Introduction
Nasopharyngeal carcinoma (NPC) is the most common
head and neck malignancy with a unique geographical and ethnic
distribution, being particularly prevalent in Southern China and
Southeast Asia (1). As with most
other types of cancers, the prognosis for NPC is strongly
associated with the presenting stage (2). Although NPC is extremely
radiosensitive (3), metastasis is
found in 7% of patients at initial diagnosis and 20% or more
develop metastasis following treatment (4). Moreover, the appearance of local or
distant relapse determines a less favorable prognosis for these
patients (5). Chemotherapy is a
crucial treatment for late stage NPC patients; however, the regimen
is often not completed due to the severe side effects (6). Chinese traditional medicines have a
mild pharmaco logical action and have been utilized in tumor
therapy via naturally occurring drugs (7–10).
Cepharanthine hydrochloride (CH) is a biscoclaurine
(bisbenzylisoquinoline) amphipathic alkaloid that is isolated from
Stephania cepharantha Hayata (11). CH has several pharmacological
actions, including anti-inflammatory (12), anti-allergic (13) and immunomodulatory activities
(14) in vivo. In anticancer
investigations, CH exhibited multiple pharmacological actions,
including potentiating the effects of antitumor agents (15), inducing apoptosis (16,17)
and radiation sensitization (18,19),
and reversing the multidrug resistance (20–22).
However, to the best of our knowlege, no studies have focused on
the antitumor effects of CH on NPC.
In the present study, we initially investigated the
effects of CH in human NPC cell lines, including CNE-1 and CNE-2,
on cell growth and apoptosis in vitro, and demonstrated, to
the best of our knowledge, for the first time that, CH exerts a
potent anti-NPC effect by inhibiting cell growth and inducing
apoptosis. To gain a global and deep insight into the molecular
mechanisms for the anti-NPC action of CH, we performed cDNA
microarray analysis to identify differentially expressed genes in
response to CH in CNE-2 cells, which was confirmed by reverse
transcriptase-quantitative PCR (RT-qPCR).
Materials and methods
Cell culture and CH treatment
The human CNE-1 and CNE-2 NPC cell lines were grown
at 37°C in 5% CO2 atmosphere in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% fetal bovine serum
(both from Gibco, Grand Island, NY, USA), 100 μg/ml
penicillin and 100 U/ml streptomycin (Harbin Pharmaceutical Group
Co., Ltd., Harbin, China). CH was purchased from the College of
Life Science and Technology of Jinan University (Guangzhou, China)
and dissolved in a physiologic saline as a stock solution. CNE-1
and CNE-2 cells were treated with various concentrations (5, 10,
20, 40, 60 and 80 mg/l) of CH for the indicated durations (24, 48,
72 and 96 h). Control cells were treated with saline only.
MTT assay
CNE-1 and CNE-2 cells were seeded at a density of
2×104 cells/well in 96-well plates overnight and treated
with various concentrations of CH for different periods of time as
indicated. Then, 20 μl of 5 mg/ml of 3-(4,5-dimethyl
thiazol-2yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma, St.
Louis, MO, USA) was added to each of the culture wells, and the
plates were incubated for another 4 h. Following medium removal,
100 μl of DMSO was added to each well and plates were gently
agitated for 15 min to dissolve all crystals. Optical absorbance
was determined at 490 nm using an ELISA microplate reader (Tecan,
Männedorf, Swiss).
ATP-tumor chemosensitivity assay
Cells (100 ml) (2×104 cells/ml) were
seeded in 96-well plates for 24 h at 37°C with 5% CO2,
and treated with phosphate-buffered saline (PBS) as the control or
50% inhibiting concentration (IC50) of CH (12.5 and 20.0
mg/l for CNE-1 and CNE-2 cells, respectively) for 24–96 h. At the
end of the culture period, 50 ml with ATP extraction reagent (DCS
Innovative Diagnostik Systeme, Hamburg, Germany) was added to the
cells. The 100 μl aliquots of the lysates from each well
were transferred into the corresponding wells of the new 96-well
plates, and then 20 μl luciferin-luciferase reagent (DCS
Innovative Diagnostik Systeme) was added into each well. In
addition, we added 120 ml PBS in the blank well as the blank
control. The light output corresponding to the level of ATP present
was measured in a luminometer (Berthold Technologies, Bad Wildbad,
Germany) and the growth inhibition was calculated using the
formula: Growth inhibition percentage (%) = (Mc –
Mt)/(Mc – Mb) × 100, where
Mc, Mt and Mb stand for results of
the control, test and blank groups, respectively.
Cell cycle analysis
CNE-1 and CNE-2 cells were treated with saline or
the IC50 of CH for 48 h, and then collected by
centrifugation and rinsed twice with PBS. The cells were fixed in
70% ethanol and centrifuged again. The cells were stained with
propidium iodide solution (50 mg/l) containing 1 mg/l of RNase A
(both from Sigma) for 30 min at 37°C. After incubation, the stained
cells were rapidly analyzed using a FACScan flow cytometer (BD
Biosciences, San Jose, CA, USA).
Electron microscopic analysis of cell
apoptosis
Briefly, CNE-1 and CNE-2 cells, which were treated
with saline or the IC50 of CH for 48 h, were collected
and washed twice in 0.1 mol/l phosphate buffer (pH 7.4) by
centrifugation. The cell agglomerate of CNE-1 and CNE-2 cells were
immersed in 0.1 mol/l phosphate buffer (pH 7.4) with 2.5%
glutaraldehyde for 2 h at least, then post-fixed with 1% osmium
tetroxide in 0.1 mol/l phosphate buffer (pH 7.0) for 30 min. After
dehydration in a graded series of acetonum, the samples were
embedded in Epon 812 at room temperature overnight. Ultrathin
sections were cut, mounted on copper grids, and stained with uranyl
acetate and lead citrate using standard methods. Stained grids were
examined and photographed using a transmission electron microscope
(TEM) (Hitachi, Toyko, Japan).
cDNA microarray analysis
cDNA microarray construction
The cDNA microarray was manufactured by the Shanghai
Biochip Co., Ltd., China. Briefly, the human genes assessed in the
present study, contained 16,450 unigenes (including 10-positive
control and 6-negative control cDNAs), which were associated with
various cell functions and signaling pathways, involving cell
cycle, DNA repair, apoptosis, metabolism, cytoskeleton,
proinflammatory effect, signaling transduction, and transcription
factor.
RNA preparation, reverse
transcription, probe construction and hybridization
CNE-2 cells were exposed to saline or
IC50 CH for 24 h, and total RNA was extracted using a
Qiagen RNeasy® kit (Qiagen, Hilden, Germany) according
to the manufacturer's instructions. The quality of total RNA was
assessed by agarose gel electrophoresis.
