Introduction
The a disintegrin and metalloproteinase (ADAM)
belong to a proteinase family consisting of 40 putative
membrane-bound cell surface glycoproteins (1). These zinc-dependent proteases contain
an amino terminal metalloproteinase and a disintegrin domain, a
cysteine-rich and an EGF-like sequence, a transmembrane and a
cytoplasmic domain (2). It has been
proven that 13 members of ADAM family are catalytically active
through their metalloproteinase domain. These catalytic members
play an important role in mediating extracellular matrix protein
degradation and shedding of growth factors, cytokines and adhesion
molecules. Besides, the disintegrin domain of ADAMs is also
involved in binding to integrins to mediate cell-cell and
cell-matrix interactions (3). ADAMs
have been shown to play important roles in cancer progression
(4). ADAM9, ADAM10 and ADAM17 are
suggested to cleave epidermal growth factor receptor (EGFR) ligands
and HB-EGF to activate EGFR signaling pathway in tumor cells
(5–7). Furthermore, ADAMs can cleave many cell
adhesion molecules to mediate tumor metastasis. For example, it has
been reported that cell adhesion molecule cadherins are substrates
for ADAM10 and ADAM15 (8–10). The cleavage of E-cadherin and
N-cadherin in tumor cells supports cancer cell migration, invasion
and metastasis (11).
As a catalytic active member of ADAM family, ADAM15
is the only ADAM containing an arginineglycine-aspartic acid (RGD)
motif in the disintegrin domain (12). The RGD motif of ADAM15 interacts
with integrins αvβ3 and
α5β1 to regulate cell adhesion and motility
(13). ADAM15 is widely expressed
in normal tissues and cancer cell lines. Overexpression of ADAM15
has been reported in many solid tumors including breast, lung,
colorectal, ovarian and prostate cancer (14–16).
Dependent on the tumor type, ADAM15 either promotes or suppresses
cancer progression. For example, in prostate cancer, Najy et
al demonstrated that ADAM15 promoted cancer cell migration and
metastatic progression (17). While
Toquet et al reported that downregulation of ADAM15 promoted
colon cancer metastasis and was corrected with poor prognosis of
colon cancer patients (18).
Lung cancer is the most common cancer worldwide and
is also the leading cause of malignancy-related diseases worldwide
(19). Approximatly 85–90% of lung
cancer cases are non-small cell lung cancer (NSCLC) (20). In China, the incidence of lung
cancer has constantly increased each year and the mortality rate
has markedly increased by 465% over the past 30 years (21). Although ADAM15 has been reported to
be overexpressed in lung carcinomas (22), the prognostic value of ADAM15 in
lung cancer is not yet clear, and its function in lung cancer is
still unknown. In the present study, we investigated the prognostic
value of ADAM15 in Chinese NSCLC cancer patients and studied the
roles of ADAM15 in lung cancer cell migration and invasion.
Materials and methods
Reagents
Antibodies to ADAM15 (EPR5619), EGFR, phospho-EGFR,
ERK1/2, phospho-ERK1/2, AKT and phosphor-AKT (S473) were purchased
from Abcam (Cambridge, MA, USA). Antibodies to β-actin were from
Santa Cruz Biotechnology (Santa Cruz, CA, USA). Matrigel was from
Life Technologies (Thermo Fisher Scientific, New York, NY, USA).
Recombinant MMP9 was purchased from R&D Systems (Minneapolis,
MN, USA). The MMP activity assay kit (cat no. ab112146) was from
Abcam.
Patients
A total of 121 lung carcinoma samples were collected
at the Hainan General Hospital from January 2004 to December 2008.
Before specimen collection, none of the patients had received
radiotherapy and chemotherapy. All of the patients diagnosed as
NSCLC received cytoreductive surgery before cisplatin-based
adjuvant chemotherapies. According to the 2009 WHO/IASLC (23), histopathological classification
(Table I) was performed by an
experienced pathologist. Disease-free survival (DFS) and overall
survival (OS) were analyzed as previously described (24). The present study was approved by the
Research Ethics Committee of Hainan General Hospital, Hainan,
China. Informed consent was obtained from all the patients.
 | Table IPrimers used in real-time PCR
assay. |
Table I
Primers used in real-time PCR
assay.
|
Oligonucleotide | Sequence
(5′→3′) | Purpose |
|---|
| Human MMP1 | F
ctggccacaactgccaaatg | RT-PCR |
| Human MMP1 | R
ctgtccctgaacagcccagtactta | RT-PCR |
| Human MMP2 | F
tctcctgacattgaccttggc | RT-PCR |
| Human MMP2 | R
caaggtgctggctgagtagatc | RT-PCR |
| Human MMP3 | F
attccatggagccaggctttc | RT-PCR |
| Human MMP3 | R
catttgggtcaaactccaactgtg | RT-PCR |
| Human MMP7 | F
tgagctacagtgggaacagg | RT-PCR |
| Human MMP7 | R
tcatcgaagtgagcatctcc | RT-PCR |
| Human MMP9 | F
ttgacagcgacaagaagtgg | RT-PCR |
| Human MMP9 | R
gccattcacgtcgtccttat | RT-PCR |
| Human MMP10 | F
gtcacttcagctcctttcct | RT-PCR |
| Human MMP10 | R
atcttgcgaaaggcggaact | RT-PCR |
| Human β-actin | F
tccctggagaagagctacg | RT-PCR |
| Human β-actin | R
gtagtttcgtggatgccaca | RT-PCR |
Immunohistochemistry
Serial paraffin sections were incubated with
anti-ADAM15 antibody (1:100 dilution in 5% BSA in PBS) at 4°C
overnight. Rabbit IgG was used as a negative control. After washing
three times with PBS, the slides were incubated with biotinylated
secondary antibody (1:200 dilution) at room temperature for 30 min.
After washing three times with PBS, the slides were stained with
the Elite ABC kit (Vector Labs, USA). Finally, the slides were
counterstained with hematoxylin, dehydrated, cleared and then
mounted with Permount mounting medium (Fisher Scientific, USA).
