtained by all subjects.
All samples were collected and classified using histopathological
evaluation and stored at −80°C until used.
Cell culture and treatment
Human renal tubular epithelial cells (HKC) and all
human renal carcinoma cell lines, including ACHN, Caki-1, 786-O,
SN12C, A704, A498 and TK10, were purchased from the American Type
Culture Collection (ATCC; Manassas, VA, USA). All cells were
cultured in RPMI-1640 medium (Life Technologies, Grand Island, NY,
USA) supplemented with 10% fetal bovine serum (FBS) under standard
conditions (5% CO2, 37°C). The cells were transfected
with lentiviruses that expressed pre-miR-19a, anti-miR-19a or
negative control (NC) using Lipofectamine 3000 (Invitrogen,
Carlsbad, CA, USA) at a final concentration of 50 nM. The
lentiviruses for knockdown of Notch1 and Jagged1 were designed and
purchased from Genechem (Shanghai, China). Following a 48-h
transfection, the expression of miR-19a was detected by real-time
PCR and the expression levels of PIK3CA, Notch1, Jagged1 and MMP9
were assessed by western blot analysis.
In situ hybridization for miR-19a
The paraffin-embedded tissue samples were serially
cut at 4 µm. Slides were deparaffinized in dimethylbenzene
and dehydrated in gradient alcohol. Expression of miR-19a was
detected by in situ hybridization with probes for miR-19a
according to the manufacturer’s protocol provided in the microRNA
ISH Optimization kit (Exiqon Inc., Vedbaek, Denmark). The probe
sequence was designed as UGUGCAAAUCUAUGCAAAACUGA. The slides were
visualized with 3,3′-diaminobenzidine (DAB) for 5 min, and
counterstained with haematoxylin for 1 min. Results of the slides
were evaluated, and images were captured with an Olympus microscope
(Olympus C-7070; Tokyo, Japan).
Quantitative real-time polymerase chain
reaction (qPCR)
Total RNA was extracted from the indicated cells
using the Ultrapure RNA kit (CWBiotech, Beijing, China) according
to the manufacturer’s instructions. The expression of miR-19a was
detected by qPCR using miRcute miRNA (Qiagen, Valencia, CA, USA).
The specific primer sets for miRNA-19a (HmiR0201) and U6
(miRQP9001) were purchased from geneCopoeia. miR-19a expression was
normalized to U6. The expression of PIK3CA was detected by qPCR
using the FastLane Cell SYBR® Green kit (Qiagen). The
primers for PIK3CA and β-actin were: PIK3CA sense, TGCTAAAGA
GGAACACTGTCCA and antisense, GGTACTGGCCAAA GATTCAAAG; β-actin
sense, AGGGGCCGGACTCGT CATACT and antisense, GGCGGCACCACCATGTACCCT.
The 2−ΔΔCT method was used to analyze the data, and
β-actin expression was used as internal control.
Western blot analysis
The total protein was extracted from the indicated
cells using RIPA lysis buffer (Boster, Wuhan, China). Protein
concentrations were determined using the BCA protein assay kit
(Thermo, Waltham, MA, USA). A total of 60 µg total protein
was separated using SDS-PAGE and then transferred to nitrocellulose
membranes. The membranes were blocked in 8% non-fat milk diluted in
TBST and incubated with the indicated primary antibody [polyclonal
antibody anti-PIK3CA and anti-MMP9 (from ABZOOM; rabbit, 1:200);
polyclonal antibody anti-Notch1 and anti-Jagged1 (from Immunoway;
rabbit, 1:100); monoclonal antibody anti-β-actin (from Boster;
mouse, 1:3,000)] overnight at 4°C. The membranes were washed with
TBST for three times for 15 min each and incubated with the
secondary antibody for 60 min at 37°C. The signals on the membrane
were detected by enhanced chemiluminescence reagent. β-actin was
used as as internal control expression. Data were analyzed by
densitometry using Image-Pro plus software 6.0.
Dual luciferase report system
The wild-type (wt) and mutant (mut)
3′-untranscriptional region (3′-UTR) of PIK3CA were designed by
GeneCopoeia, and were inserted into the dual luciferase reporter
vector. For the luciferase assay, 105 cells were plated
and cultured in 12-well plates to reach ~70% confluency. 786-o and
Caki-1 cells were co-transfected with miR-19a mimic or miR-19a
inhibitors and wt/mut 3′-UTR of PIK3CA dual luciferase reporter
vector, respectively. Following a 48-h transfection, the luciferase
activities were detected using a dual luciferase reporter gene
assay kit (BioVision, Milpitas, CA, USA) on a luminometer (Roche,
Mannheim, Germany). Renilla luciferase activity was
normalized to firefly luciferase activity.
Transwell assay
Starved cells were resuspended in serum-free medium
and added to the upper chamber of a Transwell chamber. The lower
chamber was filled with medium containing 10% FBS. Following a 24-h
culture, the cells that attached to the bottom were fixed and
stained with crystal violet for 45 min and dried in air. The
optical density (OD) at 570 nm of the crystal violet dissolved in
10% acetic acid was detected using an enzyme immunoassay
analyzer.
CCK-8 assay
Five thousand cells were seeded in each 96-well
plate. Then, following the indicated treatment, the cells were
further incubated for 0, 24, 48 and 72 h, respectively. Ten
microliters of CCK-8 reagents (Solarbio, Beijing, China) were added
to the well at 1 h before the end of the incubation. The OD at 570
nm of each well was detected by enzyme immunoassay analyzer.
Scratch assay
Cells in each group were collected and resuspended
in complete medium containing 10% FBS. Then, 5×104 cells
were seeded in 24-well plates and cultured at 37°C in 5%
CO2 until reaching ~100% confluency. The cells were
scratched with the head of a 10-µl tip and the cells were
gently washed with serum-free medium. Following a 24-h culture in
serum-free medium at 37°C in 5% Co2, the cells were
cultured in complete medium containing 10% FBS for three days.