During the reaction of cDNA, Cy3/Cy5-dUTP was
incorporated into the cDNA of the control and treatment sample,
respectively. Briefly, total 10 μl solution including 3
μg random primer and 50 μg of total RNA from
untreated and CH-treated cells was added into the PCR tube and
degenerated, respectively, at 70°C for 10 min. Then, 1.0 μl
Cy3/Cy5-dUTP (1 mM) (Amersham Biosciences, Piscataway, NJ, USA),
1.0 μl SuperScript II (200 U/μl) (Invitrogen-Life
Technologies, Carlsbad, CA, USA) were added into the tube to
construct the reverse transcription reaction system, which was
incubated for 2 h at 42°C, degenerated for 5 min at 70°C in the
dark and terminated by 2 μl NaOH (2.5 M). The probe was
purified according to the QIAquick Nucleotide Removal kit (Qiagen),
quantified and stored at −20°C in vacuum.
For hybridization, 30 pmol Cy3/Cy5 labeling probe
with a total volume up to 9 μl was added into the tube and
degenerated for 3 min at 94°C. Then, 2 μg human Cot-1 DNA
(Invitrogen-Life Technologies) was added into the tube and
incubated for 45 min at 70°C. The previously prepared solution was
mixed with 10 μl 4X hybridization solution and 20 μl
methylamine to a final volume of 40 μl. Hybridization was
titrated in microarray and carried out at 42°C for 16–18 h in the
dark and moist box. Following completion of hybridization, the
microarray was washed with 1X SSC/0.2% SDS wash buffer for 10 min
at 55°C, 0.1X SSC/0.2% SDS wash buffer for 10 min at 55°C twice,
0.1X SSC wash buffer for 5 min twice and ddH2O for 2 min
at room temperature, and was dried by centrifugation at 1,500 rpm
for 5 h.
Microarray data analysis
Microarray images from two-color fluorescent
hybridization were scanned with an Agilent scanner (Agilent
Technologies, Inc., Santa Clara, CA, USA). The scanning results
were analyzed using ImaGene software (BioDiscovery, Hawthorne, CA,
USA), and normalized using GeneSpring software; Silicon Genetics,
Redwood City, CA, USA). Fluorescent images were gridded to locate
the spot corresponding to each gene. Raw gene expression data were
generated for each gene. The fold-change of each probe was
calculated from the two dye-swap arrays, and the probes with
≥2-fold change in one assay and ≤0.5-fold in its dye-swap were
considered to have a statistically significant change in gene
expression.
Clustering analysis was carried out to classify the
differentially expressed genes based on the similarity of the
functions using GeneSpring software. The order can be observed
according to the functions of differentially expressed genes.
RT-qPCR
The same batches of total RNA from untreated and
CH-treated CNE-2 cells for cDNA microarray were utilized for the
synthesis of first-strand cDNA. The specific primer sequences and
the expected amplicon size for GAPDH, CDKN1A, NR4A1 and DAXX are
shown in Table I. RT-qPCR was
performed using a SYBR-Green reaction mixture in the ABI 7300
detection system (Applied Biosystems, Foster City, CA, USA). The
amplification program used was: one cycle at 50°C for 2 min and
95°C for 10 min; followed by 40 cycles at 95°C for 15 sec and 60°C
for 1 min. The gene expression data were normalized to the
endogenous control GAPDH. The relative gene expression levels were
measured according to the formula 2−ΔΔCt. The samples
were run in triplicate and repeated twice.
 | Table IThe primer sequences of each checked
gene and its PCR production length. |
Table I
The primer sequences of each checked
gene and its PCR production length.
| Gene name | Primer
sequence | Length (bp) |
|---|
| GAPDH | F:
TGACTTCAACAGCGACACCCA
R: CACCCTGTTGCTGTAGCCAAA | 121
(954–1,074) |
| CDKN1A | F:
CAGCGACCTTCCTCATCCA
R: GACTCCTTGTTCCGCTGCTAA | 71
(1,697–1,767) |
| NR4A1 | F:
