Introduction
Prostate cancer represents a major cause of
cancer-related mortality and morbidity (1). The majority of prostate cancers behave
in an indolent manner, but a subset is highly aggressive. Despite
recent advances in research, the only established pretreatment
prognostic parameters currently include Gleason grade and tumor
extent on biopsies, preoperative PSA and clinical stage. As these
data are statistically powerful but not sufficient for optimal
individual treatment decisions, it can be hoped, that a better
understanding of the biology of the disease will eventually lead to
better prognostic biomarkers.
DNA double-strand breaks (DSBs) are highly cytotoxic
lesions that typically occur during cell division but can also be
induced by exogenous noxes including ionizing radiation and various
DNA-damaging chemicals (2). Cells
typically repair DSBs by homologous recombination (HR) or
non-homologous DNA end-joining (NHEJ) pathways (3). NHEJ deficiencies and unrepaired DSBs
can result in severe cellular consequences ranging from death to
neoplastic transformation (3–9).
DNA ligase IV (LIG4) plays an essential role in the
NHEJ machinery, the major DSB repair mechanism (10,11).
Hypomorphic LIG4 mutations lead to LIG4 syndrome characterized by
immunodeficiency, microcephaly, growth retardation, unusual facial
features, developmental delay and acute radiosensitivity, genomic
instability and malignancy (12).
Moreover, polymorphic variants (SNP) in LIG4 has been identified in
several malignancies (13–17) including prostate cancers (18,19).
In a recent study we identified LIG4 as a
potentially relevant gene in prostate cancer, which can be
inactivated by mutation and genomic deletion (20,21).
The present study was undertaken to further investigate LIG4
protein expression in prostate cancer and its association with
tumor phenotype, outcome and key genomic alterations. For this
purpose a tissue microarray containing 11,152 prostate cancer
specimens with follow-up information and attached molecular
information was utilized. Our data identify a tight link of
increased LIG4 expression to tumor phenotype, early PSA recurrence,
ERG fusion and PTEN deletions in prostate cancer.
Materials and methods
Patients
Radical prostatectomy specimens were available from
11,152 patients, undergoing surgery between 1992 and 2011 at the
Department of Urology, and the Martini Clinics at the University
Medical Center Hamburg-Eppendorf. Follow-up data were available for
a total of 9,695 patients with a median follow-up of 36.8 months
(range, 1–228 months, Table I).
Prostate-specific antigen values were measured following surgery
and recurrence was defined as a postoperative PSA of 0.2 ng/ml and
increasing at first of appearance. All prostate specimens were
analyzed according to a standard procedure, including a complete
embedding of the entire prostate for histological analysis
(22). The TMA manufacturing
process was described previously in detail (23). In short, one 0.6-mm core was taken
from a representative tumor area from each patient. The tissues
were distributed among 24 TMA blocks, each containing 144–522 tumor
samples. Presence or absence of cancer tissue was validated by
immunohistochemical AMACR and 34BE12 analysis on adjacent TMA
sections. For internal controls, each TMA block also contained
various control tissues, including normal prostate tissue. The
molecular database attached to this TMA contained results on ERG
expression in 9,628, ERG break apart by fluorescence in situ
hybridization (FISH) analysis in 6,106 [expanded from (24)], and deletion status of 5q21 in 3,037
[expanded from (25)], 6q15 in
3,528 [expanded from (26)],
PTEN in 6,130 [expanded from (27)], and 3p13 in 1,290 [expanded from
(28)] tumors. Analysis of patient
and corresponding histopathological data for research purposes, as
well as construction of tissue micro-arrays from archived
diagnostic left-over tissues, was approved by the local laws
(HmbKHG, §12,1) and by the local ethics committee (Ethics
Commission of Hamburg, WF-049/09 and PV3652). All study was carried
out in compliance with the Helsinki Declaration.
 | Table IComposition of the prognosis tissue
microarray containing 11,152 prostate cancer specimens. |
Table I
Composition of the prognosis tissue
microarray containing 11,152 prostate cancer specimens.
| No. of patients
|
|---|
| Study cohort on
tissue microarray (n=11,152) | Biochemical among
categoriesrelapse (n=1,824) |
|---|
| Follow-up
(month) |
| Mean | 53.4 | – |
| Median | 36.8 | – |
| Age (years) |
| <50 | 318 | 49 |
| 50–60 | 2,768 | 460 |
| 60–70 | 6,548 | 1,081 |
| >70 | 1,439 | 232 |
| Pretreatment PSA
(ng/ml) |
| <4 | 1,407 | 142 |
| 4–10 | 6,735 | 827 |
| 10–20 | 2,159 | 521 |
| >20 | 720 | 309 |
| pT stage (AJCC
2002) |
| pT2 | 7,370 | 570 |
| pT3a | 2,409 | 587 |
| pT3b | 1,262 | 618 |
| pT4 | 63 | 49 |
| Gleason grade |
| ≤3+3 | 2,859 | 193 |
| 3+4 | 6,183 | 849 |
| 4+3 | 1,565 | 573 |
| ≥4+4 | 482 | 208 |
| pN stage |
| pN0 | 6,117 | 1,126 |
| pN+ | 561 | 291 |
| Surgical
margin |
| Negative | 8,984 | 1,146 |
| Positive | 1,970 | 642 |
Immunohistochemistry
Freshly cut TMA sections were analyzed in one day
and in one experiment. Primary antibody specific for LIG4 (rabbit,
at 1:150 dilution; Sigma) was applied, slides were deparaffinized
and exposed to heat-induced antigen retrieval for 5 min in an
autoclave at 121°C in 7.8 Tris-EDTA-citrate buffer. Bound antibody
was then visualized using the EnVision kit (Dako). LIG4 expression
was homogeneous within individual cancer tissue samples. Assessment
of immunostaining was thus limited to recording the staining
intensity in 4 categories: Negative, weak, moderate and strong
immunostaining. Staining levels in cancer cells were defined by
estimating each spot by one person experienced in TMA analyses.
Statistics
Statistical calculations were performed with JPM 9
software (SAS Institute Inc., Cary, NC, USA). Contingency tables
and the chi-square (x2) test were performed to search
for associations between molecular parameters and tumor phenotype.
Survival curves were calculated according to Kaplan-Meier. The
log-rank test was applied to detect significant survival
differences between groups. COX proportional hazards regression
analysis was performed to test the statistical independence and
significance between pathological, molecular and clinical
variables.
Results
Technical aspects
A total of 2,493 of 11,152 arrayed tissue samples
(22%) were non-informative for LIG4 analysis due to the complete
lack of tissue or absence of unequivocal cancer cells.