Cells in each group were photographed.
Statistical analysis
Data analysis was performed using Graphpad Prism5
software and SPSS 16.0 software. Depending on the experimental
conditions, Student’s t-tests or one-way ANOVA were used. Data are
expressed as mean ± SD. P-values <0.05 were considered
statistically significant compared to the controls.
Results
miR-19a is upregulated in renal carcinoma
tissues and cell lines
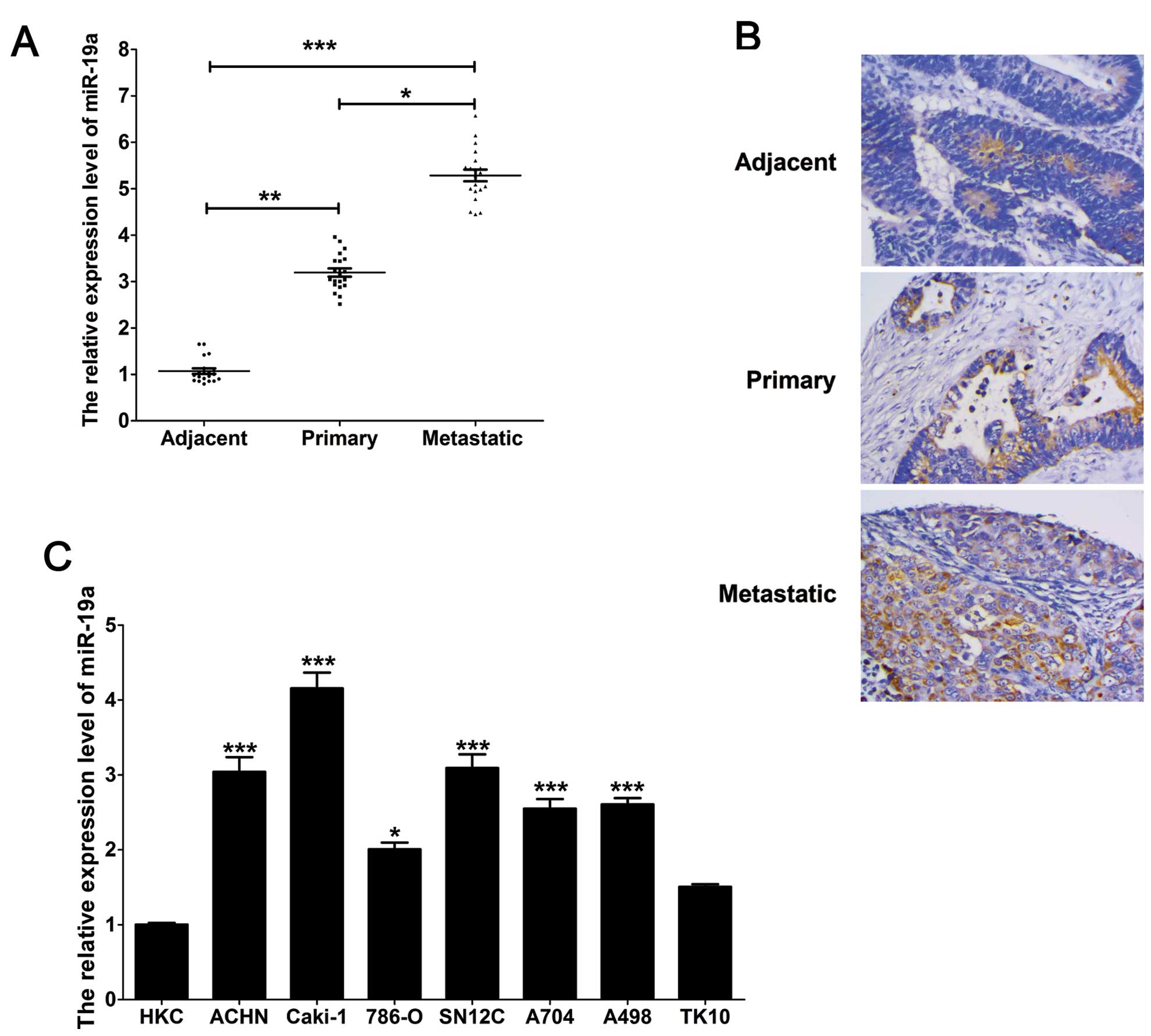
The average expression level of miR-19a was
significantly upregulated (P<0.001) in the renal carcinoma
tissue samples, including primary and metastatic clear cell renal
carcinoma tissues, compared with the 20 adjacent controls, as
indicated by qPCR. In addition, the expression of miR-19a in the
metastatic clear cell renal carcinoma tissues was higher than that
in the primary tissues (Fig. 1A).
Furthermore, these results were confirmed by in situ
hybridization (Fig. 1B). Moreover,
similar results were observed in the renal carcinoma cell lines.
The expression of miR-19a in the renal carcinoma cell lines was
significantly higher than that in the HKC cells. Moreover, miR-19a
expression in the ACHN and Caki-1 cells derived from a metastatic
site was higher than that in cells derived from a primary clear
cell adenocarcinoma, including 786-O, SN12C, A704, A498 and TK10
cells (Fig. 1C).
