Introduction
Patients with locally advanced rectal cancer are
routinely treated with preoperative chemoradiotherapy (CRT).
Randomized phase III trials have previously shown that preoperative
CRT significantly improves the local control of cancer as compared
with preoperative radiation alone or postoperative CRT (1–3). We
previously performed a phase I clinical study and determined the
recommended dose (80 mg/m2) of
tegafur/gimeracil/oteracil (S-1) for preoperative CRT (4). We then undertook a phase II clinical
study to evaluate the efficacy and toxicity of preoperative CRT
using tegafur-uracil (UFT; 300 mg/m2/day) vs. S-1 (80
mg/m2/day) in 59 patients with locally advanced rectal
cancer (5). This is the first study
to use S-1 as a single agent for CRT of locally advanced rectal
cancer. Using the Response Evaluation Criteria in Solid Tumors
(RECIST) guidelines, we found no statistically significant
difference in terms of the patient response rate (p=0.52) between
the S-1 and UFT groups, with no difference in the downstaging rate,
resection of tumor, sphincter preservation and marginal invasion.
The only significant finding was that the incidence of grade 3
diarrhea was significantly more frequent in the S-1 group (7%) as
compared with the UFT group (0%) (p=0.02).
Although this is an overall positive result, patient
responses to preoperative CRT differ between individuals. Some
studies have therefore sought to investigate the gene expression
profiles in patients with rectal cancer to predict their response
to preoperative CRT (6–9). However, as yet, there are no specific
biomarkers with which to predict these responses. Therefore, it is
becoming increasingly important to identify predictive biomarkers
of preoperative CRT.
Fluorouracil-based regimens have been generally used
for preoperative CRT in patients with rectal cancer and some
studies have reported the efficacy of oral fluorouracil pro-drugs,
such as UFT, S-1 and capecitabine (10–13).
However, the best agent for preoperative CRT in patients with
rectal cancer has not yet been determined.
Here, we employed cDNA and miRNA microarray analyses
to screen for biomarkers that could be useful in predicting the
responses of patients with rectal cancer to preoperative CRT
following treatment with S-1 or UFT, with the later hope that these
biomarkers can be used to tailor therapies for patients with rectal
cancer.
Materials and methods
Patients
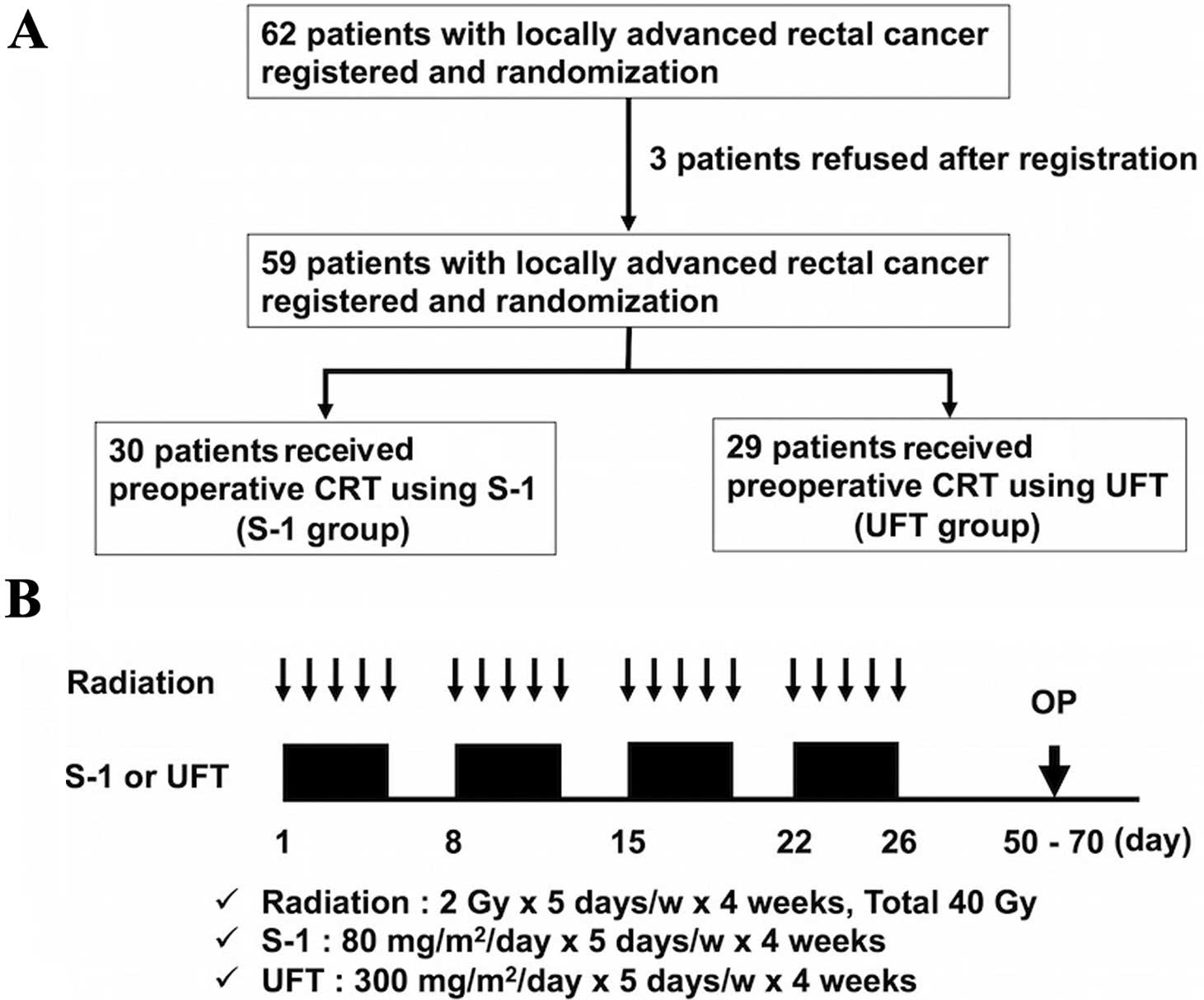
From April 2008 to October 2010, 62 patients with
locally advanced rectal cancer within 10 cm from the anal verge
were enrolled in the present study across multiple institutions.