CTCATATGCCACCCCATGTG
R: TGGAGTCATTCTGCAGCTCAA | 159
(2,207–2,165) |
| DAXX | F:
AAGAGCCCCATGTCCTCACTAC
R: CTCCCCTGAAGATCTGCTGATC | 81
(1,595–1,675) |
Statistics analysis
SPSS software version 18.0 (SPSS, Inc., Chicago, IL,
USA) was used for all statistical analyses by means of t-test,
factor analysis and one-factor analysis of variance. The data in
the tables and figures were publicized in the form of mean or mean
± standard deviation (SD). P<0.05 was considered to indicate a
statistically significant result.
Results
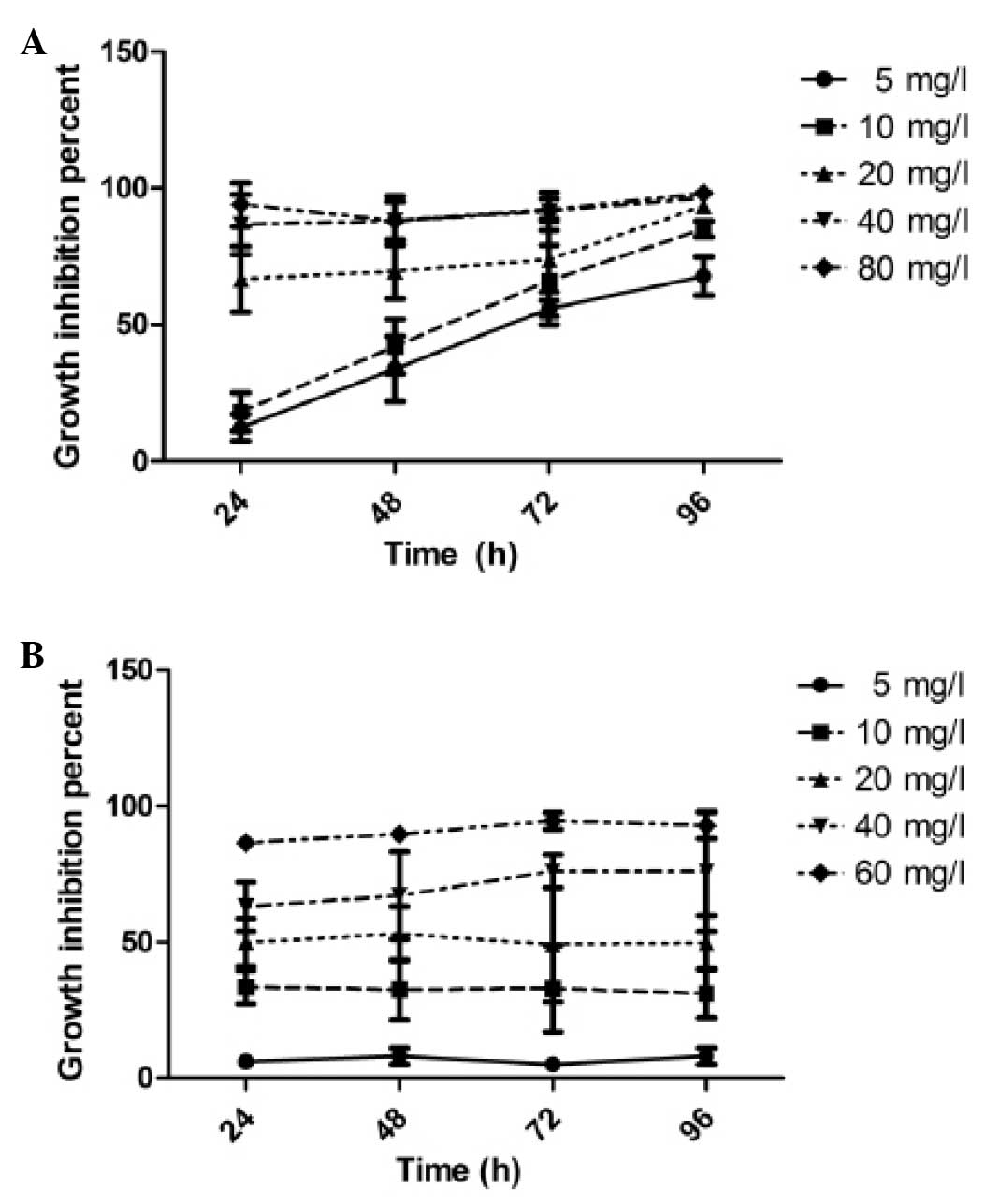
Effects of CH on cell growth
After CNE-1 and CNE-2 cells were exposed to CH at
various concentrations ranging from 5.0 to 80 mg/l for 24–96 h,
cell viability was determined using the MTT assay. We observed that
CNE-1 cells treated with CH presented a dose- and time-dependent
decrease in cell viability when compared with the untreated cells
(Fig. 1A). CNE-2 cells exhibited a
dose- but not time-dependent effect in response to CH ranging from
5.0 to 60.0 mg/l (Fig. 1B). The
median IC50 values of CH for CNE-1 and CNE-2 cells
calculated after 48 h treatment were 12.5 and 20.0 μg/ml,
respectively. To confirm the cytotoxic effect of CH, we performed
an ATP-tumor chemosensitivity assay to determine the inhibition
ratio (IR) of CNE-1 and CNE-2 cells in response to IC50
of CH for 24–96 h. The data (Table
II) were in accordance with the MTT results. These results
indicated that CH effectively inhibited CNE-1 and CNE-2 cell
growth.
 | Table IIThe inhibition ratio (IR) of CNE-1
and CNE-2 cells in response to IC50 of CH. |
Table II
The inhibition ratio (IR) of CNE-1
and CNE-2 cells in response to IC50 of CH.
| Cells | 24 h IR (%) | 48 h IR (%) | 72 h IR (%) | 96 h IR (%) |
|---|
| CNE-1 | 27.7±5.2 | 50.1±9.5 | 64.9±4.3 | 73.7±3.0 |
| CNE-2 | 44.5±4.6 | 50.9±2.9 | 55.0±4.8 | 54.4±11.4 |
Effects of CH on cell cycle
progression
To determine whether CH impaired the cell cycle of
CNE-1 and CNE-2 cells, flow cytometry was employed to examine the
distribution of the cell cycle. As shown in Table III, 48 h after treatment with
IC50 of CH apparently retarded CNE-1 and CNE-2 cells to
enter the S phase. This result clearly suggested that the growth
inhibitory effect of CH in CNE-1 and CNE-2 cells resulted from cell
cycle arrest in the G1 phase.
 | Table IIIThe cell cycle influence of
IC50 CH on CNE-1 and CNE-2 cells. |
Table III
The cell cycle influence of
IC50 CH on CNE-1 and CNE-2 cells.
| Groups | Cell cycle
| Proliferation index
(%) |
|---|
|
G0/G1 (%) | S (%) | G2/M
(%) |
|---|
| Control group of
CNE-1 | 42.0±12.7 | 46.0±10.1 | 12.0±2.6 | 58.00 |
| Experimental group
of CNE-1 | 70.9±10.7a | 22.9±7.9a | 6.1±2.1a | 29.03 |
| Control group of
CNE-2 | 40.3±6.7 | 45.7±3.8 | 13.9±3.0 | 59.66 |
| Experimental group
of CNE-2 | 74.4±5.7a | 18.2±6.2a | 7.3±2.2a | 25.53 |
Effects of CH on cell apoptosis
TEM, which is one of the best methods to observe
cell morphology including cytoplasm, organelle and nuclei, was used
to confirm apoptosis of CNE-1 and CNE-2 cells treated with PBS or
with IC50 of CH for 48 h. As shown in Fig. 2A and C, there were no typical
morphological changes indicative of apoptosis in the controls of
CNE-1 and CNE-2 cells. By contrast, when CNE-1 and CNE-2 cells were
exposed to IC50 of CH for 48 h, typical apoptotic
morphological changes were observed (Fig. 2B and D).
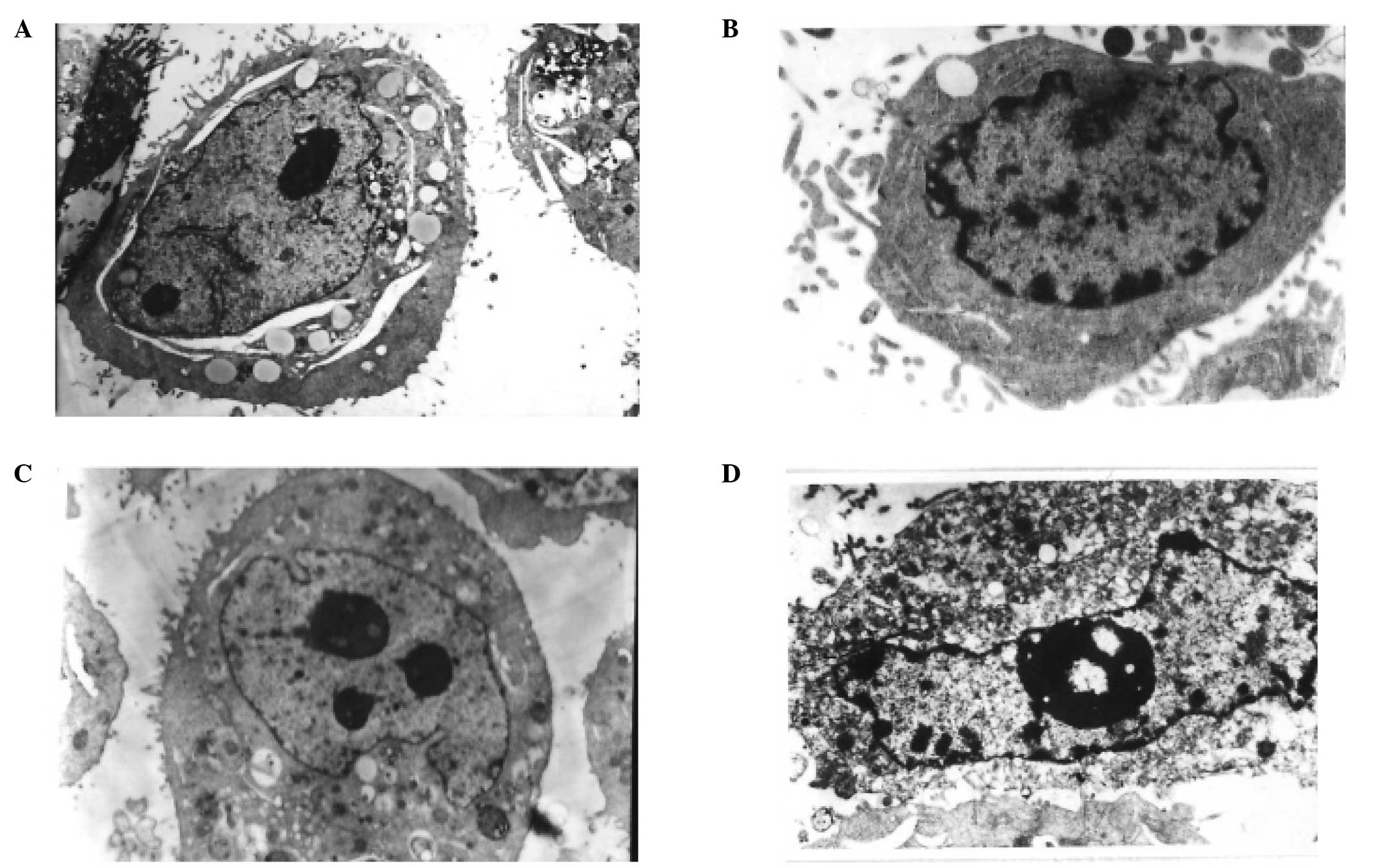 | Figure 2TEM micrographs of CNE-1 and CNE-2
cells treated (A and C) without and with (B and D) IC50
CH value for two days. (A and C) Untreated CNE-1 (magnification,
×5,000) and CNE-2 cells (magnification, ×5,000), showing typical
ultrastructure characterized by a well-preserved plasma membrane.