miR-19a regulates PIK3CA expression at
the transcriptional and translational levels by directly targeting
its 3′-UTR
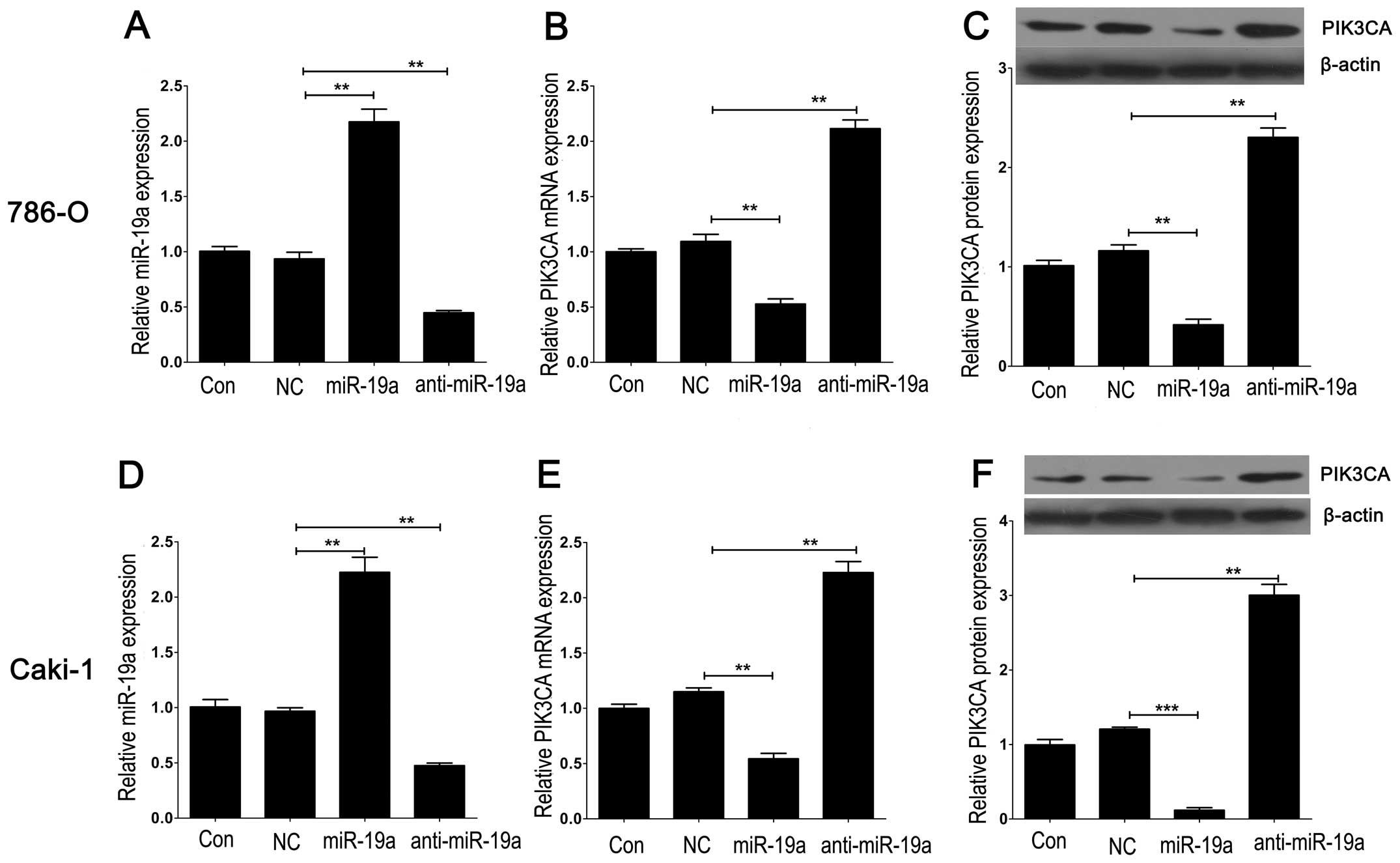
To further investigate the downstream molecules
targeted by miR-19a, we transfected Lv-pre-miR-19a or
Lv-anti-miR-19a into 786-O and Caki-1 cells to induce or knock down
the expression of miR-19a. We found that miR-19a expression was
markedly upregulated or downregulated in the 786-O and Caki-1 cells
following Lv-pre-miR-19a or Lv-anti-miR-19a transfection,
indicating that the efficiency of transfection was satisfied for
further analysis (Fig. 2A and D).
Furthermore, mRNA and protein expression levels of PIK3CA, a
putative target of miR-19a screened by a bioinformatic tool
(Targetscan), were markedly downregulated by Lv-pre-miR-19a
transfection and upregulated by Lv-anti-miR-19a transfection,
compared with the control in the 86-O and Caki-1 cell lines
(Fig. 2B, C, E and F). We then
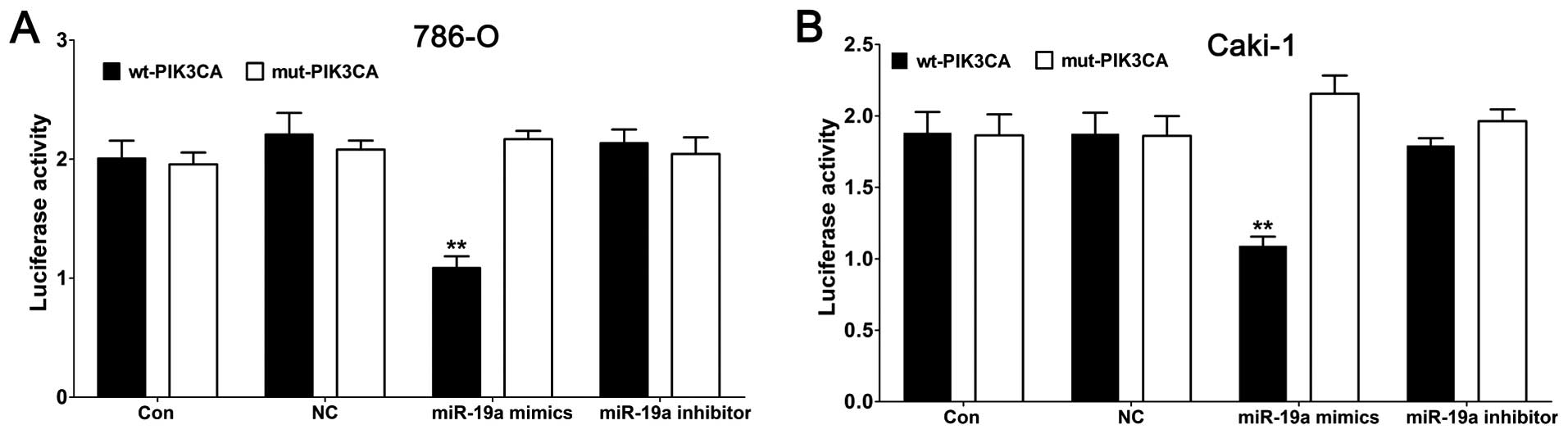
aimed to ascertain whether the 3′-UTR of PIK3CA had a direct target
site for miR-19a. Vectors encoding the wt or mut 3′-UTR of PIK3CA
were constructed into a dual luciferase reporter gene. By dual
luciferase reporter assay, we found that the luciferase activity
was significantly repressed in the miR-19a mimic transfectant
compared to the NC transfectant. Moreover, miR-19a-mediated
repression of luciferase activity was abolished by the mutant-type
3′-UTR of PIK3CA (Fig. 3A and B).