Patients were randomly assigned to receive preoperative CRT with
S-1 (S-1 group) or preoperative CRT with UFT (UFT group). Despite
signing the consent form, three patients chose to leave the study
before treatment, and they were treated surgically without CRT
(Fig. 1). Finally, 59 patients (S-1
group, n=30; UFT group, n=29) were confirmed, each fulfilling the
following criteria: i) Eastern Cooperative Oncology group (ECOG)
performance status of 0–2; ii) white blood count
≥4,000/mm3; iii) platelet count ≥100,000/mm3;
iv) serum total bilirubin <1.5 mg/dl; v) creatinine <1.5
mg/dl; and vi) heart function (stable cardiac rhythm, no active
angina, no clinical evidence of congestive heart failure). Informed
consent was obtained from all patients included in this study, and
the present study protocol was approved by our local ethics
committees. There was no statistically significant difference in
clinicopathological features between the S-1 and UFT group except
for gender (data not shown).
For gene expression profiling, biopsy specimens were
prospectively collected during colonoscopic examination before
commencing preoperative CRT. These samples were used for RNA
extraction when parallel specimens contained at least 70% tumor
cells. Samples were snap frozen immediately in liquid nitrogen and
stored at −80°C until RNA extraction.
Preoperative CRT
Preoperative CRT is routinely prescribed for
patients with locally advanced cancers (≥T3 and/or ≥N1) or for
those who have very distal T2N0 cancers close to or involving the
sphincter. Patients received CRT with a total dose of 40 Gy of
pelvic irradiation, which was administered five times per week,
with a daily fraction of 2 Gy using a four-field technique. The
superior margin of the radiation field was the bifurcation point of
the aorta, whereas the inferior margin was marked at least 4 cm
below the tumor. Radiation was delivered concomitantly with S-1 (80
mg/m2) or UFT (300 mg/m2). If grade 3
toxicity occurred, the dose of S-1 or UFT was reduced to 75% of the
previous dose. If the neutrophil count decreased to <1,500/l or
the platelet count decreased to <100,000/l, chemotherapy was
stopped until recovery. Surgery was performed 6–8 weeks after
completion of preoperative CRT. The design and timeline of
preoperative CRT administration is shown in Fig. 1.
cDNA microarray
cDNA and miRNA microarray (described below) were
performed at Hokkaido System Science Co., Ltd. (Hokkaido, Japan).
High-quality RNA (50–200 ng) was amplified and cyanine 3-CTP
labeled with the One Color Low Input Quick Amp Labeling kit
(version 6.5; Agilent Technologies, Santa Clara, CA, USA) according
to the manufacturer's instructions. Labeled and amplified RNA was
purified with Qiagen RNeasy Mini spin columns (Qiagen, Valencia,
CA, USA). RNA labeling efficiency was controlled using the NanoDrop
spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Labeled
cRNA (600 ng) was fragmented and hybridized to the Whole Human
Genome Expression Array G4851A (8×60K; Agilent) according to the
manufacturer's instructions. In brief, 600 ng of labeled cRNA was
combined with 5 µl 10X blocking agent, brought to 24
µl with ddH2O, and then combined with 1 µl
of 25X fragmentation buffer. The cRNA was then fragmented for
exactly 30 min at 60°C, cooled on ice for 1 min, and then combined
with 25 µl of 2X GEx Hybridization Buffer HI-RPM by
pipetting. The mix was centrifuged briefly and hybridized
immediately to the microarrays. Arrays were rotated overnight for
17 h at 6°C at a high speed of rotation. Arrays were disassembled
in an ozone-filtered room using GE Wash Buffer I, washed for 1 min
at room temperature and then washed for 1 min with pre-warmed
(37°C) GE Wash Buffer II. The arrays were scanned immediately using
an Agilent Microarray Scanner (Agilent).
miRNA microarray
Cyanine-3-labeled miRNA was prepared from 100 ng
total RNA using Agilent miRNA Complete Labeling and Hyb kit
(Agilent) according to the manufacturer's instructions. Labeled
miRNA was hybridized to Agilent Human miRNA Microarrays version 3,
release 12.0 (P/N G4471A, AMADID 021827) annotated against the
Sanger miRbase 12.0 database of miRNAs, according to the
manufacturer's instructions. The arrays were scanned immediately
using an Agilent Microarray Scanner.