LIG4 immunohistochemistry
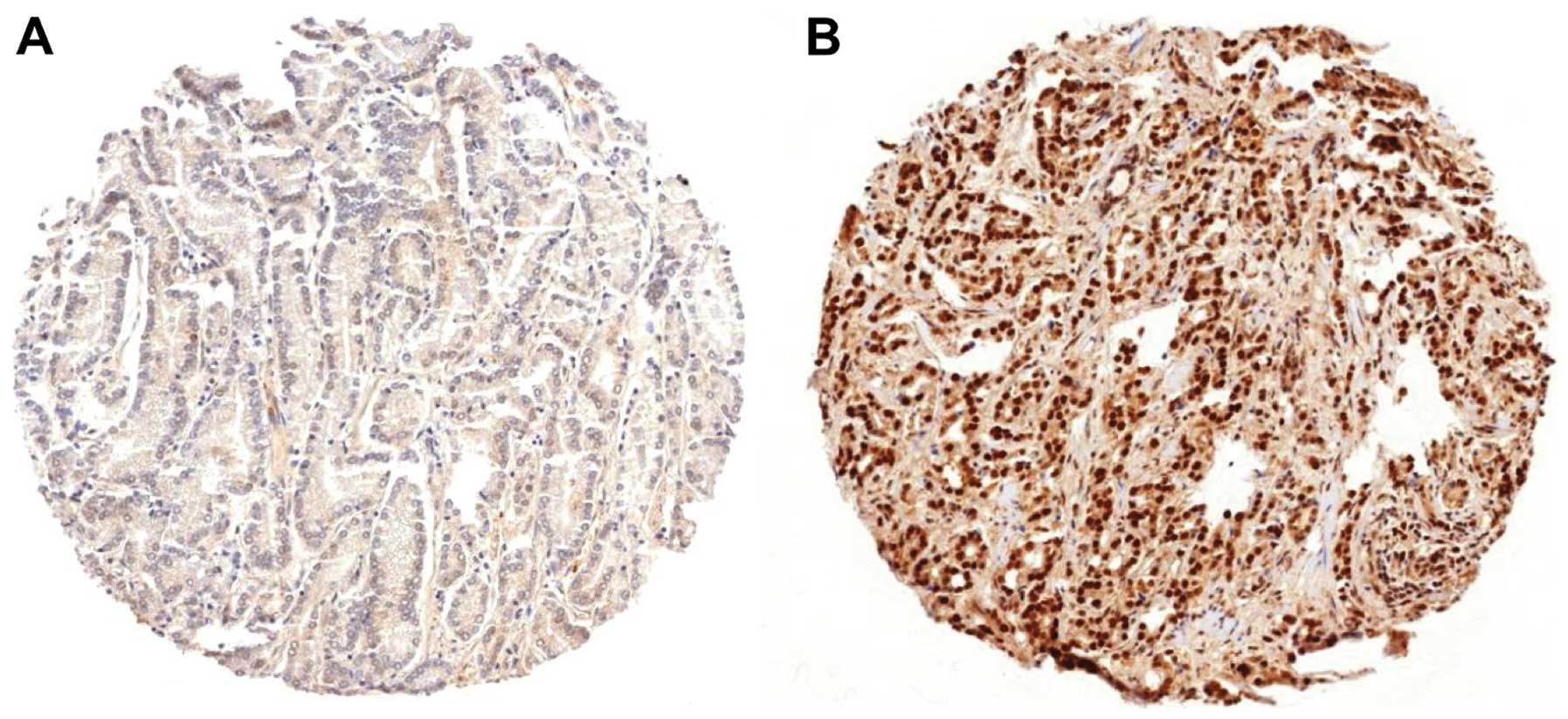
LIG4 expression was localized in the nucleus of the
cells with increased intensities in malignant as compared to benign
prostate epithelium. A total of 7,905 of our 8,663 interpretable
prostate cancers (91%) showed positive LIG4 expression, which was
considered weak in 12%, moderate in 23% and strong in 56% of cases.
Representative images are given in Fig.
1. Strong LIG4 expression was positive significantly linked to
high Gleason score (P<0.0001) and positive nodal involvement
(P=0.03), but not to high preoperative PSA levels, advanced tumor
stage and positive surgical margin when all cancers were
analyzed.
LIG4 versus ERG status
To evaluate whether LIG4 expression is linked to
ERG status, we used our pre-existing database including data
on TMPRSS2:ERG fusion status obtained by FISH in 4,980
patients and by IHC in 7,769 tumors for which LIG4 immunostaining
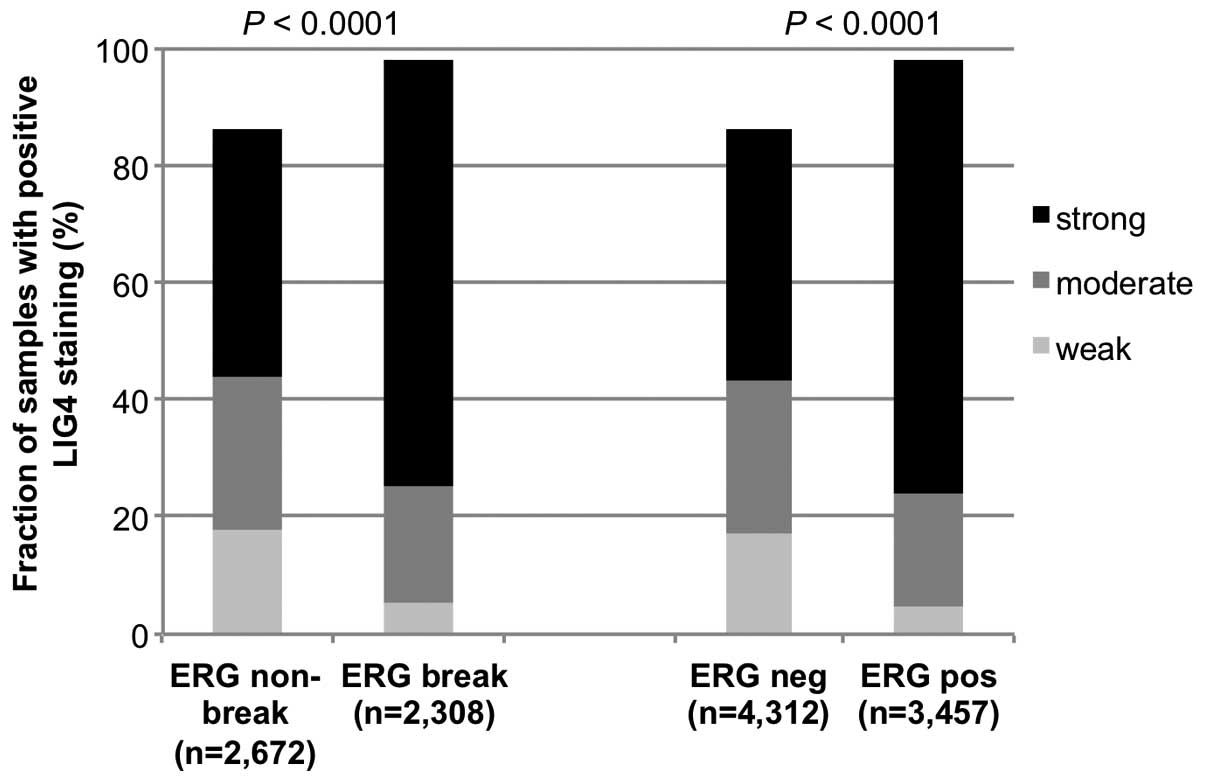
was also available. Strong LIG4 expression was significantly more
frequent in fusion-type (1,677/2,308, 73%) than in non-fusion-type
prostate cancers (1,121/2,672, 42%, P<0.0001, Fig. 2) as analyzed by FISH. Accordingly,
strong LIG4 staining occurred more often in ERG expression positive
(2,569/3,457, 74%) than in ERG expression negative prostate cancers
(1,849/4,312, 43%, P<0.0001, Fig.