These results demonstrated that miR-19a directly targets PIK3CA and
regulates its expression at the transcriptional and translational
levels.
Effects of miR-19a on cell proliferation,
migration and invasion in 786-O and Caki-1 cells
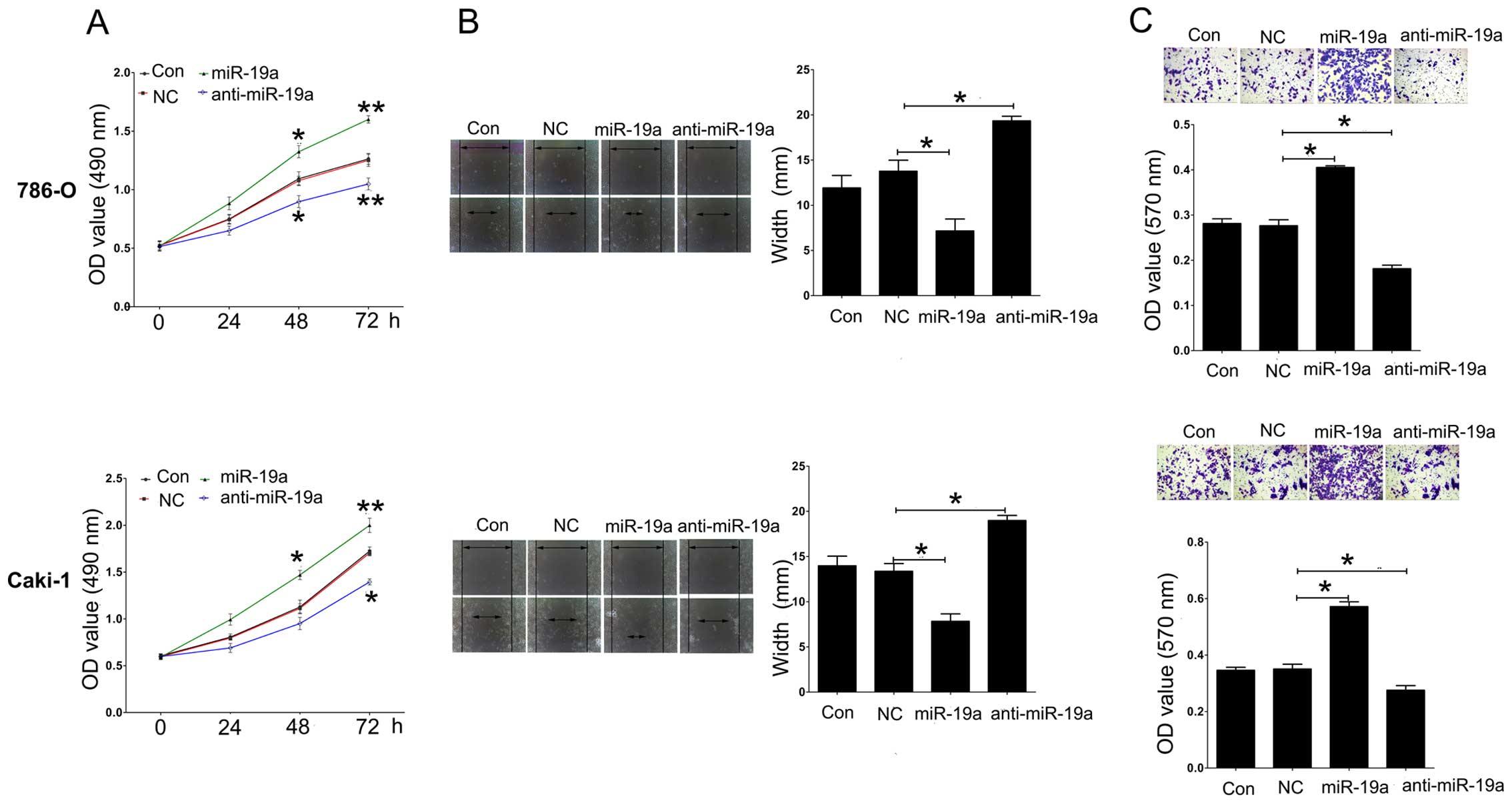
786-O and Caki-1 cell proliferation was measured
using CCK-8 assay following overexpression or knockdown of miR-19a.
We found that overexpression of miR-19a induced promotion of
proliferation, while downregulation of miR-19a inhibited cell
proliferation in the 786-O and Caki-1 cell lines (Fig. 4A). In addition, a scratch assay was
used to analyze cell migration after overexpression or knockdown of
miR-19a. Upregulation of miR-19a significantly induced cell
migration compared to NC, while knockdown of miR-19a repressed cell
migration in the 786-O and Caki-1 cell lines (Fig. 4B). Furthermore, using a Transwell
assay, we found that introduction of miR-19a expression
significantly induced cell invasion compared to NC, while knockdown
of miR-19a had an opposite effect on the 786-O and Caki-1 cell
lines (Fig. 4C).