Data analysis
Data were analyzed using GeneSpring version 10
(Agilent). Among the 30 training samples in the S-1 group, 18 were
classified as responders and 12 as the non-responders. Among the 29
training samples in the UFT group, 15 were classified as responders
and 14 as non-responders. Gene expression profiles were compared
and fold-change values calculated to compare the responders and
non-responders in both groups and identify predictive markers for
patient responses to preoperative CRT. Genes that were
differentially expressed were clustered.
Ingenuity pathway analysis
An ingenuity pathway analysis (IPA) (Ingenuity
Systems, Mountain View, CA, USA) was used to identify biological
and molecular networks that were related to the efficacy of the use
of preoperative CRT. A detailed description is given in the online
repository (http://www.ingenuity.com).
Statistical analysis
A total of 60 patients were required to have a power
of 90 and a 5% single-sided significance level for the detection of
a 25% increase in response rate in the combined experimental arm.
The expression patterns in both the cDNA and miRNA microarrays were
compared using unpaired t-tests. All differences were considered
statistically significant at a p-value <0.05. All statistical
analyses were performed using GeneSpring version 10.
Results
Response to CRT
Patient responses to CRT were evaluated with RECIST
guidelines. One patient (3%) in the S-1 group was determined to
have had a complete response (CR); no patients in the UFT group
showed a complete response. A partial response (PR) was noted in 17
patients (57%) in the S-1 group and 15 patients (52%) in the UFT
group. Stable disease (SD) was determined in 12 patients (40%) in
the S-1 group and 14 patients (48%) in the UFT group. No patient
demonstrated progressive disease (PD). There was no statistically
significant difference between the two groups in regard to response
rate (CR and PR; p=0.52). For the purposes of the present study,
patients were classified as responders (CR and PR) or