2). Since increased LIG4 expression was more frequent in
fusion-type prostate cancers, the associations of LIG4 expression
with tumor phenotype and clinical cancer features were separately
analyzed in both non-fusion and fusion-type prostate cancers
(Tables II and III). In both non-fusion and fusion-type
prostate cancers, strong LIG4 expression was significantly linked
to advanced Gleason grade (P<0.001). There was only a marginal
relationship between LIG4 expression and pT stage which reached
significance in ERG-negative cancers (P=0.0005, Table II).
 | Table IIAssociations between LIG4 expression
and clinicopathological parameters in the subgroup of ERG-negative
prostate cancer. |
Table II
Associations between LIG4 expression
and clinicopathological parameters in the subgroup of ERG-negative
prostate cancer.
| Evaluable (N) | LIG4 IHC result (%)
| P-value |
|---|
| Negative | Weak | Moderate | Strong |
|---|
| All cancers | 4,312 | 14 | 17 | 26 | 43 | |
| Tumor stage |
| pT2 | 2,872 | 15 | 18 | 25 | 41 | 0.0005 |
| pT3a | 887 | 12 | 15 | 28 | 45 | |
| pT3b | 513 | 9 | 16 | 26 | 49 | |
| pT4 | 28 | 21 | 11 | 29 | 39 | |
| Gleason grade |
| ≤3+3 | 987 | 20 | 25 | 23 | 32 | <0.0001 |
| 3+4 | 2,036 | 13 | 16 | 27 | 44 | |
| 4+3 | 711 | 10 | 12 | 28 | 50 | |
| ≥4+4 | 238 | 8 | 12 | 25 | 55 | |
| Lymph node
metastasis |
| N0 | 2,469 | 13 | 16 | 26 | 45 | 0.006 |
| N+ | 226 | 7 | 13 | 26 | 54 | |
| Surgical
margin |
| Negative | 3,443 | 14 | 17 | 26 | 42 | 0.36 |
| Positive | 791 | 12 | 17 | 27 | 44 | |
 | Table IIIAssociations between LIG4 expression
and clinicopathological parameters in the subgroup of ERG-positive
prostate cancer. |
Table III
Associations between LIG4 expression
and clinicopathological parameters in the subgroup of ERG-positive
prostate cancer.
| Evaluable (N) | LIG4 IHC result (%)
| P-value |
|---|
| Negative | Weak | Moderate | Strong |
|---|
| All cancers | 3,457 | 2 | 5 | 19 | 74 | |
| Tumor stage |
| pT2 | 2,039 | 2 | 5 | 18 | 74 | 0.19 |
| pT3a | 951 | 1 | 5 | 18 | 76 | |
| pT3b | 430 | 2 | 3 | 23 | 72 | |
| pT4 | 20 | 0 | 10 | 15 | 75 | |
| Gleason grade |
| ≤3+3 | 792 | 3 | 8 | 19 | 70 | 0.0005 |
| 3+4 | 2,038 | 2 | 4 | 18 | 76 | |
| 4+3 | 490 | 1 | 4 | 18 | 76 | |
| ≥4+4 | 114 | 2 | 3 | 24 | 72 | |
| Lymph node
metastasis |
| N0 | 1,932 | 2 | 4 | 18 | 76 | 0.01 |
| N+ | 204 | 3 | 3 | 25 | 68 | |
| Surgical
margin |
| Negative | 2,707 | 2 | 5 | 19 | 74 | 0.69 |
| Positive | 691 | 1 | 5 | 19 | 74 | |
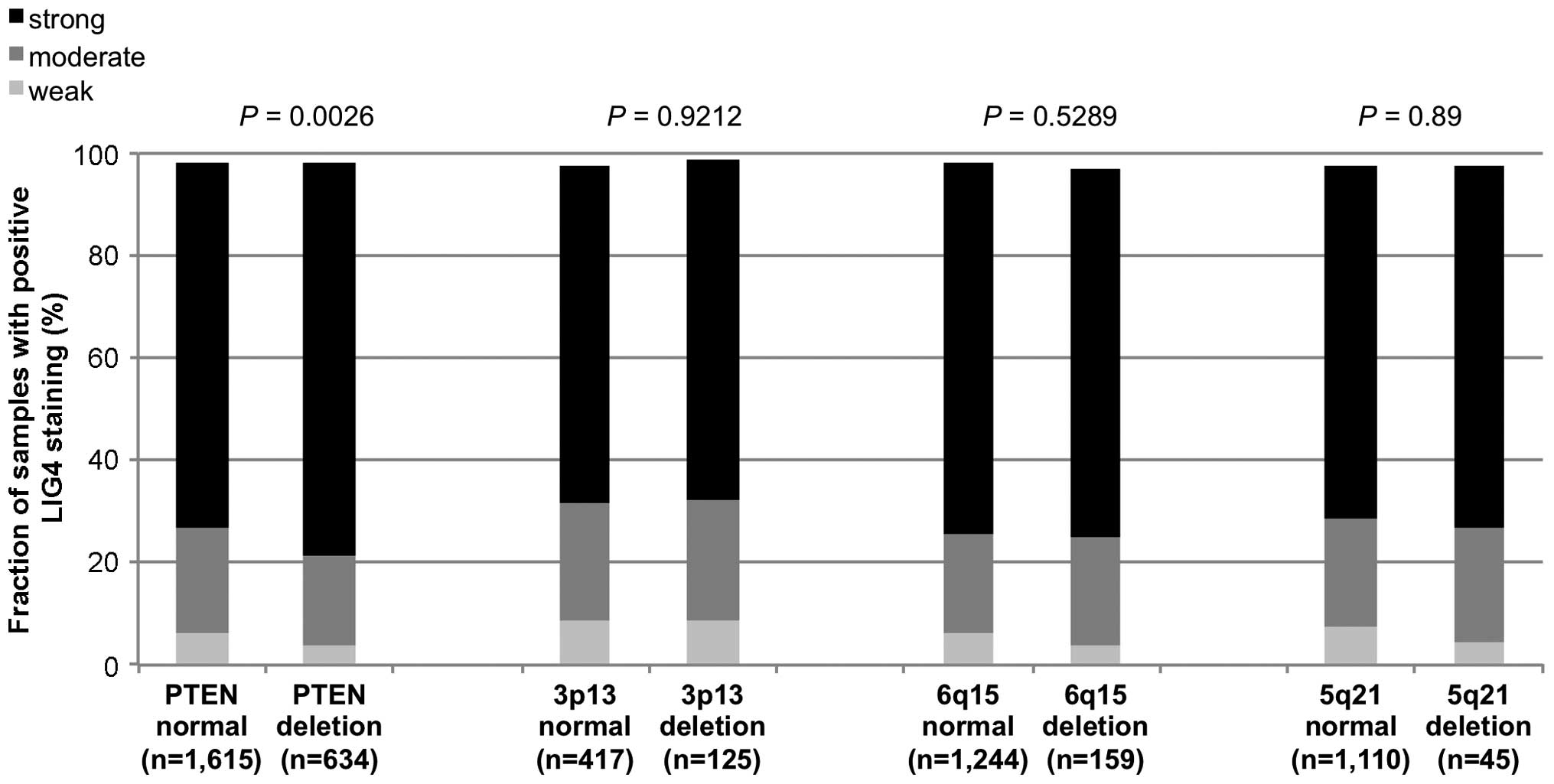
Associations with genomic deletions in
non-fusion and fusion-type prostate cancers
Previous studies provided evidence for distinct
molecular subgroups of prostate cancers defined by
TMPRSS2:ERG fusions and several genomic deletions. We and
others have described a strong link of deletions at 5q21 and 6q15
to non-fusion type and of deletions at PTEN and 3p13 to
fusion-type prostate cancers (25–27,29–31).