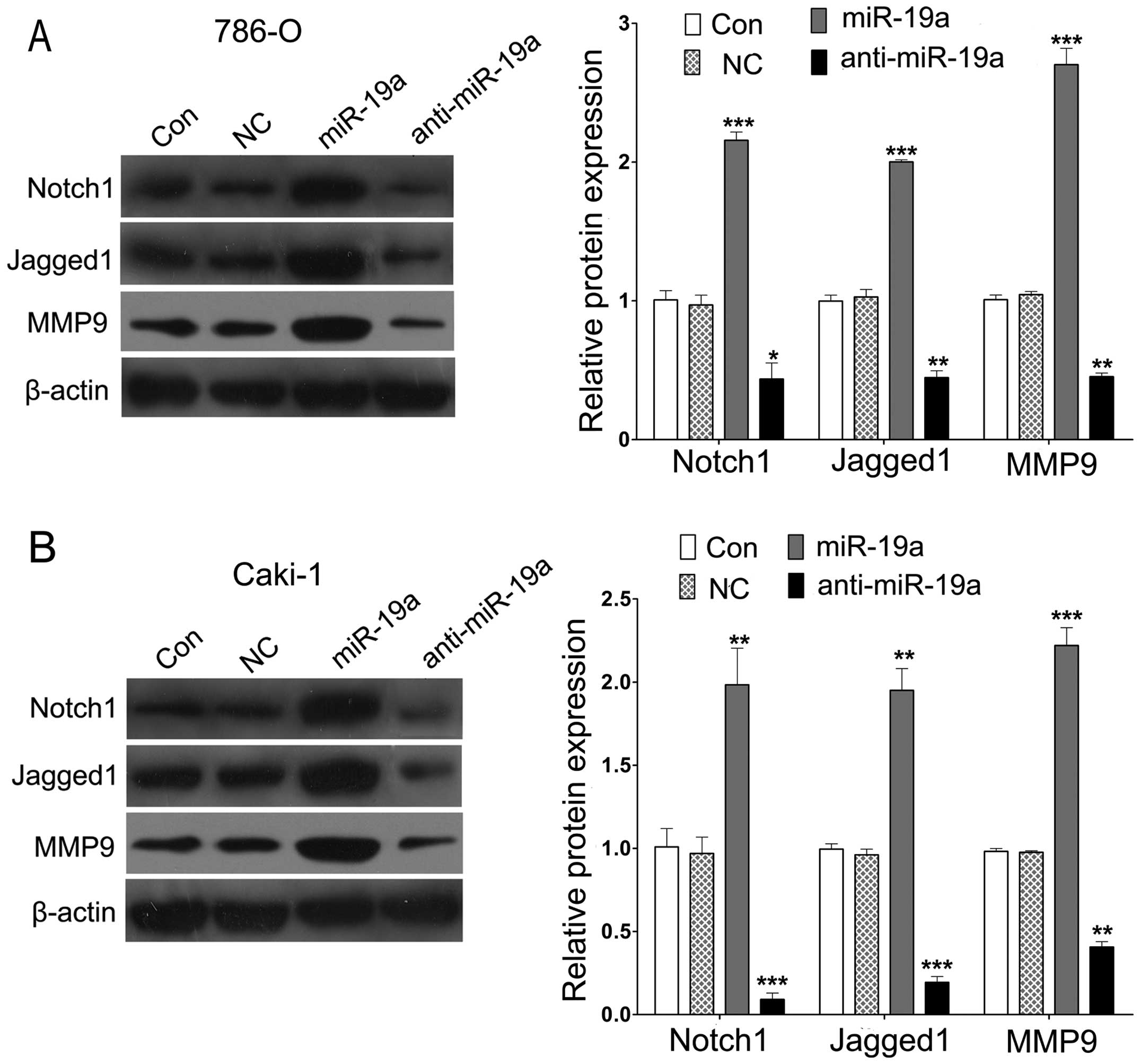
miR-19a regulates Notch1, Jagged1 and
MMP9 expression
To explore potential downstream molecular pathways
underlying miR-19a targeting to PIK3CA, we assessed the expression
of proliferation- and invasion-related genes including Notch1,
Jagged1 and MMP9 by western blot analysis in the 786-O and Caki-1
cells following Lv-pre-miR-19a or Lv-anti-miR-19a transfection. A
significant increase in the expression of Notch1, Jagged1 and MMP9
proteins was observed in the cells treated with Lv-pre-miR-19a.
Inversely, knockdown of miR-19a significantly reduced the
expression of Notch1, Jagged1 and MMP9 proteins in the 786-O and
Caki-1 cells compared with that in the control (Fig. 5A and B).