non-responders (SD and PD).
DNA microarray
Gene expression profiling was performed using
customized and focused DNA microarrays. In the S-1 group, 184 genes
were identified that were significantly (p<0.05) differentially
expressed between the responders and the non-responders (data not
shown). In the UFT group, 193 genes were identified that were
significantly (p<0.05) differentially expressed between the
responders and the non-responders (data not shown). TBX18
upregulation was common among the responders in both groups,
whereas reduced expression of BTNL8, LOC375010, ADH1B, HRASLS2,
LOC284232, GCNT3 and ALDH1A2 was common to responders in both the
S-1 and UFT groups (Table I). The
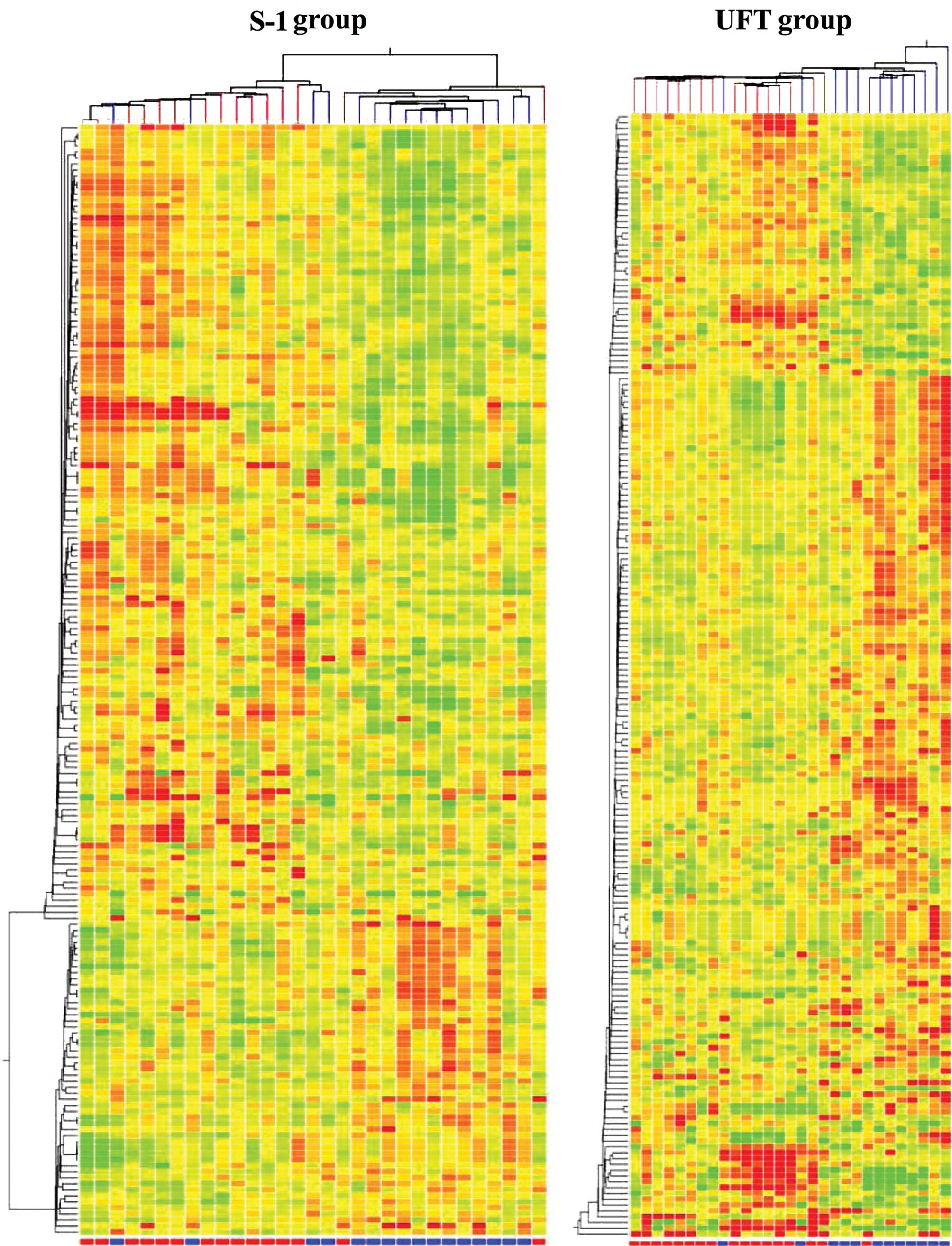
results of a hierarchical cluster analysis are presented in
Fig. 2. The responders and
non-responders were not clustered in both groups.
 | Table IGenes expressed in the responders in
both the S-1 and UFT groups with cDNA microarray. |
Table I
Genes expressed in the responders in
both the S-1 and UFT groups with cDNA microarray.
| Probe name | Gene symbol | Genbank accession
no. | Regulation | P-value | S-1 group (n=30)
fold-change | P-value | UFT group (n=29)
fold-change |
|---|
| A_23_P134041 | TBX18 | NM_001080508 | Up | 0.013 | 2.28 | 0.018 | 2.72 |
| A_23_P7412 | BTNL8 | NM_001040462 | Down | 0.015 | 4.98 | 0.011 | 5.58 |
| A_24_P187056 | LOC375010 | AK090412 | Down | 0.007 | 2.73 | 0.029 | 2.19 |
| A_33_P3353737 | ADH1B | NM_000668 | Down | 0.009 | 2.53 | 0.049 | 2.37 |
| A_23_P105012 | HRASLS2 | NM_017878 | Down | 0.030 | 2.53 | 0.023 | 3.04 |
| A_33_P3282359 | LOC284232 | NR_027995 | Down | 0.008 | 2.40 | 0.037 | 2.34 |
| A_23_P420209 | GCNT3 | NM_004751 | Down | 0.041 | 2.27 | 0.011 | 4.18 |
| A_32_P13151 | LOC284232 | NR_027995 | Down | 0.028 | 2.20 | 0.031 | 2.24 |
| A_24_P73577 | ALDH1A2 | NM_170697 | Down | 0.050 | 2.13 | 0.010 | 2.32 |
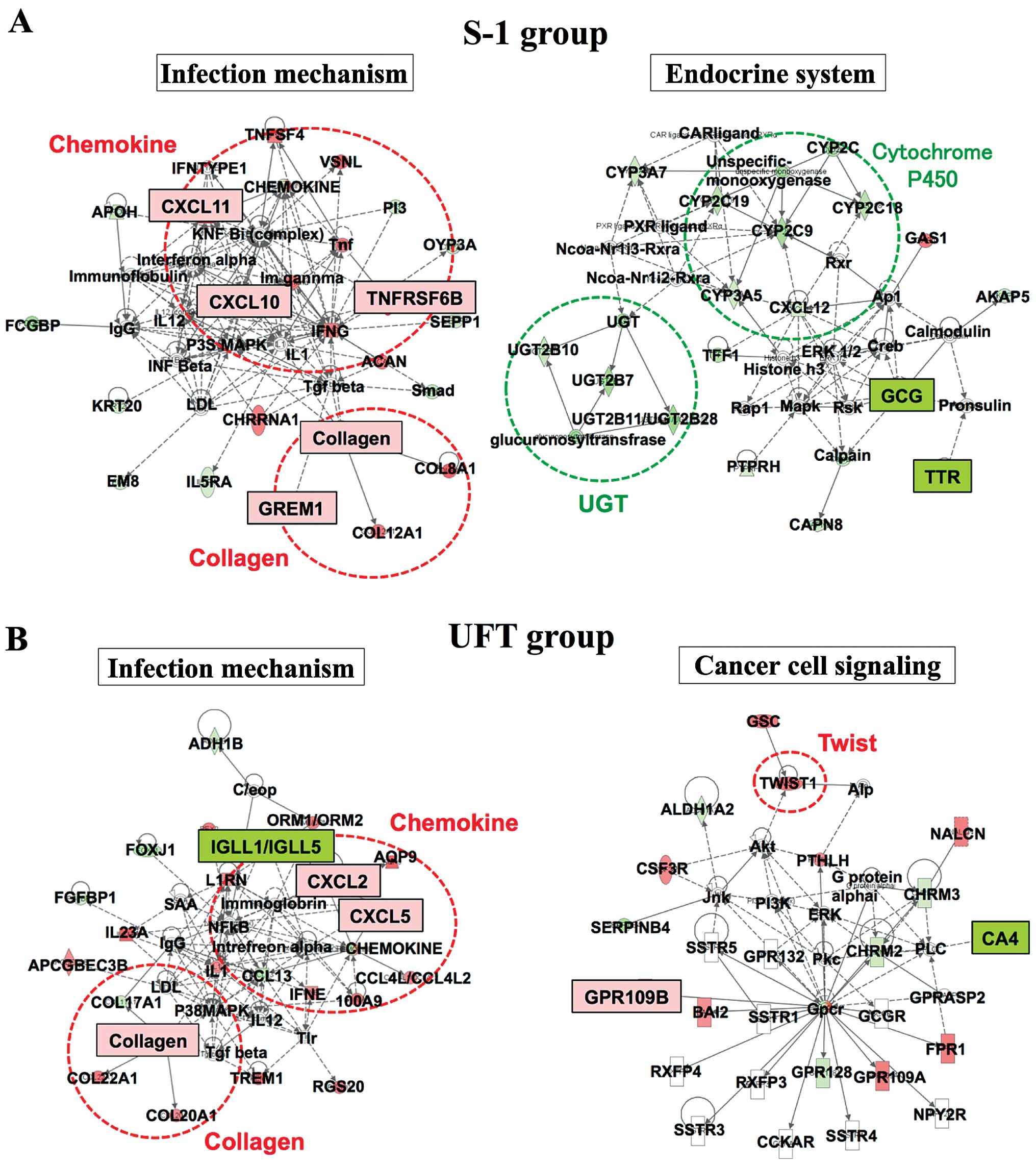
Ingenuity pathway analysis
To identify predictive genes for preoperative CRT,