Histological images were captured from the microscope (Carl Zeiss,
AX10; Germany) with an objective magnification of ×20. ADAM15
staining was independently scored by two pathologists and was
calculated using a previously defined scoring system (24). Briefly, the proportion of the
positive tumor cell was scored as: 0, <5%; 1+, 5–20%; 2+,
21–50%; 3+, 50–70% and 4+, 70–100%. The intensity was arbitrarily
scored as 0, weak (no color or light blue); 1, moderate (light
yellow); 2, strong (yellow brown) and 3, very strong (brown). The
overall score was calculated by multiplying the two scores obtained
from each sample. A score of ≥8 was defined as a high ADAM15
expression and scores of <8 defined as a low ADAM15
expression.
Cell lines
The human lung cancer cell lines A549, NCI-H1648 and
NCI-H647 were purchased from ATCC and cultured at 37°C with 5%
CO2 in Dulbecco's modified Eagle's medium (DMEM)
(Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS),
100 U/ml penicillin G, 100 µg/ml streptomycin and 2 mM
L-glutamine.
Generating shRNA or siRNA knockdown
cells
The lentivirus silencing vector expressing shRNA
targeting human ADAM15 (TRC no. TRCN0000371222) was obtained from
Sigma-Aldrich (St. Louis, MO, USA). This silencing and the empty
vectors alone (TRC2-pLKO-puro, as control) along with pxPAX2 and
pCMV-VSVG were transfected into 293T cells to produce lentivirus.
After 24 h of transfection, supernatant was collected and this
lentiviral preparation was used to infect cells. Stably infected
cells were isolated in selection media containing puromycin (2
µg/ml).
For siRNA treatments, A549 cells were plated in a
3.5 cm dish and transfected with 50 pm MMP9 or non-targeting siRNA
(Human On-TARGETplus siRNA pools of four oligos; Dharmacon, USA)
using Lipofectamine RNAi Max (Invitrogen). After transfection 48 h,
cells were used for invasion assay.
Plasmid and transfection
ADAM15 full-length sequence (NM_207191) was
amplified from reverse transcripted cDNA from A549 cells. The
ADAM15 sequence was cloned into the mammalian expression vector
pCMV-3Tag-3 (Agilent Technologies, USA). DNA sequencing confirmed
the sequence fidelity. Then, A549 cells were transfected with
pCMV-3Tag-3-ADAM15 plasmid or empty control vector using
Lipofectamine 2000 (Invitrogen). After 24 h, cells were collected
and used for the experiment.
Wound-healing migration assay
Cells were starved in DMEM containing 0.5% FBS for 6
h to inactivate cell proliferation and then wounded by pipette tips
(25). The final concentration of
10% FBS was added into the culture medium to stimulate cell
migration. After 8 h of migration, cells were labeled with 2 mg/ml
calcein-AM (Invitrogen) for 30 min and visualized using an inverted
microscope (magnification, ×10; Olympus, Tokyo, Japan). Migrated
cells were manually quantified. Three independent experiments were
performed.
Transwell migration assay
A Transwell migration assay with 6.5-mm-diameter
polycarbonate filters (8-µm pore size) was used. The filter
of the Transwell plate (BD Biosciences) was coated with Matrigel.
The bottom chambers were filled with 500 µl of DMEM
containing 10% FBS. Cells (4×104) suspended in 100
µl of DMEM containing 0.5% FBS were seeded in the top
chambers. Cells were allowed to migrate for 24 h. Non-migrated
cells were removed with cotton swabs, and migrated cells were fixed
with cold 4% paraformaldehyde and stained with 1% crystal violet.
Images were captured using an inverted microscope (magnification,
×10; Olympus), and migrated cells were quantified by manual
counting.
Western blotting and
immunoprecipitation
Cells were lysed in M2 lysis buffer [150 mM NaCl, 50
mM Tris-HCl (pH 8.0), 5 mM EDTA, 1% Nonidet P-40] containing a
protease inhibitor mixture (Roche Applied Science) and a
phosphatase inhibitor mixture (St. Louis, MO, USA). The equal
amount of total protein was subjected to SDS-PAGE analysis and
immunoblotting with the appropriate antibodies.
For immunoprecipitation, lysates were precipitated
with antibody and protein G-agarose beads by incubation at 4°C
overnight. Beads were washed four times with 1 ml M2 buffer, and
the bound proteins were removed by boiling in SDS buffer and
resolved in 4–20% SDS-polyacrylamide gels for western blot
analysis.
Real-time PCR
TRIzol reagent (Invitrogen) was used to isolate
total RNA. The cDNA was prepared using reverse transcriptase
SuperScript II (Invitrogen) with 2 µg of DNase I-treated
total RNA. Then, 2 µl of the cDNA was mixed with an 18
µl PCR assay mixture containing 0.5 M each primer and 1
µl Brilliant SYBR®-Green QPCR Master Mix
(Stratagene, USA). PCR was conducted with the MyiQ Single-Color
Real-Time PCR Detection System (Bio-Rad, USA) using the following
conditions: 95°C for 10 min followed by 40 cycles of 95°C for 15
sec, 55°C for 30 sec and 72°C for 30 sec. The primers are listed in
Table I. The threshold cycle number
for each MMP was normalized to that of β-actin, and the resulting
value was converted to a linear scale. All assays were performed at
least three times from independent RNA preparations.
MMP activity assay
The MMP activity was determined using an assay kit
from Abcam according to the manufacturer's protocol. Briefly,
5×105 cells were seeded into 6-well plates and allowed
to attach overnight, then starved with serum-free media for another
12 h. Cells were then stimulated with 10% FBS and 50 µl of
medium was removed at different time points. The conditioned medium
was mixed with 50 µl of 2 mM APMA working solution and
incubated for 15 min at 20°C followed by addition of 100 µl
of the fluorogenic peptide substrate solution in the kit. A
microplate reader with a filter set of Ex/Em = 490/525 nm was used
to measure the fluorescence activity.