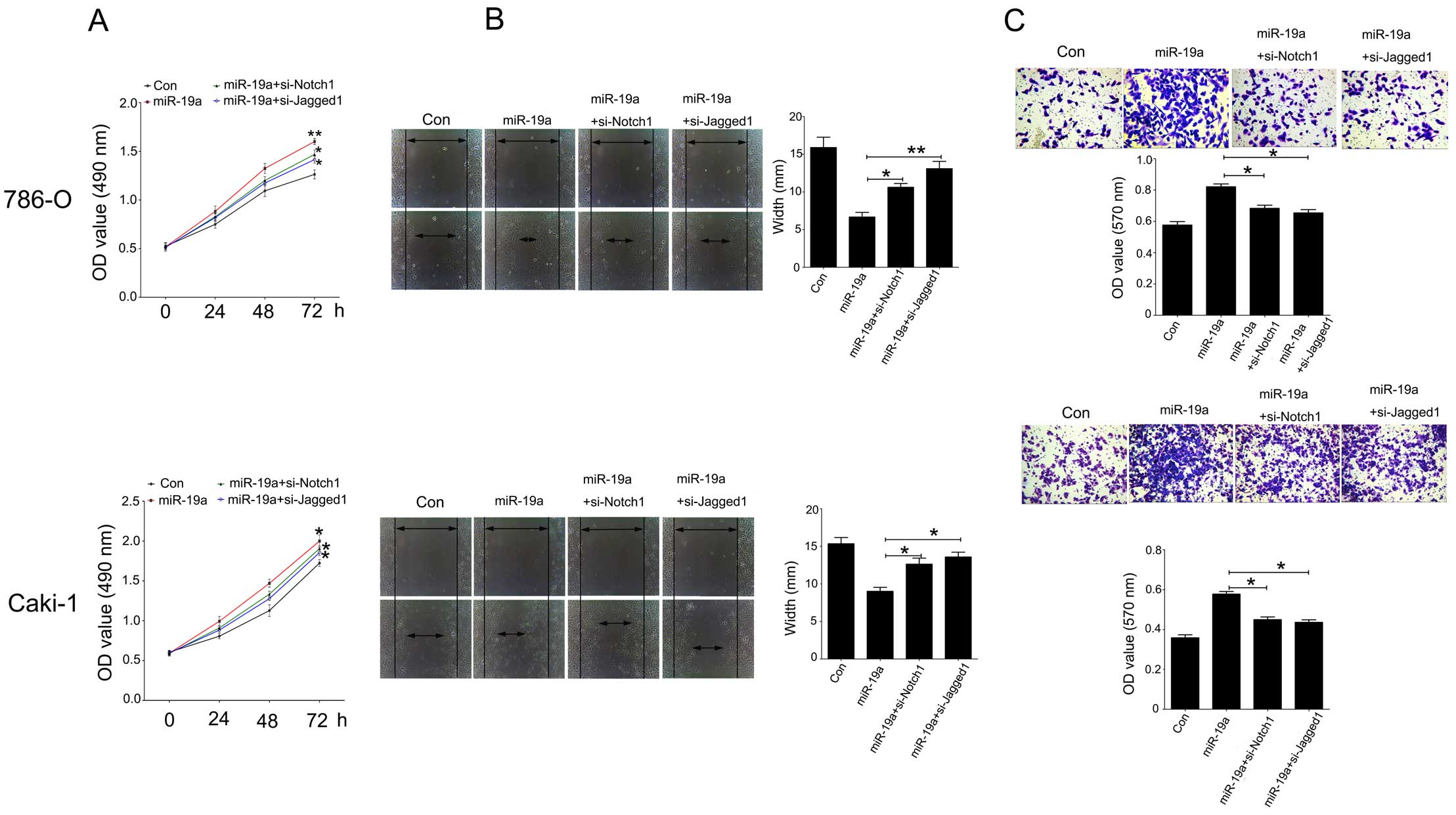
Notch1 and Jagged1 siRNAs attenuate the
effects of miR-19a on cell proliferation, migration and invasion in
786-O and Caki-1 cells
To investigate the role of Notch1 and Jagged1
signaling in the miR-19a/PIK3CA pathway in renal carcinoma cells, a
loss-of-function experiment was performed. As shown in Fig. 6A, CCK-8 assay was used to determine
the proliferation of the 786-O and Caki-1 cells following
overexpression of miR-19a and transfection with siRNA-Notch1 or
siRNA-Jagged1. Knockdown of Notch1 or Jagged1 inhibited the cell
proliferation promoted by miR-19a in the 786-O and Caki-1 cells. In
addition, a scratch assay was used to analyze cell migration after
knockdown of Notch1 or Jagged1 in the cells treated with
Lv-pre-miR-19a. Downregulation of Notch1 or Jagged1 significantly
suppressed cell migration compared to the Lv-pre-miR-19a group in
the 786-O and Caki-1 cell lines (Fig.
6B). Furthermore, using a Transwell assay, we found that
knockdown of Notch1 or Jagged1 significantly attenuated cell
invasion induced by miR-19a in the 786-O and Caki-1 cell lines
(Fig. 6C).
Discussion
Due to metastasis, ccRCC, the most common histologic
subtype of RCC, has become the leading cause of death from adult
urologic tumors. Emerging evidence indicates that miRNAs may act as
either oncogenes or tumor suppressors and participate in the
metastasis of malignancies. It has been reported that upregulation
of miR-19a has an oncogenic role in gastric cancer (6) and in supraglottic carcinoma tissues
(15), and high expression of
miR-19a was significantly associated with a poor patient survival
rate in human astrocytoma (16) and
correlates with more aggressive phenotypes of bladder cancer
(7). In addition, overexpression of
miR-19a was noted in early-stage breast cancer patients and the
level was significantly decreased after chemo/radiotherapy; in the
case of high-risk breast cancer patients serum levels of miR-19a
were significantly more frequent in comparison to a low-risk group
(17). Consistent with a previous
study, we found that miR-19a was significantly increased in ccRCC
tissues and renal carcinoma cell lines, with a higher expression in
metastatic than in localized tissues, suggesting that miR-19a is
involved in renal carcinoma progression. Furthermore, by gain- and
loss-of-function experiments, we found that enforced miR-19a
expression facilitated renal cancer cell proliferation, migration
and invasion, whereas knockdown of miR-19a expression exhibited an
opposite effect in vitro. Thus, collectively, upregulation
of miR-19a in renal cancer may promote cancer cell metastasis and
the poor outcome of patients.
Using a bioinformatic tool, we screened the putative
targets of miR-19a and found that miR-19a conversely targets
PIK3CA, an indispensable molecule of the mammalian target of
rapamycin mTOR signaling and PI3K/AKT pathway, which is deregulated
in most types of cancer (18).
PIK3CA mutations have been associated with a worse survival rate in
metastatic colorectal (19) and
breast cancer (20). Using a dual
luciferase reporter gene system, we confirmed the results from the
bioinformatic tool, that miR-19a regulates PIK3CA expression at the
translational and transcriptional levels by targeting its 3′-UTR.