Abundant microvilli are visible on the surface, and the nucleus
became larger and contained a nucleolus and euchromatin. (B) Early
stage of apoptosis CNE-1 (magnification, ×5,000). Most organelles
in the cytoplasm lost some of their individual characteristics, few
microvilli are visible on the cell surface, and the main
ultra-microstructural changes are chromatin aggregation are visible
on the periphery of the nucleus. (D) Apoptotic characteristics of
CNE-2 (magnification, ×5,000). Few microvilli are visible on the
cell surface, with more extensive nuclear chromatin condensation,
fragmentation of nuclei and formation of apoptotic bodies being
observed. TEM, transmission electron microscope; CH, cepharanthine
hydrochiorid. |
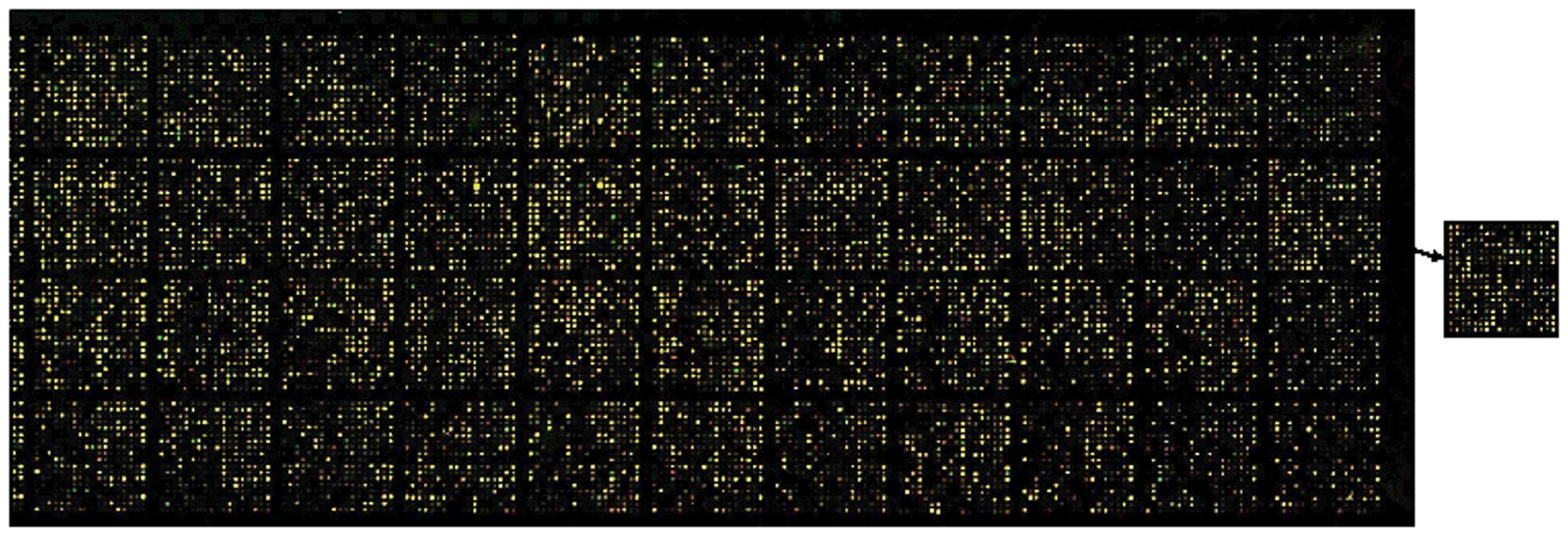
cDNA microarray profile of CNE-2 cells in
response to CH
To gain a global understanding of the
pharmacological mechanism of CH in NPC cells, the cDNA microarray
technique was used to establish the gene expression profiles of
CNE-2 cells untreated or treated with CH. Signals were invisible in
the blank spots, while the housekeeping gene density was similar,
indicating that the results were credible. Preliminary results
showed that the expression levels of 499 genes were significantly
altered by CH in CNE-2 cells (Fig.
3). After rejecting some complicated data, we found that there
were 144 upregulated and 63 downregulated genes in the CH-treated
CNE-2 cells. According to their attributes and functions in the
biological processes, these differentially expressed genes may be
classified as: i) cell cycle-related, ii) DNA repair-related, iii)
apoptosis-related genes, and iv) nuclear factor-κB (NF-κB)
transcription factors signal pathways. The segmental different
genes associated with growth inhibition are shown in Table IV.
 | Table IVThe segmental genes associated with
growth inhibition in different genes. |
Table IV
The segmental genes associated with
growth inhibition in different genes.