an Ingenuity pathway analysis (IPA) was performed (Fig. 3). The IPA showed that the 'Infection
Mechanism' network and the 'Endocrine System' network were related
to responders in the S-1 group. The 'Infection Mechanism' network
comprises chemokine genes and collagen genes such as CXC chemokine,
TNFRSF6B, GREM1 and collagen, among others. These genes were
significantly elevated in the S-1 responders, as determined using
DNA microarray. Comparatively, the 'Infection Mechanism' network
and the 'Cancer Cell Signaling' network were related to responders
in the UFT group, with elevated expression of the CXC chemokine and
collagen from the 'Infection Mechanism' network.
miRNA microarray
The miRNA microarray showed significant (p<0.05)
differential expression of six genes between the responders and
non-responders in the S-1 group, with all six genes (miR-19-3p,
miR-866-3p, miR-923, miR-494, miR-513a-5p and miR-513b) elevated in
the responders (Table II). In
contrast, 16 genes were significantly differentially expressed
between the responders and non-responders in the UFT group, with
all 16 genes (miR-154, miR-379, miR-223, miR-1542-5p, miR-144,
miR-363, miR-31, miR-1290, miR-382, miR-193a-5p, miR-451, miR-335,
miR-486-5p, miR-1246, miR-34b* and miR-144*) elevated in the
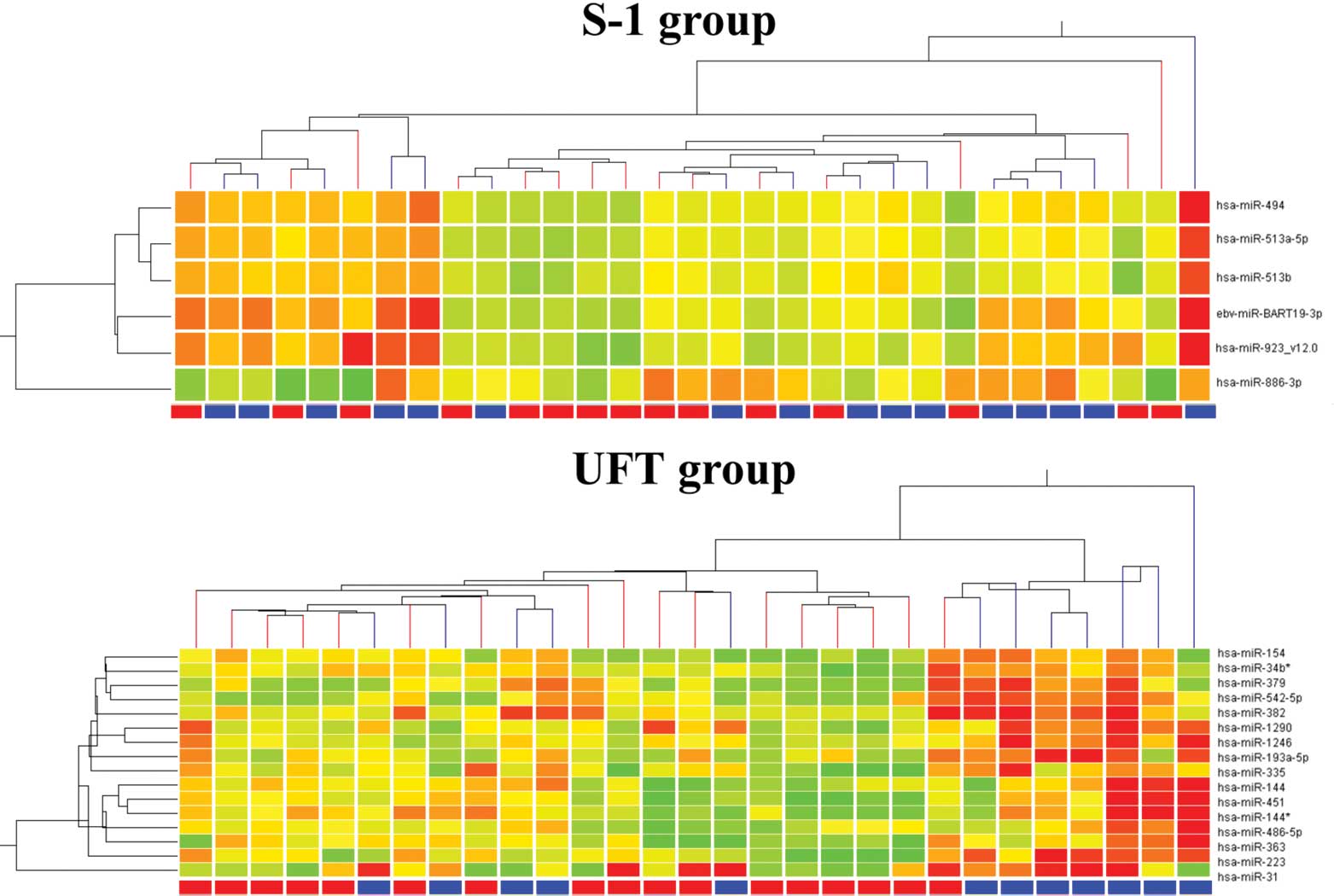
responders. The results of a hierarchical cluster analysis are
presented in Fig. 4. Again, the
responders and non-responders were not clustered in both
groups.
 | Table IIGenes expressed in the responders in
the S-1 and UFT groups with miRNA microarray. |
Table II
Genes expressed in the responders in
the S-1 and UFT groups with miRNA microarray.
| Systematic name | P-value | Fold-change | Regulation |
|---|
| S-1 group (n=30) |
| hsa-miR-494 | 0.007 | 1.74 | Up |
|
ebv-miR-BART19-3 | 0.007 | 2.20 | Up |
| hsa-miR-513a-5p | 0.010 | 1.59 | Up |
| hsa-miR-886-3p | 0.050 | 2.07 | Up |
|
hsa-miR-923_v12.0 | 0.040 | 1.80 | Up |
| hsa-miR-513b | 0.027 | 1.51 | Up |
| UFT group (n=29) |