To study, whether LIG4 expression may be particularly linked to one
of these genomic deletions, LIG4 data were compared with
pre-existing findings on PTEN, 3p13, 6q15 and 5q21
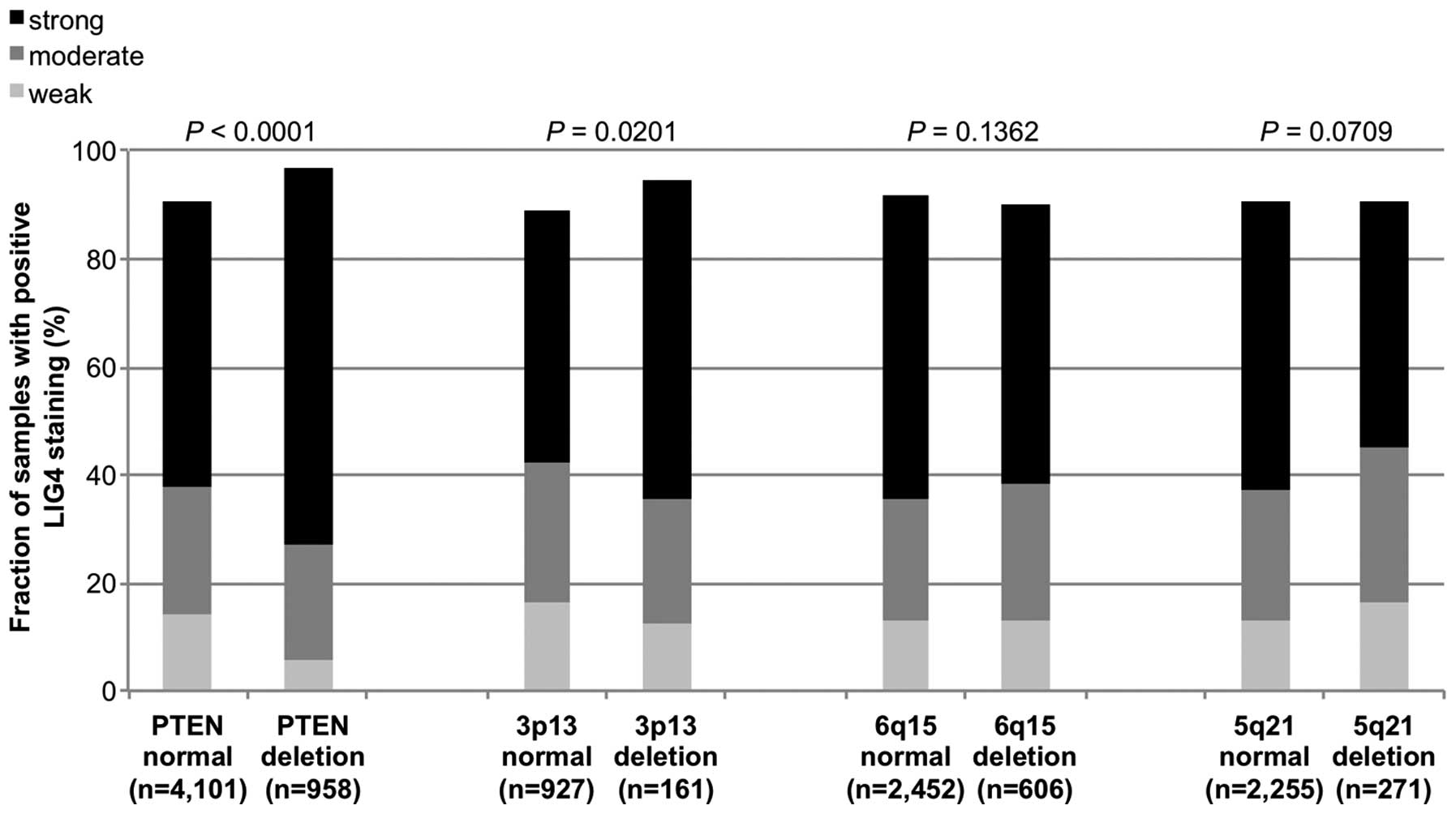
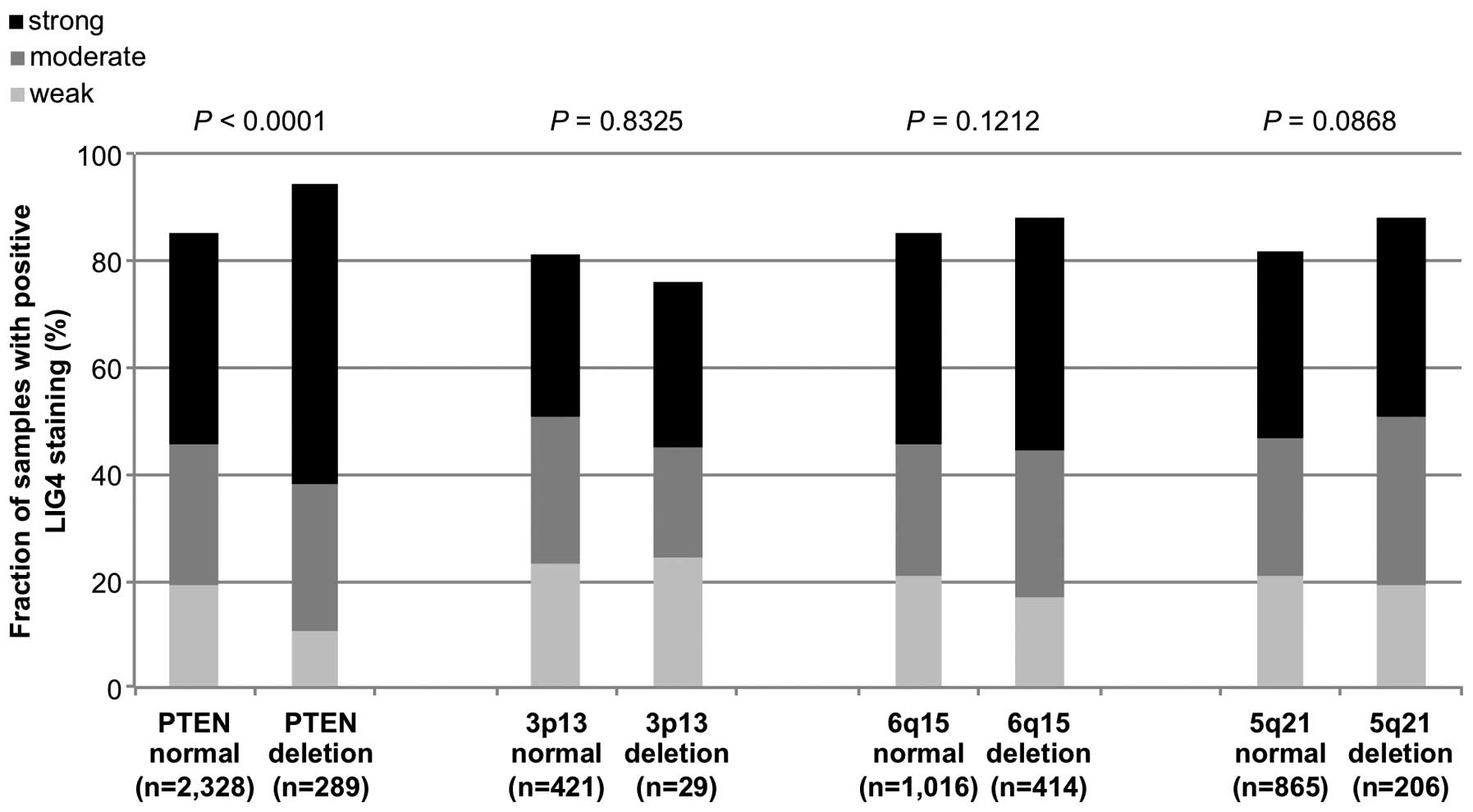
deletions. In the analysis of all tumors, LIG4 staining was
significantly associated with deletions of PTEN and 3p13
(P<0.0001 and P=0.02, Fig. 3). A
subsequent subgroup analysis of non-fusion and fusion-type prostate
cancers revealed that the significant association of increased LIG4
expression with PTEN deletions retained significance in both
non-fusion and fusion-type prostate cancers (P<0.0001 and
P=0.003, Figs. 4 and 5) while all other deletions were unrelated
or only marginally related to LIG4 expression.