Gelatin zymography
The cell culture supernatants were collected and
mixed with non-reducing SDS gel sample buffer and was loaded to a
10% polyacrylamide gel containing 0.1% SDS and 1 mg/ml gelatin.
After electrophoresis, the gels were incubated with renaturing
buffer containing 50 mmol/l Tris-HCl (pH 7.5), 0.15 mol/l NaCl, 5
mmol/l CaC12, 5 µmol/l ZnCl, 0.02%
NaN3, 0.25% Triton X-100 at room temperature for 30 min,
and then were incubated in the same buffer without Triton X-100 at
37°C overnight. Then, the gels were stained by Coomassie brilliant
blue R-250 solution.
Statistical analysis
The relationship between the expression of ADAM15
and the patients clinical characteristics were analyzed using the
χ2 or Fisher's exact tests, as appropriate. OS and DFS
curves were generated using Kaplan-Meier method and compared using
the log-rank test. Univariate and multivariate analyses were
performed using Cox regression models. P-value <0.05 was
regarded as statistically significant. Data are analyzed using SPSS
(version 20.0; IBM Corporation, Armonk, NY, USA) software program.
Results are expressed as the means ± SD and are representative of
at least three separate experiments. The two-sample t-test was used
to determine statistical differences of the means of two
groups.
Results
High expression of ADAM15 is associated
with poor prognosis in NSCLC patients
To investigate the role of ADAM15 in lung cancer, we
first examined the expression of ADAM15 in NSCLC carcinomas. The
tissue samples were collected from 121 NSCLC patients with median
age of 63 years. Patient characteristics of the population are
summarized in Table II. At the
time of last follow-up, 85% of patients had died and 8.2% had no
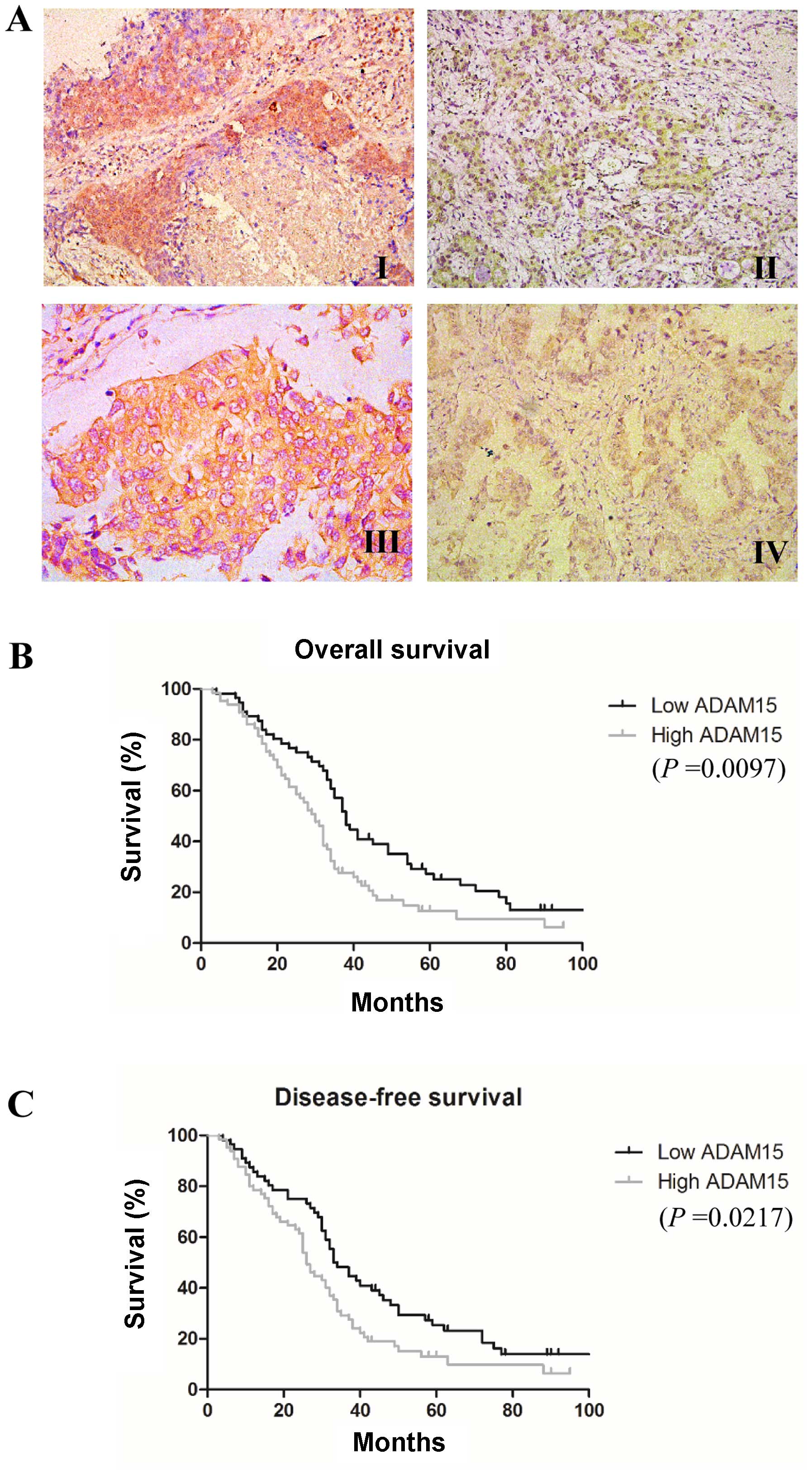
evidence of disease. As shown in Fig.