| Classification | Gene bank | Gene | Ratio |
|---|
| Cell cycle
association genes | BC014469 | CDKN2B |
2.071 |
| NM_000389 | CDKN1A |
3.374 |
| NM_000576 | IL1B |
4.933 |
| NM_000600 | IL6 |
8.1 |
| NM_002199 | IRF2 |
3.611 |
| NM_003244 | TGIF |
2.19 |
| NM_004235 | KLF4 |
2.659 |
| NM_006568 | CGRRF1 |
2.162 |
| X54489 | CXCLL | 26.42 |
| NM_002615 | SERPINF1 |
0.443 |
| NM_021147 | UNG2 |
0.452 |
| NM_001761 | CCNF |
0.391 |
| NM_002467 | MYC |
2.15 |
| NM_006190 | ORC2L |
3.187 |
| NM_003842 | TNFRSF10B |
2.064 |
| L24498 | GADD45A | 12.36 |
| DNA repair
association genes | NM_006297 | XRCC1 |
0.476 |
| NM_021147 | UNG2 |
0.452 |
| NM_014071 | NCOA6 |
0.416 |
| NM_002878 | RAD51L3 |
0.471 |
| NM_002135 | NR4A1 |
3.238 |
| NM_001350 | DAXX |
0.467 |
| L24498 | GADD45A | 12.36 |
| NM_015675 | GADD45B |
4.892 |
| Cell apoptosis
association genes | BC014469 | CDKN2B |
2.071 |
| NM_000389 | CDKN1A |
3.374 |
| NM_003842 | TNFRSF10B |
2.064 |
| NM_004760 | STK17A |
2.702 |
| NM_004341 | CAD |
2.994 |
| NM_001964 | EGR1 |
2.012 |
| IκB/NF-κB signaling
pathway | NM_006290 | TNFAIP3 |
2.991 |
| NM_006622 | PLK2 |
4.665 |
| NM_018290 | NALP12 |
3.87 |
| NM_004760 | STK17A |
2.702 |
| NM_003244 | TGIF |
2.19 |
| AL133012 | CHUK |
2.097 |
| NM_024039 | MIS12 |
2.064 |
| Z57391 | HIP14L |
2.051 |
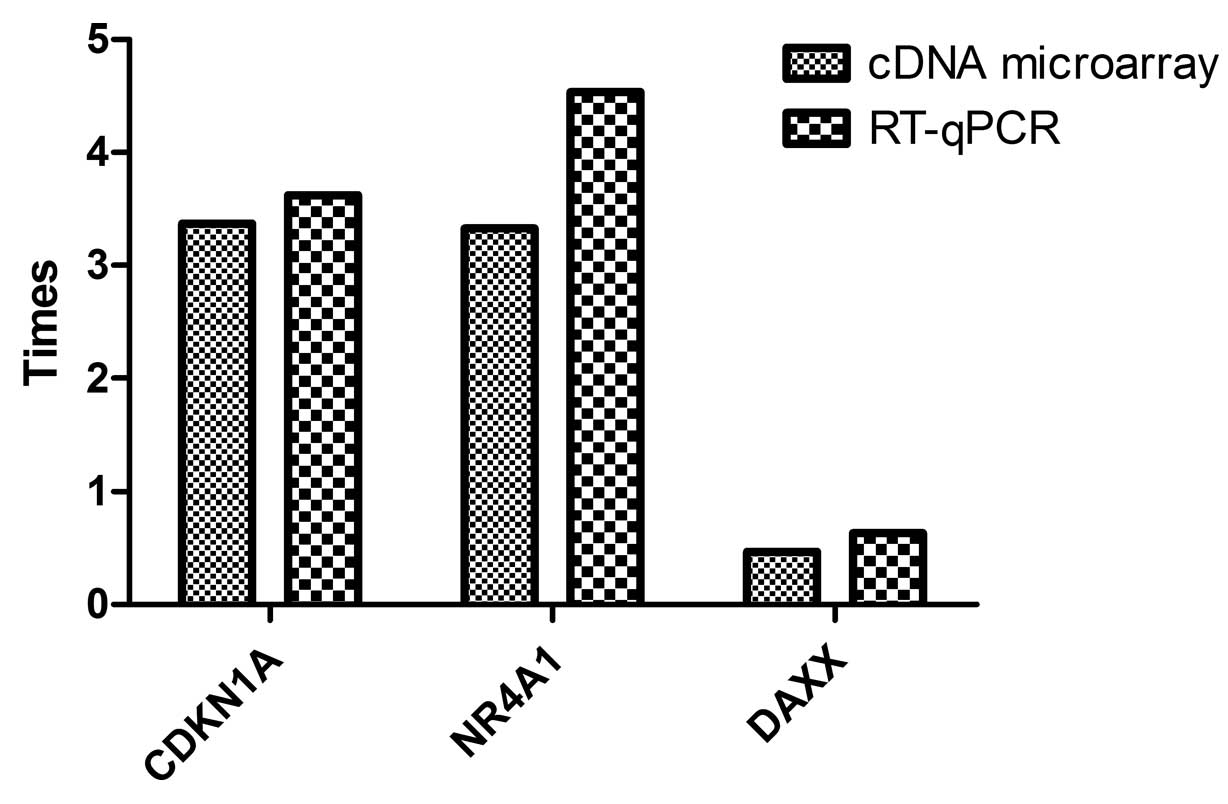
RT-qPCR analysis of several
differentially expressed genes
To verify the data gained from cDNA microarray,
three interesting genes, i.e., CDKN1A (NM_000389), NR4A1
(NM_002135) and DAXX (NM_001350) (function and mapping information
are presented in Table V), were
selected for RT-qPCR analysis. As shown in Table VI and Fig. 4, although some variability on the
individual gene expression changes existed, a consistent
differential expression trend was evident in the results from cDNA
microarray and those from RT-qPCR.
 | Table VFunction and mapping information of
the three genes selected for validation. |
Table V
Function and mapping information of
the three genes selected for validation.
| Gene name | Gene bank | Chromosome | Function |
|---|
| CDKN1A | NM_000389 | 6p21.2 | A potent
cyclin-dependent kinase inhibitor |
| NR4A1/TR3 | NM_002135 | 12q13 | A member of the
steroid/thyroid/retinoid nuclear receptor superfamily and
regulation of growth and apoptosis |
| DAXX | NM_001350 | 6p21.3 | Linking the death
receptor to the c-Jun amino-terminal kinase pathway |
 | Table VICt and ratio of each checked
gene. |
Table VI
Ct and ratio of each checked
gene.
| Genes | Control group
| Test group
| Times
(2−ΔΔCt) |
|---|
| Ct | ΔCt |
2−ΔCt | Ct | ΔCt |
2−ΔCt |
|---|
| GAPDH | 11.9584 | | | 11.9574 | | | |
| CDKN1A | 18.4303 | 6.4719 | 0.01126585 | 20.2867 | 8.3293 | 0.003109073 | 3.623540452 |
| NR4A1 | 18.8283 | 6.8699 | 0.008549762 | 21.0091 | 9.0517 | 0.001884373 | 4.537192908 |
| DAXX | 19.9025 | 7.9441 | 0.004060576 | 19.2424 | 7.285 | 0.006412044 | 0.63327323 |
Discussion
Nasopharyngeal carcinoma (NPC) is the most common
head and neck malignancy within southern China and southeast Asia.
Chemotherapy is crucial for late stage NPC patients. Nevertheless,
treatment is often not completed due to the serious side effects
caused (6). Chinese traditional
medicines, which possess an anticancer function have been utilized
in tumor therapy via naturally occurring drugs. Cepharanthine
hydrochloride (CH), which is known as a membrane-interacting agent
that has membrane-stabilizing activity, is a biscoclaurine alkaloid
extracted from the roots of Stephania cepharantha Hayata and
possesses plenty pharmacological activities (12–14).