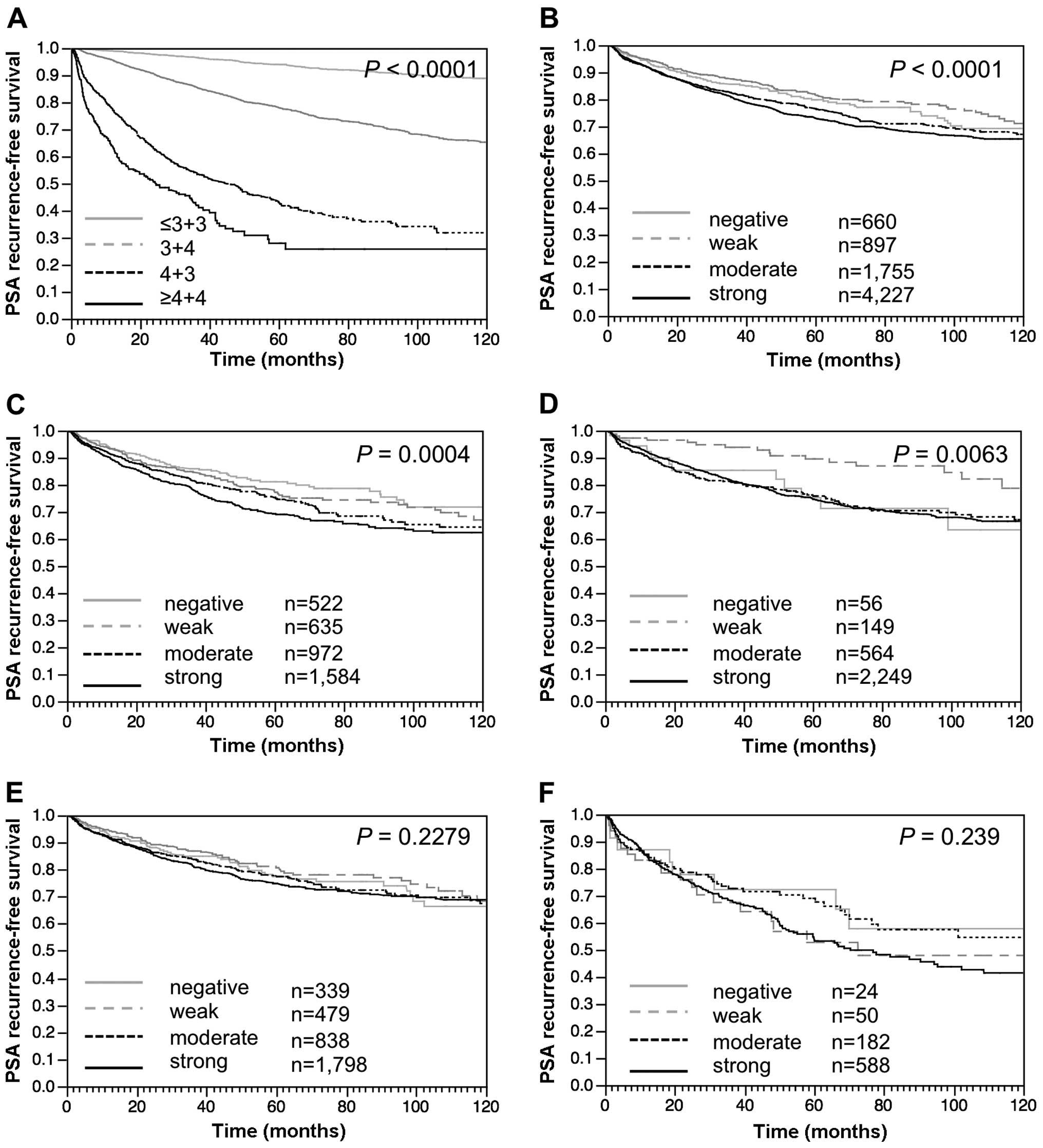
Clinical impact of LIG4
Follow-up data were available for 7,539 patients
with informative LIG4 data. Strong prognostic impact of the Gleason
grade provides indirect evidence for the overall validity of our
follow-up data (P<0.0001, Fig.
6A). Increased LIG4 staining was tightly linked to early
biochemical recurrence when all cancers (P<0.0001, Fig. 6B) or the subset of ERG-negative
(P=0.0004, Fig. 6C) and
ERG-positive (P=0.006, Fig. 6D)
prostate cancers were analyzed. Given the strong link of LIG4
expression to PTEN deletions which are known to be
prognostically relevant (27),
further analysis was performed according to the PTEN
deletion status. These results demonstrated that LIG4 expression
lost its prognostic impact in both subgroups of
PTEN-non-deleted (P=0.23, Fig.
6E) and PTEN-deleted prostate cancers (P=0.24, Fig. 6F).
Multivariate analysis
Four independent multivariate analyses were
performed evaluating the clinical relevance of LIG4 expression in
different scenarios (Table IV).
Scenario 1 was utilizing all post-operatively available parameters
including pT, pN, margin status, preoperative PSA value and Gleason
grade obtained on the resected prostate. Scenario 2 was utilizing
all postoperatively available parameters with the exception of
nodal status. The rational for this approach was that
lymphadenectomy is not a routine procedure in the surgical therapy
of prostate cancer and that excluding pN in multivariate analysis
increases case numbers. The next 2 scenarios tried to better model
the preoperative situation. Scenario 3 included the LIG4
expression, preoperative PSA, clinical stage (cT) and the Gleason
grade obtained on the prostatectomy specimen. However, the
preoperative determination of the Gleason grade in tumors is
subject to sampling errors and therefore results in under grading
in more than one third of cases. Because the postoperative Gleason
grade thus varies from the preoperative Gleason grade, another
multivariate analysis was added as scenario 4. In this scenario,
the preoperative Gleason grade obtained on the original biopsy was
combined with preoperative PSA, clinical stage and LIG4 expression.
The analyses demonstrate a tendency towards independent prognostic
relevance of LIG4 expression in 'preoperative' scenarios,
especially in ERG-fusion-positive prostate cancers (Table IV).
 | Table IVMultivariate analysis including LIG4
expression status in all prostate cancers, the ERG-negative and
ERG-positive subset. |
Table IV
Multivariate analysis including LIG4
expression status in all prostate cancers, the ERG-negative and
ERG-positive subset.