1A, ADAM15-positive staining was localized to the cytoplasm as
well as on the cell surface in tumor cells, which is similar to a
previous study (22). We further
analyzed the association of ADAM15 expression with
clinicopathological characteristics of the patients. We found that
ADAM15 expression was significantly associated with lymph node
metastasis (Table II). However, no
statistical correlations were observed between ADAM15 expression
and the patients age, gender, histologic types, tumor
differentiation stages or TNM stages (Table II).
 | Table IIClinicopathological characteristics
and results of ADAM15 immunohistochemistry. |
Table II
Clinicopathological characteristics
and results of ADAM15 immunohistochemistry.
|
Characteristics | No. of
patients | ADAM15 expression
| P-value |
|---|
| Low/no | High |
|---|
| Age (years) | | | | 0.7161 |
| ≤60 | 54 | 24 | 30 | |
| >60 | 67 | 32 | 35 | |
| Gender | | | | 0.7552 |
| Male | 76 | 36 | 40 | |
| Female | 45 | 20 | 25 | |
| Histological
type | | | | 0.8626 |
| Squamous cell
carcinoma | 68 | 31 | 37 | |
|
Adenocarcinoma | 53 | 25 | 28 | |
| Lymph node
metastasis | | | | 0.0003 |
| Negative | 48 | 32 | 16 | |
| Positive | 73 | 24 | 49 | |
|
Differentiation | | | | 0.6142 |
| Well | 64 | 31 | 33 | |
| Moderate/poor | 57 | 25 | 32 | |
| TNM stage | | | | 0.7209 |
| I | 25 | 13 | 12 | |
| II | 42 | 20 | 22 | |
| III | 54 | 23 | 31 | |
We then evaluated the prognostic significance of
ADAM15 expression in NSCLC patients. We found that high ADAM15
expression was significantly associated with decreased OS (median
30 vs. 38 months, P=0.0097) and DFS (median 26 vs. 33 months,
P=0.0217) (Fig. 1B and C).
Furthermore, a multivariate Cox regression analysis was applied to
all of the clinicopathological characteristics with ADAM15
expression levels. As shown in Table
III, high expression of ADAM15 were independently associated
with poor prognosis in NSCLC patients.
 | Table IIIMutivariate analyses for all the
patients (n=121). |
Table III
Mutivariate analyses for all the
patients (n=121).
|
Characteristics | DFS
| OS
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (years) | 1.6 (0.7–2.0) | 0.2551 | 1.5 (0.7–1.7) | 0.5431 |
| ≤60 vs.
>60 | | | | |
| Gender | 1.3 (0.6–1.5) | 0.4124 | 0.9 (0.4–1.8) | 0.2356 |
| Male vs.
female | | | | |
| Histological