Evidence suggests that CH potentiates the activity of certain
anticancer agents (15–17) and restores the effect of anticancer
drugs in multidrug/radioresistant cancer cells (18–22).
In the present study, we investigated whether CH affected CNE-1 and
CNE-2 cell growth. The results show that there was a dose- and
time-dependent decrease on the CNE-1 cell treated with CH, whereas
only dose-dependent relationships were identified with CNE-2. At
the same time, we assessed the effects of CH on the growth of CNE-1
and CNE-2 cells by means of an ATP-tumor chemosensitivity assay,
the results of which were consistent with those of the MTT assay.
Carcinomas are known to undergo an uncoordinated proliferation.
Thus, we carried out cell cycle analysis and cell apoptosis
detection. The results show that CH retarded the cell cycle and
promoted the apoptosis of CNE-1 and CNE-2 cells. Tumor
proliferation is associated with numerous growth regulatory genes
in a multistep process of carcinogenesis and neoplasm progression.
Gene expression is able to characterize the adaptation of cells to
changes in an external environment and is also a sensitive
indicator of toxicant exposure. To examine the intimate
pharmacological molecular mechanism of the anti-NPC of CH, using
cDNA microarray we identified that CH functions by cell
cycle-related genes such as CDKN1A, ID1, CCNF, UNG2 and CBX7; DNA
repaired-related genes such as XRCC1, RAD51L3 and NCOA6; and
apoptosis-related genes such as NR4A1, DAXX, GADD45α, TNFRSF10b and
DFFB. The results of RT-qPCR are consistent with the results of the
cDNA microarray in the expression of differential genes (including
CDKN1A, NR4A1 and DAXX) and show that the results of cDNA
microarray are credible.
Cell cycle association genes
The cell division cycle is an important factor
regulating cancer growth. In normal cells, the division cycle has
been divided into G1, S, G2 and the M phases.
Cells that do not divide are deemed to be in the G0
phase. When cells receive external stimulation to initiate
division, they changed from the quiescent state into the cell
division. A basic strategy when evaluating anticancer drugs is to
arrest the cell cycle. In the present study, we found that CH of
the 48 h IC50 arrested the CNE-1 and CNE-2 cells from
entering S phase and interfering with the proliferation of CNE-1
and CNE-2 cells when compared to the untreated cells. The cell
cycle control system includes the cyclin-dependent kinases (CDKs),
cyclins and cycle-dependent kinases inhibitors (CDKIs). The
interactions of CDKs and cyclins can induce cell division; however,
CDKIs inhibit the cell cycles. As shown in Table IV, we found upregulation of the
proliferation stimulator (for example CDKN1A and CDKN2B) and
downregulation of the proliferation inhibitor (for example ID1,
CCNF, UNG2 and CBX7) in our investigation plays a direct role in
CNE-1 cell proliferation following CH treatment.
p21, which is known as a cyclin-dependent kinase
inhibitor 1 or CDK-interacting protein 1, is a protein that is
encoded by the CDKN1A gene located on chromosome 6 (6p21.2)
in humans. The p21 protein binds to and inhibits the activity of
cyclin-CDK2 or -CDK4 complexes, and thus functions as a regulator
of cell cycle progression at G1 phase (22,23).
The expression of this gene is closely controlled by the tumor
suppressor protein p53, through which this protein mediates the
p53-dependent cell cycle G1 phase arrest in response to
a variety of stress stimuli (22,23).
In our experiment, we identified the overexpression of p21 by cDNA
microarray in NPC cells treated with CH. Thus, p21 inhibits the
proliferation of NPC cells treated with CH.
ID1 gene is a type of downregulation of the
proliferation inhibitor that was found by cDNA microarray. The ID1
gene encodes a type of positive regulator, a helix-loop-helix (HLH)
protein that forms heterodimers with members of the basic HLH
family of transcription factors and inhibits the DNA binding and
transcriptional activation ability of basic HLH proteins with which
it interacts. Evidence has shown that ID1 is able to promote cell
proliferation and cell cycle progression through the inactivation
of tumor suppressor pathways (24).
In addition, the overexpression of ID1 suppresses the expression of
p21 by means of the bond with its promoter-E box (25). The downregulation of ID1 is able to
block the proliferation of NPC cells treated with CH. The
mechanisms of p21 under the control of ID1 and E box may play a
vital role in the arrest of NPC cells treated with CH.
DNA repair association genes
In the process of cell division, the human stable
genome guarantees smooth cell proliferation. On the other hand,
unstable genome is characteristic of cancer cells and accelerates
broken and deformed chromosomes in the process of cancer cell
division. If genetic abnormalities cannot be repaired, cell cycle
restraint or apoptosis is triggered in the process of cancer cell
division. DNA repair-related genes are employed to correct such
drawbacks. In the present study, we found CH induces the
downregulation of DNA repair genes such as XRCC1, RAD51L3 and
NCOA6. The XRCC1 gene encoded a protein that is involved in the
efficient repair of DNA single-strand breaks formed by exposure to
ionizing radiation and alkylating agents. This protein interacts
with DNA ligase III, polymerase β and poly(ADP-ribose) polymerase
to participate in the DNA repair pathway (26). The RAD51L3 gene is another
downregulated gene derived from DNA repair genes. It encodes a
member protein of the RAD51 protein family that is known to be
involved in the homologous recombination and repair of DNA. This
protein forms a complex with several other members of the RAD51
family, including RAD51L1, RAD51L2 and XRCC2. The protein complex
formed with this protein has been shown to catalyze homologous
pairing between single- and double-stranded DNA, and is thought to
play a role in the early stage of recombinational repair of DNA
(27). We consider that
downregulation of the DNA repair genes such as XRCC1, RAD51L3 and
NCOA6, is associated with the functions of CH in NPC cells.
Following treatment of NPC cells with CH, upregulation of the
proliferation stimulator (for example CDKN1A and CDKN2B) and
downregulation of the proliferation inhibitor (for example ID1,
CCNF, UNG2 and CBX7) may play a direct role in the inhibition of
NPC cell proliferation and block NPC cell cycles by their signal
pathway. In the process of cell cycle arrest, the damaged DNA of
NPC cells induced by CH need to be repaired and escape the
checkpoint of cell cycles. However, the downregulation DNA repair
association genes inhibits DNA repair and proliferation, and cell
apoptosis association genes via CH induce NPC cell apoptosis.