| hsa-miR-154 | 0.021 | 2.01 | Up |
| hsa-miR-379 | 0.013 | 2.41 | Up |
| hsa-miR-223 | 0.017 | 2.84 | Up |
| hsa-miR-542-5p | 0.010 | 2.33 | Up |
| hsa-miR-144 | 0.018 | 2.69 | Up |
| hsa-miR-363 | 0.022 | 2.28 | Up |
| hsa-miR-31 | 0.025 | 3.37 | Up |
| hsa-miR-1290 | 0.012 | 2.24 | Up |
| hsa-miR-382 | 0.034 | 2.01 | Up |
| hsa-miR-193a-5p | 0.021 | 2.14 | Up |
| hsa-miR-451 | 0.006 | 3.30 | Up |
| hsa-miR-335 | 0.030 | 2.46 | Up |
| hsa-miR-486-5p | 0.008 | 2.18 | Up |
| hsa-miR-1246 | 0.002 | 2.31 | Up |
| hsa-miR-34b* | 0.028 | 2.02 | Up |
| hsa-miR-144* | 0.026 | 2.69 | Up |
Discussion
Changes in gene expression patterns have been used
to predict patient outcomes in multiple types of cancer. In the
present study, 184 and 193 genes were differentially expressed
between the responders and the non-responders in the patients
treated with preoperative CRT using S-1 (S-1 group) and UFT (UFT
group), respectively. TBX18 upregulation and BTNL8, LOC375010,
ADH1B, HRASLS2, LOC284232, GCNT3 and ALDH1A2 downregulation were
common to the responders from both treatment groups. Previous
studies have also reported significant differential gene expression
between responders and non-responders using preoperative CRT
(6,8), yet few common genes could be
identified between different treatment regimes. Furthermore,
although some studies have reported the use of microarrays to
predict the responses of patients with rectal cancer to CRT using
preoperative biopsy tissue samples, this is the first study to date
to use both cDNA and miRNA microarray analyses to predict patient
responses. Therefore, from our results, we recommend the use of
preoperative CRT in patients who show a differential expression in
any of these eight common differentially expressed genes in biopsy
specimens collected during colonoscopic examination for rectal
cancer. Moreover, the use of S-1 or UFT is preferred if genes
specific to that treatment identified in the present study are also
identified from the patient biopsies.