type | 1.4 (0.7–1.5) | 0.2097 | 1.3 (0.5–1.5) | 0.1093 |
| Squamous vs.
adenocarcinoma | | | | |
| Lymph node
metastasis | 3.9 (0.9–5.0) | 0.0016 | 4.0 (0.8–6.4) | 0.0024 |
| Positive vs.
negative | | | | |
|
Differentiation | 1.0 (0.3–1.2) | 0.2632 | 1.5 (0.7–1.8) | 0.1736 |
| Well vs. moderate
to poor | | | | |
| TNM stage | 0.8 (0.6–1.8) | 0.4862 | 0.9 (0.5–1.2) | 0.2156 |
| I vs. II vs.
III | | | | |
| Level of ADAM15
expression | 4.1 (0.7–7.3) | 0.0042 | 6.1 (1.8–9.5) | 0.0233 |
| High vs. low | | | | |
Knockdown of ADAM15 in lung cancer cells
attenuates cell migration and invasion
To further investigate the function of ADAM15 in
lung cancer progression, we used a shRNA-mediated approach to
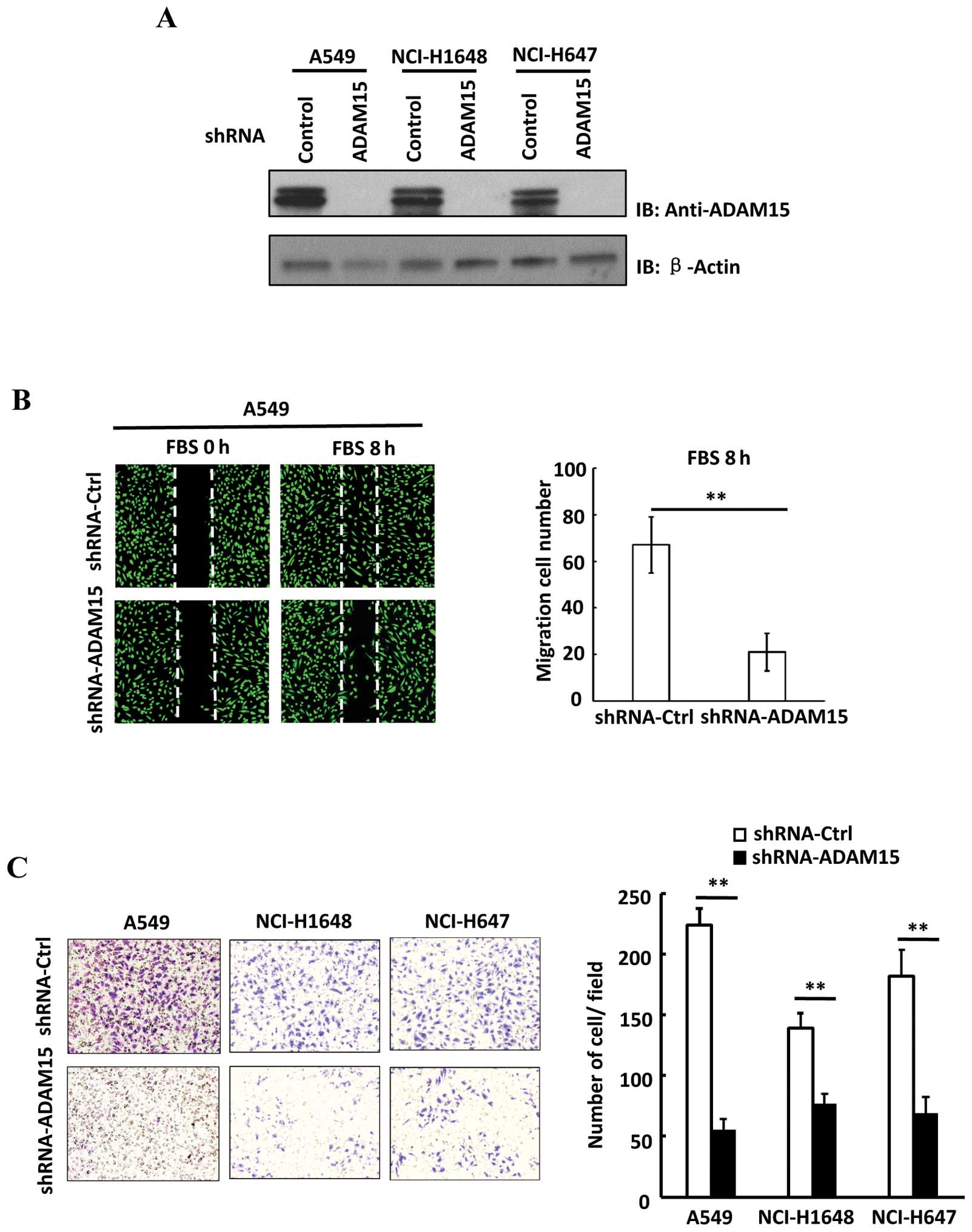
suppress the expression of ADAM15 in lung cancer cells. As shown in
Fig. 2A, the shRNA specific for
ADAM15 efficiently eliminated its expression in multiple metastatic
lung cancer cell lines including A549, NCI-H1648 and NCI-H647
cells. Since we found that ADAM15 overexpression is correlated with
lymph node metastasis, we then asked whether ADAM15 downregulation
influence cell migration and invasion. To measure the cell
migration ability, a wound-healing migration assay was performed in
both vector control and shRNA-ADAM15 A549 cells. Notably, we found
that knockdown of ADAM15 significant delayed wound closure rate in
A549 cells (Fig. 2B). Then, a
Matrigel Transwell assay was performed to evaluate the cell
invasion ability. As shown in Fig.
2C, knockdown of ADAM15 attenuated the invasion ability of lung
cancer cells. In addition, no change of cell cycle or cell
proliferation was observed in shRNA-ADAM15 A549 cells (data not
shown). Thus, our data suggest that ADAM15 plays an important role
in mediating lung cancer cell migration and invasion.
ADAM15 upregulates MMP activity and the
expression of MMP9 in lung cancer cells
Tumor cell migration and metastasis are controlled
by the interactions between surface adhesion molecules and
surrounding microenvironment. In this scenario, matrix
metalloproteinases (MMPs) play a critical role in tumor invasion
and metastasis by modulating cell-cell and cell-extracellular
matrix (ECM) interactions. We then asked whether shRNA-mediated
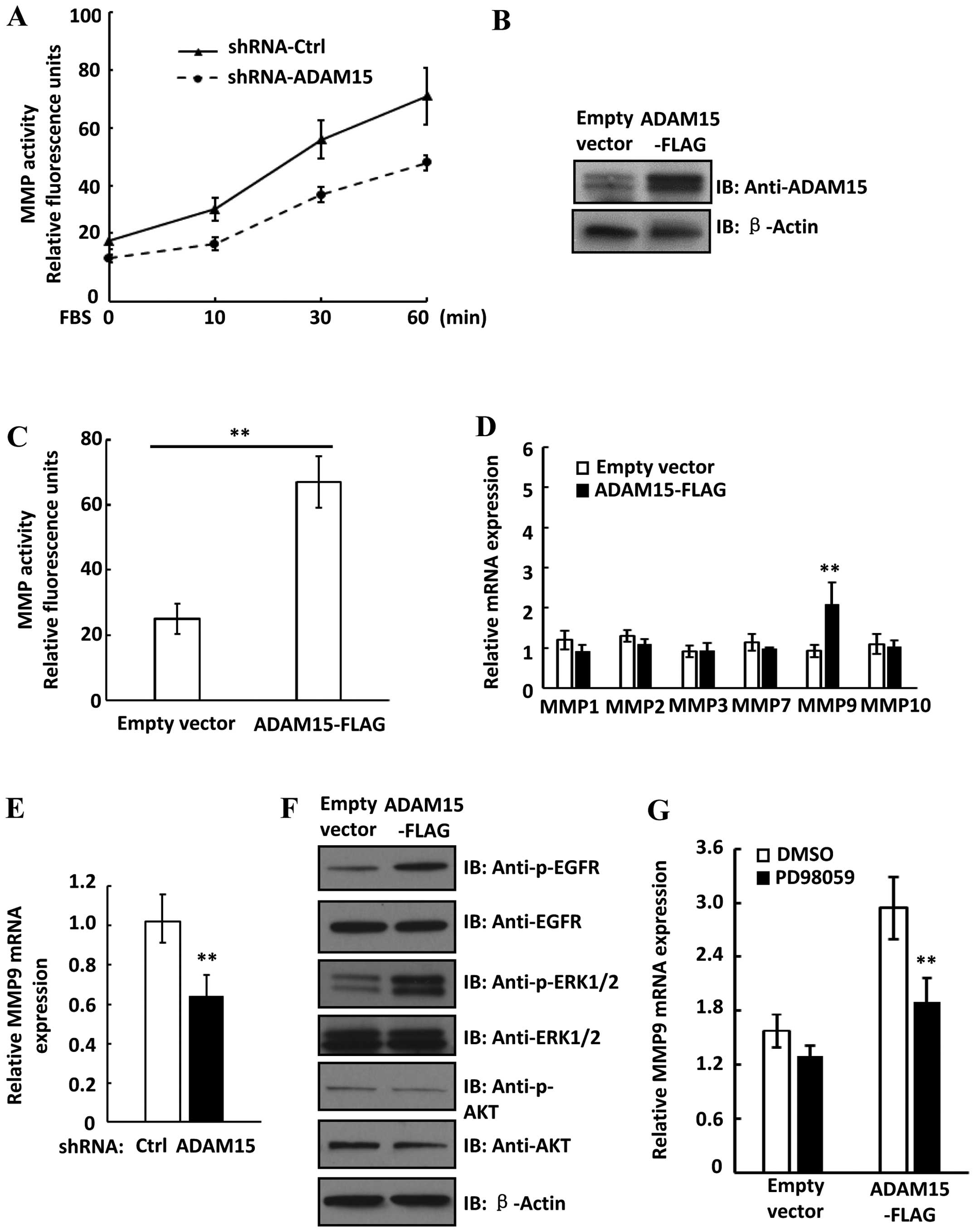
knockdown of ADAM15 influences MMP activity in lung cancer cells.