Cell apoptosis association genes
The word 'apoptosis' comes form the Greek word
meaning 'falling leaves' and was first used to describe a new form
of cell death distinct from necrosis. Apoptosis is the morphologic
appearance of programmed cell death which is an important mechanism
in embryonic development, neurodegenerative diseases and
homeostasis (28). However, the
dysregulated apoptosis is a crucial step in tumorigenesis (28). Concerning cancer therapy, the
ultimate purpose of cytotoxic therapies is to induce apoptosis or
death of tumor cells. In the present study, typical apoptotic
morphological changes were observed in these cells when the cells
were exposed to IC50 CH for 48 h. This showed the
ability of CH to induce apoptosis. The process of apoptosis
involves numerous genes. We found CH induces the expression of
apoptotic genes (for example NR4A1, DAXX, GADD45α and
TNFRSF10b).
NR4A1 is a member of the steroid/thyroid/retinoid
nuclear receptor superfamily. During apoptosis, NR4A1 expression is
rapidly induced and plays roles in regulating growth and apoptosis
in cancer cells. NR4A1 binding to the Bcl-2 N-terminal loop region
and resulting in a conformational change in Bcl-2 can convert Bcl-2
from a protector to a killer protein (29). In the present study, we found that
the expression of NR4A1 in NPC cells treated with CH was 3.238- and
4.537-folds, respectively, compared with the control group in the
cDNA microarray test and RT-qPCR. The overexpression of NR4A1 plays
an important role in the apoptosis of NPC cells treated with
CH.
DAXX, which links the death receptor to the c-Jun
amino-terminal kinase pathway, is another protein associated with
apoptosis. DAXX negatively regulates apoptosis at the early
embryonic stages (30) and cancer
cells (31) via the p53 pathway.
The downregulation of DAXX induced by CH may be one of the pathways
leading to apoptosis in NPC cells.
GADD45, which responds to environmental stresses and
the anticancer activity of chemotherapeutic agents, is an
apoptosis-related gene mediated by p53 and the p53 homologues in
our study. After DNA is damaged, the GADD45 protein family members
are induced rapidly, participate actively in DNA repair mechanisms,
and result in cell cycle arrest and/or apoptosis (32). Evidence has also been provided that
drug therapies act to directly or indirectly upregulate GADD45α and
GADD45β and promote cancer cell apoptosis. In the present study, we
found that CH induces the upregulation of GADD45α and GADD45β in
NCP cells. As GADD45α and GADD45β are essential components of
numerous metabolic pathways that control proliferation and induce
cancer cell apoptosis, they may be regarded as emerging drug
targets in NPC cells treated with CH.
Nuclear factor-κB transcription factor
signaling pathways
Nuclear factor-κB (NF-κB) transcription factor
signaling pathways are central components of immune responses and
inflammatory. Evidence suggests that NF-κB signaling pathways that
are involved in its activation are also important for tumor
development. NF-κB is a negative regulator of cell proliferation
(33) and a positive activator of
anti-apoptotic genes (34). TNFAIP3
is a key negative regulator of the NF-κB signal, through a wide
variety of cell surface receptors and viral proteins. Previous
findings have shown that TNFAIP3 reduces cell proliferation
(35) and induces the apoptosis of
cancer treated with chemotherapeutics (36–38).
In the present study, similar results were obtained whereby TNFAIP3
was upregulated by CH, reduced proliferation and induced NPC cell
apoptosis.
In conclusion, to the best of our knowledge, we
report for the first time that CH inhibited cell proliferation and
induced apoptosis in NPC cells. The cDNA microarray analysis
revealed that the anti-NPC action of CH may be mediated by
regulating the expression levels of a variety of genes involved in
cell cycle regulation, DNA repair, cell apoptosis and the NF-κB
signaling pathway. Our findings provide a rational and scientific
basis for the further study and development of CH as a potentially
useful agent for NPC therapy, although more in-depth investigations
are required.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (grant no. 30660203), the Applied Basic
Special Research Program of Science and Technology Department of
Guangxi Zhuang Autonomous Region (grant no. 0731062), and
Innovation Program of Guangxi Graduate Education.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang J, Huang Y, Guan Z, Zhang JL, Su HK,
Zhang W, Yue CF, Yan M, Guan S and Liu QQ: E3-ligase Skp2 predicts
poor prognosis and maintains cancer stem cell pool in
nasopharyngeal carcinoma. Oncotarget. 5:5591–5601. 2014.PubMed/NCBI
|
|
3
|
Lin JC, Jan JS, Hsu CY, Liang WM, Jiang RS
and Wang WY: Phase III study of concurrent chemoradiotherapy versus
radiotherapy alone for advanced nasopharyngeal carcinoma: Positive
effect on overall and progression-free survival. J Clin Oncol.
21:631–637. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chang YH, Wu CC, Chang KP, Yu JS, Chang YC
and Liao PC: Cell secretome analysis using hollow fiber culture
system leads to the discovery of CLIC1 protein as a novel plasma
marker for nasopharyngeal carcinoma. J Proteome Res. 8:5465–5474.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee AW, Poon YF, Foo W, Law SC, Cheung FK,
Chan DK, Tung SY, Thaw M and Ho JH: Retrospective analysis of 5037
patients with nasopharyngeal carcinoma treated during 1976–1985:
Overall survival and patterns of failure. Int J Radiat Oncol Biol
Phys. 23:261–270. 1992. View Article : Google Scholar
|
|
6
|
Lin HX, Hua YJ, Chen QY, Luo DH, Sun R,
Qiu F, Mo HY, Mai HQ, Guo X, Xian LJ, et al: Randomized study of
sinusoidal chronomodulated versus flat intermittent induction
chemotherapy with cisplatin and 5-fluorouracil followed by
traditional radiotherapy for locoregionally advanced nasopharyngeal
carcinoma. Chin J Cancer. 32:502–511. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lo LC, Chen CY, Chen ST, Chen HC, Lee TC
and Chang CS: Therapeutic efficacy of traditional Chinese medicine,
Shen-Mai San, in cancer patients undergoing chemotherapy or
radiotherapy: Study protocol for a randomized, double-blind,
placebo-controlled trial. Trials. 13:2322012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li X, Chen T, Lin S, Zhao J, Chen P, Ba Q,
Guo H, Liu Y, Li J, Chu R, et al: Valeriana jatamansi constituent
IVHD-valtrate as a novel therapeutic agent to human ovarian cancer:
In vitro and in vivo activities and mechanisms. Curr Cancer Drug
Targets. 13:472–483. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hou X, Yuan X, Zhang B, Wang S and Chen Q:
Screening active anti-breast cancer compounds from Cortex Magnolia
officinalis by 2D LC-MS. J Sep Sci. 36:706–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su CC: Tanshinone IIA potentiates the
efficacy of 5-FU in Colo205 colon cancer cells in vivo through
downregulation of P-gp and LC3-II. Exp Ther Med. 3:555–559.