An IPA was preformed to identify predictive genes
for preoperative CRT. In the present study, IPA showed that CXC
chemokine and collagen, which are part of the 'Infection Mechanism'
network, were related to responders in the S-1 and UFT groups. A
previous report indicated that CXCR4 and CXCL12 were predictive
factors of distant recurrence and poor prognosis in patients with
rectal cancer who were treated with preoperative CRT (14). This result supports our finding that
the CXC chemokine may be a predictive factor of response to
preoperative CRT in advanced rectal cancer.
Using microRNA analyses, we found 16 genes to be
significantly elevated in the responders as compared with the
non-responders in the UFT group. miR-223 was among these elevated
genes. In the S-1 group, six genes were significantly higher in the
responders than in the non-responders and, although not
significant, miR-223 expression showed a tendency towards elevated
expression (p=0.06). Previous studies have linked miR-223 with
malignant prognosis in colorectal, gastric and breast cancers and
in esophageal squamous cell carcinoma (15–18).
Another study showed that overexpression of miR-223 downregulates
the expression of ataxia telangiectasia mutated (ATM) gene and
sensitizes U87 cells to radiation in vitro and in
vivo (19). We also previously
reported that miR-223 was higher in responders as compared with
non-responders in patients with rectal cancer who were treated with
preoperative CRT using S-1 (20).
Collectively, these results support our hypothesis that miR-223 may
be a predictive factor in ascertaining the response of patients
with rectal cancer to preoperative CRT. In cases where miR-223
expression changes are noted in biopsy specimens, we also recommend
the use of preoperative CRT. Moreover, if certain miRNAs are
upregulated (miR-494, miR-BART19-3, miR-513a-5p, miR-886-3p,
miR-923 and miR-513b), it is better to use S-1 as an agent for CRT.
The reciprocal is true for the identification of certain miRNAs
(miR-154, miR-379, miR542-5p, miR-144, miR-363, miR-31, miR-1290,
miR-382, miR-193a-5p, miR-451, miR-335, miR-486-5p, miR-1246,
miR-34b and miR-144) that were upregulated in the UFT group.
In conclusion, cDNA and miRNA expression patterns
offer the opportunity to identify potential new biomarkers to
predict the response of patients with rectal cancer to preoperative
CRT and will likely be useful in establishing individualized
therapies for these patients.
Acknowledgments
The present study was partly financed by the
Research Support Foundation of the University of Tokushima and
Taiho Pharmaceutical, Co., Ltd. (research funding provided to
Professor Mitsuo Shimada).
References
|
1
|
Sauer R: Adjuvant and neoadjuvant
radiotherapy and concurrent radiochemotherapy for rectal cancer.
Pathol Oncol Res. 8:7–17. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gérard JP, Conroy T, Bonnetain F, Bouché
O, Chapet O, Closon-Dejardin MT, Untereiner M, Leduc B, Francois E,
Maurel J, et al: Preoperative radiotherapy with or without
concurrent fluorouracil and leucovorin in T3-4 rectal cancers:
Results of FFCD 9203. J Clin Oncol. 24:4620–4625. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bosset JF, Collette L, Calais G, Mineur L,
Maingon P, Radosevic-Jelic L, Daban A, Bardet E, Beny A and Ollier
JC; EORTC Radiotherapy group Trial 22921: Chemotherapy with
preoperative radiotherapy in rectal cancer. N Engl J Med.
355:1114–1123. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Morimoto S, Shimada M, Kurita N, Sato H,
Iwata T, Nishioka M, Yoshikawa K, Miyatani T, Kashihara H, Takasu
C, et al: Preoperative radiotherapy combined with S-1 for advanced
lower rectal cancer: Phase I trial. Hepatogastroenterology.