Notably, we found that FBS-induced MMP activation was decreased in
ADAM15 knockdown cells compared to shRNA control cells (Fig. 3A). Moreover, overexpression of
ADAM15 in A549 cells significantly increased MMP activity, further
suggesting that ADAM15 upregulates MMP activity in lung cancer
cells (Fig. 3B and C). To determine
whether upregulation of MMP expression contribute to increased MMP
activity in ADAM15 overexpressed cells, we examined expression of
several MMP members in both control and ADAM15 overex-pressed cells
by real-time PCR. Notably, we found that only MMP9 expression was
significantly increased in ADAM15 overexpressed cells (Fig. 3D). Moreover, knockdown of ADAM15
downregulated MMP9 expression in A549 cells, suggesting ADAM15
upregulated MMP9 expression in lung cancer cells (Fig. 3E). It was reported that the MEK-ERK
and PI3K-Akt pathways upregulated MMP9 expression in cancer cells
(26,27). We examined the activation of these
pathways in ADAM15 overexpressed cells. As shown in Fig. 3F, overexpression of ADAM15 in A549
cells increased phosphorylation levels of EGFR and ERK, while
phosphorylation of Akt was not changed in ADAM15 overexpressed
cells. We then treated cells with an ERK inhibitor PD98059 and we
found that PD98059 significantly attenuated ADAM15-induced MMP9
expression (Fig. 3G). Thus, these
data suggested that ADAM15 upregulated expression of MMP9 via
MEK-ERK pathway in lung cancer cells.
ADAM15 activates and interacts with MMP9
in lung cancer cells
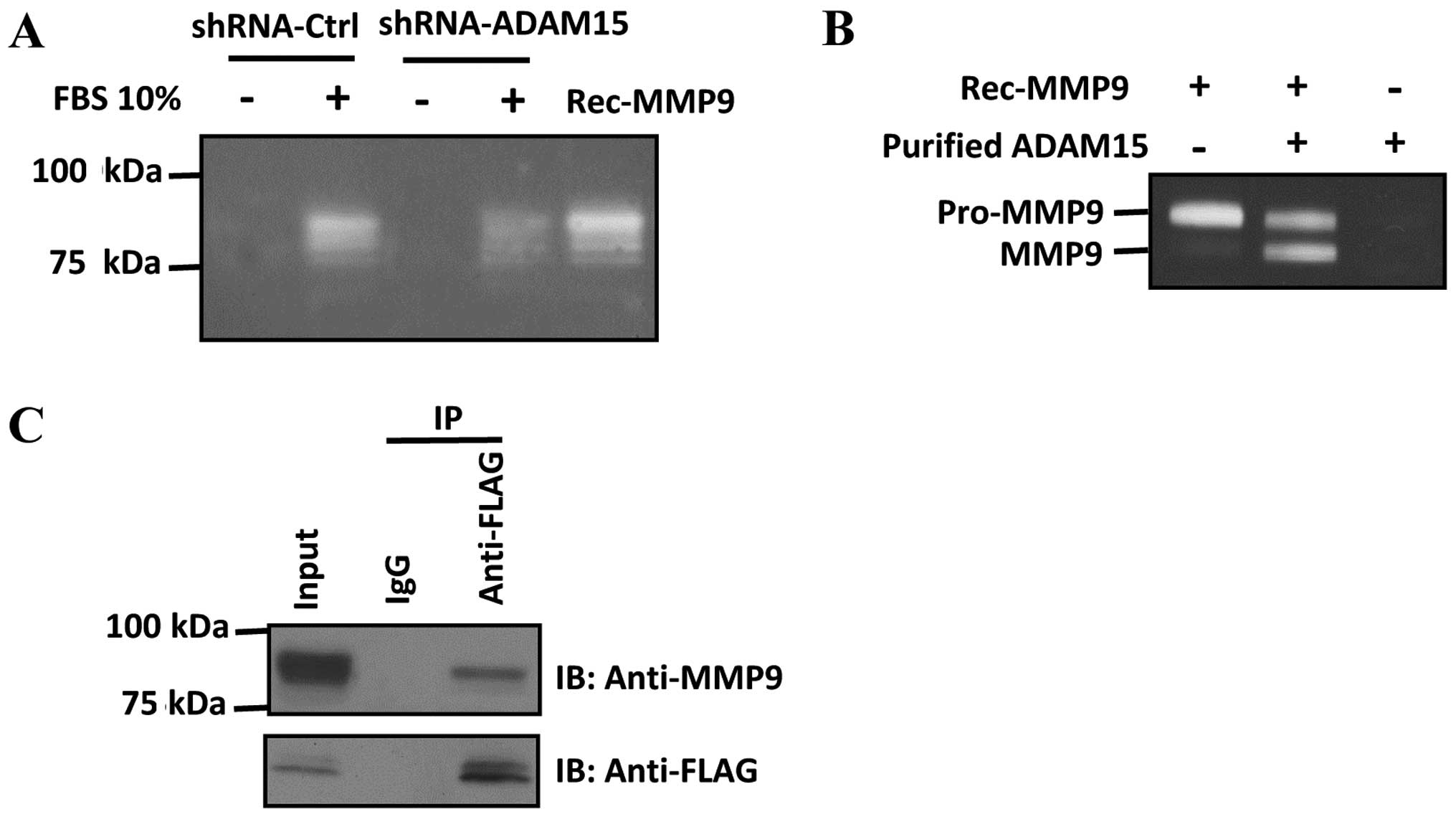
To study ADAM15-dependent MMP9 activity in lung
cancer cells, we used gelatin zymography to measure the activity of
MMP9 in ADAM15-shRNA knockdown cells. We found that FBS-induced
MMP9 activation was decreased in ADAM15 knockdown cells (Fig. 4A). We used purified enzymes to
examine the role of ADAM15 in MMP9 activation. Notably, we found
that incubation of ADAM15 and pro-MMP9 converts pro-MMP9 to lower
molecular weight species corresponding to its active form,
suggesting that ADAM15 proteolytically cleave and activate pro-MMP9
in vitro (Fig. 4B).