| Group | Scenario | P-value
|
|---|
| Evaluable (N) | Preoperative PSA
level | pT stage | cT stage | Gleason grade
prostatectomy | Gleason grade
biopsy | pN stage | R status | LIG4
expression |
|---|
| All |
| 1 | 4,446 | <0.0001 | <0.0001 | – | <0.0001 | – | <0.0001 | <0.0001 | 0.25 |
| 2 | 7,364 | <0.0001 | <0.0001 | – | <0.0001 | – | – | <0.0001 | 0.03 |
| 3 | 7,236 | <0.0001 | – | <0.0001 | <0.0001 | – | – | – | 0.04 |
| 4 | 7,131 | <0.0001 | – | <0.0001 | – | <0.0001 | – | – | 0.03 |
| ERG-negative
subset |
| 1 | 2,248 | <0.0001 | <0.0001 | – | <0.0001 | – | <0.0001 | 0.01 | 0.8 |
| 2 | 3,626 | <0.0001 | <0.0001 | – | <0.0001 | – | – | <0.0001 | 0.65 |
| 3 | 3,592 | <0.0001 | – | 0.0002 | <0.0001 | – | – | – | 0.42 |
| 4 | 3,543 | <0.0001 | – | <0.0001 | – | <0.0001 | – | – | 0.12 |
| ERG-positive
subset |
| 1 | 1,802 | 0.0301 | <0.0001 | – | <0.0001 | – | 0.07 | 0.006 | 0.21 |
| 2 | 2,949 | 0.0009 | <0.0001 | – | <0.0001 | – | – | <0.0001 | 0.01 |
| 3 | 2,868 | <0.0001 | – | <0.0001 | <0.0001 | – | – | – | 0.02 |
| 4 | 2,821 | <0.0001 | – | <0.0001 | – | <0.0001 | – | | 0.007 |
Discussion
Our study identified increased LIG4 as a predictor
of an increased risk for early PSA recurrence in prostate
cancer.
The successful immunohistochemical analysis of
>10,000 prostate cancers revealed that LIG4 is abundantly
present in prostate cancer as it was detectable in >90% of all
cancers using the IHC protocol. Our observation of abundant LIG4
expression is consistent with its essential role for survival of
dividing cells. Impaired NHEJ, for example by loss of LIG4, is
embryonic lethal (32–34). Since DSB is an inherent side effect
of DNA replication, it occurs most frequently in rapidly dividing
cells such as developing tissues or cancer cells (35), but can also be induced
replication-independently by oxidative stress or exogenous noxes
like ionizing radiation (36).
Increased LIG4 staining intensities in cancerous compared to benign
prostate epithelium is consistent with the generally increased
proliferation rate of prostate cancer cells as compared to normal
prostatic glands.
It is a distinct advantage of the tissue microarray
technique, that hundreds or thousands of tissue samples can be
analyzed in one day in one experiment by using one set of reagents
at absolutely identical concentrations, temperatures and exposure
times. Maximal experimental standardization is a prerequisite for
the distinction of subtle expression differences of proteins that
are abundantly expressed in prostate cancer such as LIG4. The
parallel analysis of thousands of tumors typically enables the
identification of differences between subgroups although the
immunohistochemical analysis of a number of individual samples may
be impaired by either too extensive or inefficient formalin
fixation. The comparison between different LIG4 staining levels and
tumor phenotype or PSA recurrence revealed significant associations
in our study. Particularly, there was a significant association of
strong LIG4 expression with early biochemical recurrence.
Another major aim of our study was to analyze the
relationship with key genomic alterations in prostate cancer,
including gene fusions and chromosomal deletions, all of which are
potentially due to or facilitated by impaired DNA repair
mechanisms. About half of prostate cancers carry gene fusions
linking the androgen-regulated gene TMPRSS2 with
transcription factors of the ETS family (37). In consequence of the most frequent
of these rearrangements, the TMPRSS2:ERG fusion, the
expression of ERG becomes androgen-regulated and ERG massively
overexpressed. Our data demonstrate that strong LIG4 immunostaining
is significantly associated with fusion-type prostate cancer. High
LIG4 expression was almost twice as frequent in 'fusion-type' than
in 'non-fusion-type' prostate cancers. Finding this association by
two independent approaches for ERG fusion detection (IHC/FISH)
largely excludes a false positive association due to inefficient
expression for both LIG4 and ERG in a subset of damaged
non-reactive tissues.
TMPRSS2:ERG fusion is caused by either
translocation or deletion of a large (3.7 Mb) chromosome fragment
separating the TMPRSS2 and ERG gene loci. Androgen
signaling, which promotes recruitment of androgen receptor (AR) and
topoisomerase II β (TOP2B) to sites of TMPRSS2:ERG
breakpoints, has been identified as the underlying trigger driving
fusion (38). TOP2B resolves DNA
topologic constraints eventually occurring during DNA movement by a
transient DSB allowing for passing one DNA strand through another
(39). As a consequence, components
of the DSB breakage repair machinery are required for repair of
these transient DSB (40), with
LIG4 catalyzing the final step of ligation of the open DNA ends
(41). The strong association
between ERG fusion and LIG4 overexpression in our study thus
reflects the constitutive activation of the DSB repair machinery in
cancers with elevated androgen signaling activity.
Of note, structural rearrangements are not limited
to ERG fusion-positive cancers but do also occur in
ERG-negative tumors. A number of large interstitial
deletions have been identified, some of which can be linked to
ERG-positive (including for example deletions at 3p13 and
the PTEN locus at 10q23), or ERG-negative cancers
(for example deletions at 5q21, 6q15 or 2q23) (21,25–27,31,42,43).
Likewise, balanced as well as imbalanced translocations have been
detected in both ERG-positive and ERG-negative tumors
(29,20). Since interstitial deletions, like
translocations, can be caused by DSB (44), it was not surprising that also 40%
of ERG-negative cancers showed LIG4 expression indicating
activated DSB repair. The overall lack of a clear-cut association
between presence of deletions and LIG4 expression, however,
indicates that many deletions might also occur for other reasons
than topoisomerase-driven DSB. For example, deletions may also
develop by DNA breakage during chromosome segregation in dividing
cells. Berger et al (29)
reported multiple fusions connecting segments from different
chromosomes to 'closed chains'. Such fused chromosomes will
inevitably break randomly if the kinetochores are pulled into the
daughter cells by the microtubules of the spindle cell apparatus.
Also, complex rearrangements have been reported from dicentromeric
chromosomes, which undergo breakage during cell division, leaving
one daughter cell with a deletion and the other with a duplication
and inversion (45).
Deletion of PTEN was the only aberration
linked to strong LIG4 expression independently from the ERG
status. This suggests that PTEN deletion may frequently
develop through topoisomerase-mediated DSB and subsequent LIG4
recruitment. PTEN deletion is typically small, often
involving only the gene locus and small (<2 Mb) adjacent
segments (unpublished data). This is in contrast to almost all
other deletions found in prostate cancer, which are typically large
and encompass >10 Mb (31). It
is possible, that PTEN deletion is driven by a similar,
topoisomerase and LIG4-dependent DSB mechanism than ERG
fusion. The strong association between PTEN deletion and
LIG4 may, thus be attributable, to a specific mechanism requiring
increased levels of LIG4.
Interestingly, LIG4 overexpression was equally
linked to early PSA recurrence in ERG-positive and ERG-negative
cancers. This association was not surprising given that LIG4
expression indicates active DSB repair. It can be assumed that
tumors with increased repair activity have a high number of DSB,
and thus a high likelihood of repair errors eventually resulting in
alterations of cancer relevant genes and overall genetic
instability. It has been shown before that increased genetic
instability is linked to poor outcome in many solid cancer types
(46,47).