It was previously demonstrated that overexpression of miR-19a was
significantly associated with the metastasis of gastric cancer and
poor overall prognosis at the clinical tissue level and promoted
epithelial-mesenchymal transition through the PI3K/AKT pathway, and
silencing of the PI3K/AKT pathway abolished the effect of miR-19a
(6). PTEN, a critical molecule of
the PI3K/AKT pathway, was found to be a target of miR-19b in
prostate epithelial and cancer cells. Expression of miR-19b altered
the expression of key components in the PI3K/Akt pathway, including
PIK3CA resulting in acceleration of epithelial and prostate cancer
cell proliferation (21). However,
it was reported that a majority of non-small cell lung cancer
tissue samples showed low miR-1 expression and high PIK3CA
expression; miR-1 upregulation inhibited non-small cell lung cancer
cell proliferation, migration and invasion by directly targeting
PIK3CA (22). Wang et al
demonstrated that siRNA-mediated silencing of PIK3CA blocked the
inhibitory effect of miR-375 on colorectal cancer cell growth
(23). Thus, PIK3CA exhibited an
opposite effect depending on which miRNA was its target and on
tumor type. In the present study, we found that PIK3CA was
targeting by miR-19a and its upregulation was accompanied by
inhibition of renal carcinoma cell growth and invasion.
Furthermore, we next explored the downstream
molecules associated with the miR-19a/PIK3CA pathway. Suppression
of the PI3K/Akt signaling pathway was associated with the
Notch/Jagged pathway in several types of cancer (24). We found that induced expression of
miR-19a by lentivirus resulted in a significant increase in Notch
signaling and MMP9 level in renal cancer cell lines while
downregulation of miR-19a decreased the expression levels of these
genes. It was reported that high MMP9 expression levels are
associated with poor prognosis of RCC (25). Furthermore, a poor survival rate
with a high frequency of metastases in conventional RCC was found
to be associated with MMP9 (26).
Blockade of MMP9 inhibited the migration and invasion of RCC cells
(27). In the present study, we
showed that downregulation of miR-19a suppressed the MMP9
expression and subsequently suppressed migration and invasion of
RCC cells. Notch genes, including Notch1, Notch2, Notch3 and
Notch4, encode receptors for at least five different Notch ligands,
including Jagged1. The Notch proteins mediate various cellular
processes including differentiation, proliferation and apoptosis
(28). It is possible that Notch
pathway activation is a common mechanism in the pathophysiology of
acquired renal diseases (29).
Notch can act as either an oncogene or a tumor-suppressor gene in
the development of cancer depending on the tumor type (30). A previous study showed that high
expression of Notch1 and Jagged1 increased the risk of metastasis
in T1 stage ccRCC by stimulating the proliferation and migration of
tumor cells (14). The expression
of Notch1 and Jagged1 proteins was higher in RCC than in
non-neoplastic tissues, correlative with tumor size, grade, TNM
stage and disease relapse; and suppression of the Notch pathway was
found to be associated with cell proliferation inhibition, as well
as induced G2/M phase cell cycle arrest and cell apoptosis in 786–0
and Caki cell lines (31). Thus,
the Notch pathway appears to be important in the oncogenesis of
ccRCC. In the present study, we found that silencing of the Notch
pathway by siRNA blocked the promotive effect of miR-19a on renal
cancer cell growth, migration and invasion, suggesting that miR-19a
contributes to the development of RCC through the Notch pathway via
targeting PIK3CA.
In conclusion, high miR-19a expression was found to
be associated with metastatic renal carcinoma tissues. Knockdown of
miR-19a exhibited an inhibitory effect on renal cancer cell
proliferation, migration and invasion by targeting PIK3CA, while
siRNA-mediated silencing of Notch signaling attenuated the
inhibitory effects of miR-19a. Thus, our evidence suggests that
inactivation of miR-19a/Notch signaling may be therapeutically
beneficial for the treatment and prevention of kidney cancer in the
clinic.
Acknowledgments
The present study was supported by Hunan Provincial
Science and Technology Program (2013FJ3149).
References
|
1
|
Dias F, Teixeira AL, Santos JI, Gomes M,
Nogueira A, Assis J and Medeiros R: Renal cell carcinoma
development and miRNAs: A possible link to the EGFR pathway.
Pharmacogenomics. 14:1793–1803. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Redova M, Svoboda M and Slaby O: MicroRNAs
and their target gene networks in renal cell carcinoma. Biochem
Biophys Res Commun. 405:153–156. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mishra PJ: MicroRNAs as promising
biomarkers in cancer diagnostics. Biomark Res. 2:192014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hao JF, Ren KM, Bai JX, Wang SN, Shao B,
Cao N and Li X: Identification of potential biomarkers for clear
cell renal cell carcinoma based on microRNA-mRNA pathway
relationships. J Cancer Res Ther. 10(Suppl): C167–C177. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang L, Xul B, Chen S, Lu K, Liu C, Wang
Y, Zhao Y, Zhang X, Liu D and Chen M: The complex roles of
microRNAs in the metastasis of renal cell carcinoma. J Nanosci
Nanotechnol. 13:3195–3203. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu W, Xu Z, Zhang M and Zuo Y: MiR-19a
promotes epithelial-mesenchymal transition through PI3K/AKT pathway
in gastric cancer. Int J Clin Exp Pathol. 7:7286–7296.
2014.PubMed/NCBI
|
|
7
|
Feng Y, Liu J, Kang Y, He Y, Liang B, Yang
P and Yu Z: miR-19a acts as an oncogenic microRNA and is
up-regulated in bladder cancer. J Exp Clin Cancer Res. 33:672014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jia Z, Wang K, Zhang A, Wang G, Kang C,
Han L and Pu P: miR-19a and miR-19b overexpression in gliomas.