Furthermore, we found that ADAM15 interacted with MMP9 in A549
cells as examined by co-immunoprecipitation (Fig. 4C). Taken together, our data
demonstrated that ADAM15 interacts with MMP9 and promotes
maturation and activation of pro-MMP9 through a proteolytic
mechanism.
MMP9 activity is required for
ADAM15-regulated cell invasion
To study the role of MMP9 in ADAM15-regulated cell
invasion, we used a small interfering RNA targeting MMP9 to silence
MMP9 in ADAM15-overexpressed A549 cells and analyzed its invasive
ability (Fig. 4A). As shown in
Fig. 4B, overexpression of ADAM15
promoted cell invasion, while silencing MMP9 decreased
invasiveness. Moreover, both ERK inhibitor PD98059 and MMP9
inhibitor I attenuated cell invasion in A549-ADAM15 cells (Fig. 4C). Conversely, overexpression of
MMP9 in ADAM15 knockdown cells promoted cell invasion (Fig. 4D). Collectively, our data
demonstrated that ADAM15-induced MMP9 activation, at least in part,
was involved in ADAM15-regulated lung cancer cell invasion.
Discussion
It has been reported that ADAM15 was overexpressed
in lung carcinomas (22). However,
the prognosis value of ADAM15 in lung carcinomas remains elusive.
In the present study, we found that ADAM15 represents an
independent prognostic factor for the outcome in Chinese NSCLC
patients. Using a relatively large cohort of lung carcinoma
specimens, we observed that high expression of ADAM15 correlated
with decreased disease-free survival (DFS) and overall survival
(OS) in lung cancer patients. Increased expression of ADAM15 has
been reported in several types of cancer including ovary, gastric,
breast and prostatic cancers. In breast and prostatic cancers,
overexpression of ADAM15 was highly related to metastatic cancer
progression, suggesting a pro-metastatic role of ADAM15 (14). However, in colon cancer, the
decreased expression of ADAM15 was associated with cancer
metastasis and poor prognosis of colon cancer patients (18). Thus, these results reveal that
ADAM15 plays different roles in the context of various types of
cancer.
Although ADAM15 has been shown to be overexpressed
in lung carcinomas, the role of ADAM15 in lung cancer progression
is currently unknown. We found that knockdown of ADAM15 attenuated
migratory and invasive capacity in multiple lung cancer cell lines,
suggesting a pro-metastatic role of ADAM15 in lung cancer. Notably,
a similar observation was also reported in prostate cancer cells
(17). Matrix metalloproteinase
(MMPs) are a family of zinc endopeptidases that degrade
extracellular matrix components in either normal physiological
conditions or carcinogenesis (28).
Since the degradation of extracelluar matrix is one of the key
steps in cancer invasion and metastasis, we measured the MMP
activity in ADAM15 knockdown cells. Notably, we found that ADAM15
increased MMP activity in A549 cells. Furthermore, we observed that
ADAM15 activated EGFR-MEK-ERK pathway and thus upregulated MMP9
expression. As a plasma membrane-associated proteinase, ADAM15
sheds a variety of cell surface molecules including cytokines and
adhesion molecules. It has been reported that ADAM15 cleaved
E-cadherin to generate a soluble fragment and in turn simulates
EGFR signaling pathway in breast cancer cells (9). We also observed that overexpression of
ADAM15 promoted extracellular shedding of E-cadherin in A549 cells
(data not shown). Thus, the activation of EGFR-MEK-ERK signaling
pathway by ADAM15 in lung cancer cells could be due to the
increased shedding of cell surface molecules such as E-cadherin.
Future studies need to address whether the soluble E-cadherin or
other cell surface molecules contribute to EGFR pathway activation
by ADAM15 in lung cancer cells.
MMP9 is first synthesized as a proenzyme (pro-MMP9)
and requires cleavage by other proteinases to achieve its catalytic
activity. For example, it has been shown that pro-MMP9 was
proteolytically processed and activated by plasmin. Other MMPs,
including MMP-2, MMP-3 and MT1-MMP, were also involved in the
activation of MMP9. We have found that ADAM15 upregulated MMP9
expression in lung cancer cells. Since regulation of MMP activities
can be achieved at multiple levels, our data further demonstrated
that ADAM15 interacted with MMP9 and proteolytically cleaved and
activated pro-MMP9 in vitro. Other members of the ADAM
family have been reported to regulate MMP activities in cancer
cells. For example, ADAM12 together with αvβ3
integrin form a ternary protein complex with MMP-14 to activate
MMP-14 in breast cancer cells (29). ADAM17 upregulates MMP-2 and MMP-9
expression to promote prostate cancer cell invasion (30). Thus, our study identified a novel
ADAM15-mediated activation mechanism of MMP-9 in lung cancer
cells.
As an important matrix proteinase, MMP9 degrades
collagen type IV on the basement membrane. Overexpression of MMP9
has been reported in different types of cancer and it is believed
to facilitate tumor invasion and metastasis (28,31).
It has been reported that EGFR signaling pathway upregulated MMP9
expression in several types of cancer, including NSCLC (26,32,33).