2012.PubMed/NCBI
|
|
11
|
Lv JJ, Xu M, Wang D, Zhu HT, Yang CR, Wang
YF, Li Y and Zhang YJ: Cytotoxic bisbenzylisoquinoline alkaloids
from Stephania epigaea. J Nat Prod. 76:926–932. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kudo K, Hagiwara S, Hasegawa A, Kusaka J,
Koga H and Noguchi T: Cepharanthine exerts anti-inflammatory
effects via NF-κB inhibition in a LPS-induced rat model of systemic
inflammation. J Surg Res. 171:199–204. 2011. View Article : Google Scholar
|
|
13
|
Goto M, Zeller WP and Hurley RM:
Cepharanthine (biscoclaurine alkaloid) treatment in endotoxic shock
of suckling rats. J Pharm Pharmacol. 43:589–591. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Furusawa S and Wu J: The effects of
biscoclaurine alkaloid cepharanthine on mammalian cells:
Implications for cancer, shock, and inflammatory diseases. Life
Sci. 80:1073–1079. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kono K, Takahashi JA, Ueba T, Mori H,
Hashimoto N and Fukumoto M: Effects of combination chemotherapy
with biscoclaurine-derived alkaloid (Cepharanthine) and nimustine
hydrochloride on malignant glioma cell lines. J Neurooncol.
56:101–108. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takahashi-Makise N, Suzu S, Hiyoshi M,
Ohsugi T, Katano H, Umezawa K and Okada S: Biscoclaurine alkaloid
cepharanthine inhibits the growth of primary effusion lymphoma in
vitro and in vivo and induces apoptosis via suppression of the
NF-kappaB pathway. Int J Cancer. 125:1464–1472. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Harada T, Harada K and Ueyama Y: The
enhancement of tumor radioresponse by combined treatment with
cepharanthine is accompanied by the inhibition of DNA damage repair
and the induction of apoptosis in oral squamous cell carcinoma. Int
J Oncol. 41:565–572. 2012.PubMed/NCBI
|
|
18
|
Tamatani T, Azuma M, Motegi K, Takamaru N,
Kawashima Y and Bando T: Cepharanthin-enhanced radiosensitivity
through the inhibition of radiation-induced nuclear factor-κB
activity in human oral squamous cell carcinoma cells. Int J Oncol.
31:761–768. 2007.PubMed/NCBI
|
|
19
|
Zahedi P, De Souza R, Huynh L,
Piquette-Miller M and Allen C: Combination drug delivery strategy
for the treatment of multidrug resistant ovarian cancer. Mol Pharm.
8:260–269. 2011. View Article : Google Scholar
|
|
20
|
Li H, Yan Z, Ning W, Xiao-Juan G, Cai-Hong
Z, Jin-Hua J, Fang M and Qing-Duan W: Using Rhodamine 123
accumulation in CD8+ cells as a surrogate indicator to
study the P-glycoprotein modulating effect of cepharanthine
hydrochloride in vivo. J Biomed Biotechnol. 2011:2816512011.
|
|
21
|
Peng YM, Wang N, Wang YF, Han L, Zhang Y,
Jiang JH, Zhou YB and Wang QD: Correlation between reversing effect
of cepharanthine hydrochloride on multidrug resistance and
P-glycoprotein expression and function of K562/ADR cells. Yao Xue
Xue Bao. 47:594–599. 2012.In Chinese. PubMed/NCBI
|
|
22
|
Abbas T and Dutta A: p21 in cancer:
Intricate networks and multiple activities. Nat Rev Cancer.
9:400–414. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu G and Lozano G: p21 stability: Linking
chaperones to a cell cycle checkpoint. Cancer Cell. 7:113–114.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Geng H, Rademacher BL, Pittsenbarger J,
Huang CY, Harvey CT, Lafortune MC, Myrthue A, Garzotto M, Nelson
PS, Beer TM, et al: ID1 enhances docetaxel cytotoxicity in prostate
cancer cells through inhibition of p21. Cancer Res. 70:3239–3248.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Prabhu S, Ignatova A, Park ST and Sun XH:
Regulation of the expression of cyclin-dependent kinase inhibitor
p21 by E2A and Id proteins. Mol Cell Biol. 17:5888–5896.
1997.PubMed/NCBI
|
|
26
|
Okano S, Lan L, Tomkinson AE and Yasui A:
Translocation of XRCC1 and DNA ligase IIIalpha from centrosomes to
chromosomes in response to DNA damage in mitotic human cells.
Nucleic Acids Res. 33:422–429. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Suwaki N, Klare K and Tarsounas M: RAD51
paralogs: Roles in DNA damage signalling, recombinational repair
and tumorigenesis. Semin Cell Dev Biol. 22:898–905. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fuchs Y and Steller H: Programmed cell
death in animal development and disease. Cell. 147:742–758. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin B, Kolluri SK, Lin F, Liu W, Han YH,
Cao X, Dawson MI, Reed JC and Zhang XK: Conversion of Bcl-2 from
protector to killer by interaction with nuclear orphan receptor
Nur77/TR3. Cell. 116:527–540. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Michaelson JS, Bader D, Kuo F, Kozak C and
Leder P: Loss of Daxx, a promiscuously interacting protein, results
in extensive apoptosis in early mouse development. Genes Dev.
13:1918–1923. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang J, Qu LK, Zhang J, Wang W, Michaelson
JS, Degenhardt YY, El-Deiry WS and Yang X: Critical role for Daxx
in regulating Mdm2. Nat Cell Biol. 8:855–862. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tamura RE, de Vasconcellos JF, Sarkar D,
Libermann TA, Fisher PB and Zerbini LF: GADD45 proteins: Central
players in tumorigenesis. Curr Mol Med. 12:634–651. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dajee M, Lazarov M, Zhang JY, Cai T, Green
CL, Russell AJ, Marinkovich MP, Tao S, Lin Q, Kubo Y, et al:
NF-kappaB blockade and oncogenic Ras trigger invasive human
epidermal neoplasia. Nature. 421:639–643. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Karin M and Lin A: NF-kappaB at the
crossroads of life and death. Nat Immunol. 3:221–227. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sakakibara S, Espigol-Frigole G, Gasperini
P, Uldrick TS, Yarchoan R and Tosato G: A20/TNFAIP3 inhibits NF-κB
activation induced by the Kaposi's sarcoma-associated herpesvirus
vFLIP oncoprotein. Oncogene. 32:1223–1232. 2013. View Article : Google Scholar :
|
|
36
|
Al-Romaih K, Somers GR, Bayani J, Hughes
S, Prasad M, Cutz JC, Xue H, Zielenska M, Wang Y and Squire JA:
Modulation by decitabine of gene expression and growth of
osteosarcoma U2OS cells in vitro and in xenografts: Identification
of apoptotic genes as targets for demethylation. Cancer Cell Int.
7:142007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Daigeler A, Chromik AM, Geisler A, Bulut
D, Hilgert C, Krieg A, Klein-Hitpass L, Lehnhardt M, Uhl W and
Mittelkötter U: Synergistic apoptotic effects of taurolidine and
TRAIL on squamous carcinoma cells of the esophagus. Int J Oncol.
32:1205–1220. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Daigeler A, Klein-Hitpass L, Chromik MA,
Müller O, Hauser J, Homann HH, Steinau HU and Lehnhardt M:
Heterogeneous in vitro effects of doxorubicin on gene expression in
primary human liposarcoma cultures. BMC Cancer. 8:3132008.
View Article : Google Scholar : PubMed/NCBI
|