Pathol Oncol Res. 19:847–853. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hao M, Zang M, Wendlandt E, Xu Y, An G,
Gong D, Li F, Qi F, Zhang Y, Yang Y, et al: Low serum miR-19a
expression as a novel poor prognostic indicator in multiple
myeloma. Int J Cancer. 136:1835–1844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X, Xie W, Xie C, Huang C, Zhu J, Liang
Z, Deng F, Zhu M, Zhu W, Wu R, et al: Curcumin modulates
miR-19/PTEN/AKT/p53 axis to suppress bisphenol A-induced MCF-7
breast cancer cell proliferation. Phytother Res. 28:1553–1560.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qian CN, Furge KA, Knol J, Huang D, Chen
J, Dykema KJ, Kort EJ, Massie A, Khoo SK, Vanden Beldt K, et al:
Activation of the PI3K/AKT pathway induces urothelial carcinoma of
the renal pelvis: Identification in human tumors and confirmation
in animal models. Cancer Res. 69:8256–8264. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vranić S, Bilalović N, Lee LM, Kruslin B,
Lilleberg SL and Gatalica Z: PIK3CA and PTEN mutations in adenoid
cystic carcinoma of the breast metastatic to kidney. Hum Pathol.
38:1425–1431. 2007. View Article : Google Scholar
|
|
13
|
Fujiki K, Inamura H and Matsuoka M:
Detrimental effects of Notch1 signaling activated by cadmium in
renal proximal tubular epithelial cells. Cell Death Dis.
5:e13782014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ai Q, Ma X, Huang Q, Liu S, Shi T, Zhang
C, Zhu M, Zhang Y, Wang B, Ni D, et al: High-level expression of
Notch1 increased the risk of metastasis in T1 stage clear cell
renal cell carcinoma. PLoS One. 7:e350222012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang T, Han G, Wang Y, Chen K and Sun Y:
MicroRNA expression profiles in supraglottic carcinoma. Oncol Rep.
31:2029–2034. 2014.PubMed/NCBI
|
|
16
|
Zhi F, Shao N, Wang R, Deng D, Xue L, Wang
Q, Zhang Y, Shi Y, Xia X, Wang S, et al: Identification of 9 serum
microRNAs as potential noninvasive biomarkers of human astrocytoma.
Neuro Oncol. Epub ahead of print. Aug 18–2014.PubMed/NCBI
|
|
17
|
Sochor M, Basova P, Pesta M, Dusilkova N,
Bartos J, Burda P, Pospisil V and Stopka T: Oncogenic microRNAs:
miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early
breast cancer in serum. BMC Cancer. 14:4482014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu M, Bardia A, Aceto N, Bersani F, Madden
MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, et al:
Cancer therapy. Ex vivo culture of circulating breast tumor cells
for individualized testing of drug susceptibility. Science.
345:216–220. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yaeger R, Cowell E, Chou JF, Gewirtz AN,
Borsu L, Vakiani E, Solit DB, Rosen N, Capanu M, Ladanyi M, et al:
RAS mutations affect pattern of metastatic spread and increase
propensity for brain metastasis in colorectal cancer. Cancer.
121:1195–1203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Deb S, Wong SQ, Li J, Do H, Weiss J, Byrne
D, Chakrabarti A, Bosma T, Fellowes A, Dobrovic A, et al kConFab
Investigators: Mutational profiling of familial male breast cancers
reveals similarities with luminal A female breast cancer with rare
TP53 mutations. Br J Cancer. 111:2351–2360. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tian L, Fang YX, Xue JL and Chen JZ: Four
microRNAs promote prostate cell proliferation with regulation of
PTEN and its downstream signals in vitro. PLoS One. 8:e758852013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu QQ, Wu H, Huang X, Shen H, Shu YQ,
Zhang B, Xiang CC, Yu SM, Guo RH and Chen L: MiR-1 targets PIK3CA
and inhibits tumorigenic properties of A549 cells. Biomed
Pharmacother. 68:155–161. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Tang Q, Li M, Jiang S and Wang X:
MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA.
Biochem Biophys Res Commun. 444:199–204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiao W, Chen X and He M: Inhibition of the
Jagged/Notch pathway inhibits retinoblastoma cell proliferation via
suppressing the PI3K/Akt, Src, p38MAPK and Wnt/β-catenin signaling
pathways. Mol Med Rep. 10:453–458. 2014.PubMed/NCBI
|
|
25
|
Sato A, Nagase H, Obinata D, Fujiwara K,
Fukuda N, Soma M, Yamaguchi K, Kawata N and Takahashi S: Inhibition
of MMP-9 using a pyrroleimidazole polyamide reduces cell invasion
in renal cell carcinoma. Int J Oncol. 43:1441–1446. 2013.PubMed/NCBI
|
|
26
|
Cho NH, Shim HS, Rha SY, Kang SH, Hong SH,
Choi YD, Hong SJ and Cho SH: Increased expression of matrix
metalloproteinase 9 correlates with poor prognostic variables in
renal cell carcinoma. Eur Urol. 44:560–566. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liang L, Li L, Zeng J, Gao Y, Chen YL,
Wang ZQ, Wang XY, Chang LS and He D: Inhibitory effect of silibinin
on EGFR signal-induced renal cell carcinoma progression via
suppression of the EGFR/MMP-9 signaling pathway. Oncol Rep.
28:999–1005. 2012.PubMed/NCBI
|
|
28
|
Li D, Masiero M, Banham AH and Harris AL:
The notch ligand JAGGED1 as a target for anti-tumor therapy. Front
Oncol. 4:2542014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Murea M, Park JK, Sharma S, Kato H,
Gruenwald A, Niranjan T, Si H, Thomas DB, Pullman JM, Melamed ML,
et al: Expression of Notch pathway proteins correlates with
albuminuria, glomerulosclerosis, and renal function. Kidney Int.
78:514–522. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leong KG and Karsan A: Recent insights
into the role of Notch signaling in tumorigenesis. Blood.
107:2223–2233. 2006. View Article : Google Scholar
|
|
31
|
Wu K, Zhang L, Lin Y, Yang K and Cheng Y:
Inhibition of γ-secretase induces G2/M arrest and triggers
apoptosis in renal cell carcinoma. Oncol Lett. 8:55–61.
2014.PubMed/NCBI
|