59:1428–1432. 2012.
|
|
5
|
Hotchi M, Okitsu H, Miura M, et al: A
phase II trial of preoperative chemoradiotherapy with oral
DPD-inhibitory fluoropyrimimidines in patients with advanced rectal
cancer. J Cancer Ther. 3:989–995. 2012. View Article : Google Scholar
|
|
6
|
Ghadimi BM, Grade M, Difilippantonio MJ,
Varma S, Simon R, Montagna C, Füzesi L, Langer C, Becker H, Liersch
T, et al: Effectiveness of gene expression profiling for response
prediction of rectal adenocarcinomas to preoperative
chemoradiotherapy. J Clin Oncol. 23:1826–1838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim IJ, Lim SB, Kang HC, Chang HJ, Ahn SA,
Park HW, Jang SG, Park JH, Kim DY, Jung KH, et al: Microarray gene
expression profiling for predicting complete response to
preoperative chemoradiotherapy in patients with advanced rectal
cancer. Dis Colon Rectum. 50:1342–1353. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rimkus C, Friederichs J, Boulesteix AL,
Theisen J, Mages J, Becker K, Nekarda H, Rosenberg R, Janssen KP
and Siewert JR: Microarray-based prediction of tumor response to
neoadjuvant radiochemotherapy of patients with locally advanced
rectal cancer. Clin Gastroenterol Hepatol. 6:53–61. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Watanabe T, Kobunai T, Akiyoshi T, Matsuda
K, Ishihara S and Nozawa K: Prediction of response to preoperative
chemoradiotherapy in rectal cancer by using reverse transcriptase
polymerase chain reaction analysis of four genes. Dis Colon Rectum.
57:23–31. 2014. View Article : Google Scholar
|
|
10
|
Pazdur R, Hoff PM, Medgyesy D, Royce M and
Brito R: The oral fluorouracil prodrugs. Oncology. 12(Suppl 7):
S48–S51. 1998.
|
|
11
|
Giralt J, Tabernero J, Navalpotro B,
Capdevila J, Espin E, Casado E, Mañes A, Landolfi S, Sanchez-Garcia
JL, de Torres I, et al: Pre-operative chemoradiotherapy with UFT
and Leucovorin in patients with advanced rectal cancer: A phase II
study. Radiother Oncol. 89:263–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Van Cutsem E, Hoff PM, Harper P, Bukowski
RM, Cunningham D, Dufour P, Graeven U, Lokich J, Madajewicz S,
Maroun JA, et al: Oral capecitabine vs intravenous 5-fluorouracil
and leucovorin: Integrated efficacy data and novel analyses from
two large, randomised, phase III trials. Br J Cancer. 90:1190–1197.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee EM, Hong YS, Kim KP, Lee JL, Kim SY,
Park YS, Choi DH, Kim JH, Lim SB, Yu CS, et al: Phase II study of
preoperative chemoradiation with S-1 plus oxaliplatin in patients
with locally advanced rectal cancer. Cancer Sci. 104:111–115. 2013.
View Article : Google Scholar
|
|
14
|
Saigusa S, Toiyama Y, Tanaka K, Yokoe T,
Okugawa Y, Kawamoto A, Yasuda H, Inoue Y, Miki C and Kusunoki M:
Stromal CXCR4 and CXCL12 expression is associated with distant
recurrence and poor prognosis in rectal cancer after
chemoradiotherapy. Ann Surg Oncol. 17:2051–2058. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Luo X, Li H, Yue X, Deng L, Cui Y
and Lu Y: MicroRNA-223 functions as an oncogene in human colorectal
cancer cells. Oncol Rep. 32:115–120. 2014.PubMed/NCBI
|
|
16
|
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li
M, Sun L, Ji G, Shi Y, Han Z, et al: miRNA-223 promotes gastric
cancer invasion and metastasis by targeting tumor suppressor
EPB41L3. Mol Cancer Res. 9:824–833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kurashige J, Watanabe M, Iwatsuki M,
Kinoshita K, Saito S, Hiyoshi Y, Kamohara H, Baba Y, Mimori K and
Baba H: Overexpression of microRNA-223 regulates the ubiquitin
ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer.
106:182–188. 2012. View Article : Google Scholar :
|
|
18
|
Pinatel EM, Orso F, Penna E, Cimino D,
Elia AR, Circosta P, Dentelli P, Brizzi MF, Provero P and Taverna
D: miR-223 is a coordinator of breast cancer progression as
revealed by bioinformatics predictions. PLoS One. 9:e848592014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang L, Zhu J, Zaorsky NG, Deng Y, Wu X,
Liu Y, Liu F, Cai G, Gu W, Shen L, et al: MicroRNA-223 enhances
radiation sensitivity of U87MG cells in vitro and in vivo by
targeting ataxia telangiectasia mutated. Int J Radiat Oncol Biol
Phys. 88:955–960. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hotchi M, Shimada M, Kurita N, Iwata T,
Sato H, Morimoto S, Yoshikawa K, Higashijima J and Miyatani T:
microRNA expression is able to predict response to
chemoradiotherapy in rectal cancer. Mol Clin Oncol. 1:137–142.
2013.PubMed/NCBI
|