Our TMA containing >10,000 prostate cancer
specimens represents a suitable system for assessing potential
prognostic markers. In earlier studies we successfully validated
all established prognostic biomarkers in prostate cancer such as
nuclear p53 accumulation (48) and
Ki67 labeling index (49) on
smaller prostate cancer TMAs, and identified several other
prognostic biomarkers such as deletions at 8p (50), 6q15 and PTEN (27) or CRISP3 expression (51). It is noteworthy, that our approach
of analyzing molecular features on one-minute tissue specimen per
patient on a TMA measuring 0.6 mm in diameter represents a close
model of molecularly analyzing core needle biopsies. Core needle
biopsies enable the molecular analysis of comparable amounts of
tissue as on a TMA. The optimal biomarker evaluation strategy would
include the molecular analysis of the original needle biopsy of a
patient and compare its prognostic value with preoperative Gleason
grade obtained on the same biopsy as well as the preoperative PSA
value. For practical purposes, this approach is not feasible
because preoperative biopsies are typically distributed over many
different centers and not available for studies. Moreover, even if
available, these precious core needle biopsies would be exhausted
after only few studies. A convoluted approach evaluating multiple
different scenarios was thus utilized in this study. Overall, these
data suggest a prognostic relevance of LIG4 expression in prostate
cancer that may be independent of clinical and histopathological
features available in a preoperative situation.
In summary, the significant link of increased LIG4
levels with aggressiveness in a series of >11,000 prostate
cancers suggests that LIG4 analysis may prove instrumental as a
prognostic biomarker either alone or in combination with other
factors. Morover, our data demonstrate, that balanced
translocations and gene fusions like TMPRSS2:ERG are
strongly linked to LIG4-dependend DSB repair, whereas 'simple'
deletions may often arise through DSB/LIG4 independent
mechanisms.
Acknowledgments
We thank Julia Schumann, Sünje Seekamp and Inge
Brandt for the excellent technical assistance.
Abbreviations:
|
DSB
|
double-strand breaks
|
|
HR
|
homologous recombinations
|
|
NHEJ
|
non-homologous DNA end-joining
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Galanty Y, Belotserkovskaya R, Coates J,
Polo S, Miller KM and Jackson SP: Mammalian SUMO E3-ligases PIAS1
and PIAS4 promote responses to DNA double-strand breaks. Nature.
462:935–939. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferguson DO, Sekiguchi JM, Chang S, Frank
KM, Gao Y, DePinho RA and Alt FW: The nonhomologous end-joining
pathway of DNA repair is required for genomic stability and the
suppression of translocations. Proc Natl Acad Sci USA.
97:6630–6633. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vanasse GJ, Halbrook J, Thomas S, Burgess
A, Hoekstra MF, Disteche CM and Willerford DM: Genetic pathsway to
recurrent chromosome translocations in murine lymphoma involves
V(D)J recombinase. J Clin Invest. 103:1669–1675. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gao N, Keane MJ, Ong T and Wallace WE:
Effects of simulated pulmonary surfactant on the cytotoxicity and
DNA-damaging activity of respirable quartz and kaolin. J Toxicol
Environ Health A. 60:153–167. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nussenzweig A, Chen C, da Costa Soares V,
Sanchez M, Sokol K, Nussenzweig MC and Li GC: Requirement for Ku80
in growth and immunoglobulin V(D)J recombination. Nature.
382:551–555. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu C, Bogue MA and Roth DB: Thymocyte
differentiation in gamma-irradiated severe-combined immunodeficient
mice: Characterization of intermediates and products of V(D)J
recombination at the T cell receptor alpha locus. Eur J Immunol.
26:2859–2865. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gu Y, Jin S, Gao Y, Weaver DT and Alt FW:
Ku70-deficient embryonic stem cells have increased ionizing
radiosensitivity, defective DNA end-binding activity, and inability
to support V(D)J recombination. Proc Natl Acad Sci USA.
94:8076–8081. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ouyang H, Nussenzweig A, Kurimasa A,
Soares VC, Li X, Cordon-Cardo C, Li W, Cheong N, Nussenzweig M,
Iliakis G, et al: Ku70 is required for DNA repair but not for T
cell antigen receptor gene recombination in vivo. J Exp Med.
186:921–929. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Garcia AM, Salomon RN, Witsell A,
Liepkalns J, Calder RB, Lee M, Lundell M, Vijg J and McVey M: Loss
of the bloom syndrome helicase increases DNA ligase 4-independent
genome rearrangements and tumorigenesis in aging Drosophila. Genome
Biol. 12:R1212011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kapusta A, Matsuda A, Marmignon A, Ku M,
Silve A, Meyer E, Forney JD, Malinsky S and Bétermier M: Highly
precise and developmentally programmed genome assembly in
Paramecium requires ligase IV-dependent end joining. PLoS Genet.
7:e10020492011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chistiakov DA, Voronova NV and Chistiakov
PA: Genetic variations in DNA repair genes, radiosensitivity to
cancer and susceptibility to acute tissue reactions in
radiotherapy-treated cancer patients. Acta Oncol. 47:809–824. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Roddam PL, Rollinson S, O'Driscoll M,
Jeggo PA, Jack A and Morgan GJ: Genetic variants of NHEJ DNA ligase
IV can affect the risk of developing multiple myeloma, a tumour
characterised by aberrant class switch recombination. J Med Genet.
39:900–905. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Y, Shete S, Etzel CJ, Scheurer M,
Alexiou G, Armstrong G, Tsavachidis S, Liang FW, Gilbert M, Aldape
K, et al: Polymorphisms of LIG4, BTBD2, HMGA2, and RTEL1 genes
involved in the double-strand break repair pathway predict
glioblastoma survival. J Clin Oncol. 28:2467–2474. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kuschel B, Auranen A, McBride S, Novik KL,
Antoniou A, Lipscombe JM, Day NE, Easton DF, Ponder BA, Pharoah PD,
et al: Variants in DNA double-strand break repair genes and breast
cancer susceptibility. Hum Mol Genet. 11:1399–1407. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sakiyama T, Kohno T, Mimaki S, Ohta T,
Yanagitani N, Sobue T, Kunitoh H, Saito R, Shimizu K, Hirama C, et
al: Association of amino acid substitution polymorphisms in DNA
repair genes TP53, POLI, REV1 and LIG4 with lung cancer risk. Int J
Cancer. 114:730–737. 2005. View Article : Google Scholar
|
|
17
|
Li R, Li Y, Fang X, Yang H and Wang J,
Kristiansen K and Wang J: SNP detection for massively parallel
whole-genome resequencing. Genome Res. 19:1124–1132. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pugh TJ, Keyes M, Barclay L, Delaney A,
Krzywinski M, Thomas D, Novik K, Yang C, Agranovich A, McKenzie M,
et al: Sequence variant discovery in DNA repair genes from
radio-sensitive and radiotolerant prostate brachytherapy patients.
Clin Cancer Res. 15:5008–5016. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Damaraju S, Murray D, Dufour J, Carandang
D, Myrehaug S, Fallone G, Field C, Greiner R, Hanson J, Cass CE, et
al: Association of DNA repair and steroid metabolism gene
polymorphisms with clinical late toxicity in patients treated with
conformal radiotherapy for prostate cancer. Clin Cancer Res.
12:2545–2554. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weischenfeldt J, Simon R, Feuerbach L,
Schlangen K, Weichenhan D, Minner S, Wuttig D, Warnatz HJ, Stehr H,
Rausch T, et al: Integrative genomic analyses reveal an
androgen-driven somatic alteration landscape in early-onset
prostate cancer. Cancer Cell. 23:159–170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barbieri CE, Baca SC, Lawrence MS,
Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van
Allen E, Stransky N, et al: Exome sequencing identifies recurrent
SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet.
44:685–689. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Erbersdobler A, Fritz H, Schnöger S,
Graefen M, Hammerer P, Huland H and Henke RP: Tumour grade,
proliferation, apoptosis, microvessel density, p53, and bcl-2 in
prostate cancers: Differences between tumours located in the
transition zone and in the peripheral zone. Eur Urol. 41:40–46.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mirlacher M and Simon R: Recipient block
TMA technique. Methods Mol Biol. 664:37–44. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Minner S, Enodien M, Sirma H, Luebke AM,
Krohn A, Mayer PS, Simon R, Tennstedt P, Müller J, Scholz L, et al:
ERG status is unrelated to PSA recurrence in radically operated
prostate cancer in the absence of antihormonal therapy. Clin Cancer
Res. 17:5878–5888. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Burkhardt L, Fuchs S, Krohn A, Masser S,
Mader M, Kluth M, Bachmann F, Huland H, Steuber T, Graefen M, et
al: CHD1 is a 5q21 tumor suppressor required for ERG rearrangement
in prostate cancer. Cancer Res. 73:2795–2805. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kluth M, Hesse J, Heinl A, Krohn A,
Steurer S, Sirma H, Simon R, Mayer PS, Schumacher U, Grupp K, et
al: Genomic deletion of MAP3K7 at 6q12-22 is associated with early
PSA recurrence in prostate cancer and absence of TMPRSS2:ERG
fusions. Mod Pathol. 26:975–983. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Krohn A, Diedler T, Burkhardt L, Mayer PS,
De Silva C, Meyer-Kornblum M, Kötschau D, Tennstedt P, Huang J,
Gerhäuser C, et al: Genomic deletion of PTEN is associated with
tumor progression and early PSA recurrence in ERG fusion-positive
and fusion-negative prostate cancer. Am J Pathol. 181:401–412.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krohn A, Seidel A, Burkhardt L, Bachmann
F, Mader M, Grupp K, Eichenauer T, Becker A, Adam M, Graefen M, et
al: Recurrent deletion of 3p13 targets multiple tumour suppressor
genes and defines a distinct subgroup of aggressive ERG
fusion-positive prostate cancers. J Pathol. 231:130–141. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Berger MF, Lawrence MS, Demichelis F,
Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger
D, Sougnez C, et al: The genomic complexity of primary human
prostate cancer. Nature. s470:214–220. 2011. View Article : Google Scholar
|
|
30
|
Lapointe J, Li C, Giacomini CP, Salari K,
Huang S, Wang P, Ferrari M, Hernandez-Boussard T, Brooks JD and
Pollack JR: Genomic profiling reveals alternative genetic pathways
of prostate tumorigenesis. Cancer Res. 67:8504–8510. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Taylor BS, Schultz N, Hieronymus H,
Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva
B, et al: Integrative genomic profiling of human prostate cancer.
Cancer Cell. 18:11–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Barnes DE, Stamp G, Rosewell I, Denzel A
and Lindahl T: Targeted disruption of the gene encoding DNA ligase
IV leads to lethality in embryonic mice. Curr Biol. 8:1395–1398.
1998. View Article : Google Scholar
|
|
33
|
Frank KM, Sekiguchi JM, Seidl KJ, Swat W,
Rathbun GA, Cheng HL, Davidson L, Kangaloo L and Alt FW: Late
embryonic lethality and impaired V(D)J recombination in mice
lacking DNA ligase IV. Nature. 396:173–177. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara
Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, et al: A
critical role for DNA end-joining proteins in both lymphogenesis
and neurogenesis. Cell. 95:891–902. 1998. View Article : Google Scholar
|
|
35
|
Gatz SA, Ju L, Gruber R, Hoffmann E, Carr
AM, Wang ZQ, Liu C and Jeggo PA: Requirement for DNA ligase IV
during embryonic neuronal development. J Neurosci. 31:10088–10100.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jeggo PA: Identification of genes involved
in repair of DNA double-strand breaks in mammalian cells. Radiat
Res. 150(Suppl 5): S80–S91. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tomlins SA, Rhodes DR, Perner S,
Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J,
Kuefer R, et al: Recurrent fusion of TMPRSS2 and ETS transcription
factor genes in prostate cancer. Science. 310:644–648. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Haffner MC, Aryee MJ, Toubaji A, Esopi DM,
Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, et al:
Androgen-induced TOP2B-mediated double-strand breaks and prostate
cancer gene rearrangements. Nat Genet. 42:668–675. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Deweese JE and Osheroff N: Coordinating
the two protomer active sites of human topoisomerase IIalpha: Nicks
as topoisomerase II poisons. Biochemistry. 48:1439–1441. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Haffner MC, De Marzo AM, Meeker AK, Nelson
WG and Yegnasubramanian S: Transcription-induced DNA double strand
breaks: Both oncogenic force and potential therapeutic target? Clin
Cancer Res. 17:3858–3864. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tomkinson AE and Mackey ZB: Structure and
function of mammalian DNA ligases. Mutat Res. 407:1–9. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Huang FC, Huang KF, Chen RH, Wu JE, Chen
TC, Chen CL, Lee CC, Chen JY, Lin JJ and Huang HS: Synthesis,
telomerase evaluation and anti-proliferative studies on various
series of diaminoanthraquinone-linked aminoacyl residue
derivatives. Arch Pharm (Weinheim). 345:101–111. 2012. View Article : Google Scholar
|
|
43
|
Liu W, Lindberg J, Sui G, Luo J, Egevad L,
Li T, Xie C, Wan M, Kim ST, Wang Z, et al: Identification of novel
CHD1-associated collaborative alterations of genomic structure and
functional assessment of CHD1 in prostate cancer. Oncogene.
31:3939–3948. 2012. View Article : Google Scholar
|
|
44
|
Varga T and Aplan PD: Chromosomal
aberrations induced by double strand DNA breaks. DNA Repair (Amst).
4:1038–1046. 2005. View Article : Google Scholar
|
|
45
|
Schlade-Bartusiak K, Tucker T, Safavi H,
Livingston J, van Allen MI, Eydoux P and Armstrong L: Independent
post-zygotic breaks of a dicentric chromosome result in mosaicism
for an inverted duplication deletion 9p and terminal deletion 9p.
Eur J Med Genet. 56:229–235. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Carter SL, Eklund AC, Kohane IS, Harris LN
and Szallasi Z: A signature of chromosomal instability inferred
from gene expression profiles predicts clinical outcome in multiple
human cancers. Nat Genet. 38:1043–1048. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Walther A, Houlston R and Tomlinson I:
Association between chromosomal instability and prognosis in
colorectal cancer: A meta-analysis. Gut. 57:941–950. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Schlomm T, Iwers L, Kirstein P, Jessen B,
Köllermann J, Minner S, Passow-Drolet A, Mirlacher M,
Milde-Langosch K, Graefen M, et al: Clinical significance of p53
alterations in surgically treated prostate cancers. Mod Pathol.
21:1371–1378. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bubendorf L, Sauter G, Moch H, Schmid HP,
Gasser TC, Jordan P and Mihatsch MJ: Ki67 labelling index: An
independent predictor of progression in prostate cancer treated by
radical prostatectomy. J Pathol. 178:437–441. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
El Gammal AT, Brüchmann M, Zustin J,
Isbarn H, Hellwinkel OJ, Köllermann J, Sauter G, Simon R, Wilczak
W, Schwarz J, et al: Chromosome 8p deletions and 8q gains are
associated with tumor progression and poor prognosis in prostate
cancer. Clin Cancer Res. 16:56–64. 2010. View Article : Google Scholar
|
|
51
|
Grupp K, Kohl S, Sirma H, Simon R, Steurer
S, Becker A, Adam M, Izbicki J, Sauter G, Minner S, et al:
Cysteine-rich secretory protein 3 overexpression is linked to a
subset of PTEN-deleted ERG fusion-positive prostate cancers with
early biochemical recurrence. Mod Pathol. 26:733–742. 2013.
View Article : Google Scholar
|