In NSCLC patients, MMP9 expression was strongly correlated with
EGFR expression and the co-expression of these two proteins was
associated with poor outcome in the patients (26). Our data showed that MMP9 was
involved in ADAM15-regulated cancer cell invasion. Knockdown of
MMP9 prevented ADAM15-induced cell invasion, while overexpression
of MMP9 promoted cell invasion in ADAM15 knockdown cells. These
results may suggest an important functional link among ADAM15, EGFR
signaling pathway and MMP9 in the metastasis and progression of
NSCLC. In future studies, it would be interesting to investigate
the association among ADAM15, EGFR and MMP9 expression in NSCLC
patients and re-evaluate their prognostic values.
In conclusion, out study explored the expression of
ADAM15 in NSCLC and suggested that high expression of ADAM15
correlates with poor outcome in NSCLC patients. We further
demonstrated that ADAM15 directly activates MMP9 to promote cell
migration and invasion in lung cancer cells. Taken together, the
present study suggests that ADAM15 serves as a potential
therapeutic target in NSCLC patients.
Abbreviations:
|
ADAM
|
A disintegrin and
metalloproteinase
|
|
MMP
|
matrix metalloproteinase
|
|
NSCLC
|
non-small cell lung cancer
|
|
DFS
|
disease-free survival
|
|
OS
|
overall survival
|
|
shRNA
|
short hairpin RNA
|
Acknowledgments
The present study was supported by funding from the
Natural Science Foundation of Hunan Province, China (grant no.
2015JJ6058), and the Science and Technology Program of Health and
Family Planning Commission of Hunan Province, China (grant no.
B2015-111).
References
|
1
|
Seals DF and Courtneidge SA: The ADAMs
family of metalloproteases: Multidomain proteins with multiple
functions. Genes Dev. 17:7–30. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reiss K and Saftig P: The 'a disintegrin
and metalloprotease' (ADAM) family of sheddases: Physiological and
cellular functions. Semin Cell Dev Biol. 20:126–137. 2009.
View Article : Google Scholar
|
|
3
|
White JM: ADAMs: Modulators of cell-cell
and cell-matrix interactions. Curr Opin Cell Biol. 15:598–606.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Duffy MJ, Mullooly M, O'Donovan N, Sukor
S, Crown J, Pierce A and McGowan PM: The ADAMs family of proteases:
New biomarkers and therapeutic targets for cancer? Clin Proteomics.
8:92011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Klessner JL, Desai BV, Amargo EV, Getsios
S and Green KJ: EGFR and ADAMs cooperate to regulate shedding and
endocytic trafficking of the desmosomal cadherin desmoglein 2. Mol
Biol Cell. 20:328–337. 2009. View Article : Google Scholar :
|
|
6
|
Ohtsu H, Dempsey PJ, Frank GD, Brailoiu E,
Higuchi S, Suzuki H, Nakashima H, Eguchi K and Eguchi S: ADAM17
mediates epidermal growth factor receptor transactivation and
vascular smooth muscle cell hypertrophy induced by angiotensin II.
Arterioscler Thromb Vasc Biol. 26:e133–e137. 2006.PubMed/NCBI
|
|
7
|
Higashiyama S and Nanba D: ADAM-mediated
ectodomain shedding of HB-EGF in receptor cross-talk. Biochim
Biophys Acta. 1751:110–117. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maretzky T, Reiss K, Ludwig A, Buchholz J,
Scholz F, Proksch E, de Strooper B, Hartmann D and Saftig P: ADAM10
mediates E-cadherin shedding and regulates epithelial cell-cell
adhesion, migration, and beta-catenin translocation. Proc Natl Acad
Sci USA. 102:9182–9187. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Najy AJ, Day KC and Day ML: The ectodomain
shedding of E-cadherin by ADAM15 supports ErbB receptor activation.
J Biol Chem. 283:18393–18401. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kohutek ZA, diPierro CG, Redpath GT and
Hussaini IM: ADAM-10-mediated N-cadherin cleavage is protein kinase
C-alpha dependent and promotes glioblastoma cell migration. J
Neurosci. 29:4605–4615. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Canel M, Serrels A, Frame MC and Brunton
VG: E-cadherin-integrin crosstalk in cancer invasion and
metastasis. J Cell Sci. 126:393–401. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Krätzschmar J, Lum L and Blobel CP:
Metargidin, a membrane-anchored metalloprotease-disintegrin protein
with an RGD integrin binding sequence. J Biol Chem. 271:4593–4596.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nath D, Slocombe PM, Stephens PE, Warn A,
Hutchinson GR, Yamada KM, Docherty AJ and Murphy G: Interaction of
metargidin (ADAM-15) with alphavbeta3 and alpha5beta1 integrins on
different haemopoietic cells. J Cell Sci. 112:579–587.
1999.PubMed/NCBI
|
|
14
|
Kuefer R, Day KC, Kleer CG, Sabel MS,
Hofer MD, Varambally S, Zorn CS, Chinnaiyan AM, Rubin MA and Day
ML: ADAM15 disintegrin is associated with aggressive prostate and
breast cancer disease. Neoplasia. 8:319–329. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carl-McGrath S, Lendeckel U, Ebert M,
Roessner A and Röcken C: The disintegrin-metalloproteinases ADAM9,
ADAM12, and ADAM15 are upregulated in gastric cancer. Int J Oncol.
26:17–24. 2005.
|
|
16
|
Ortiz RM, Kärkkäinen I and Huovila AP:
Aberrant alternative exon use and increased copy number of human
metalloprotease-disintegrin ADAM15 gene in breast cancer cells.
Genes Chromosomes Cancer. 41:366–378. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Najy AJ, Day KC and Day ML: ADAM15
supports prostate cancer metastasis by modulating tumor
cell-endothelial cell interaction. Cancer Res. 68:1092–1099. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Toquet C, Colson A, Jarry A, Bezieau S,
Volteau C, Boisseau P, Merlin D, Laboisse CL and Mosnier JF: ADAM15
to α5β1 integrin switch in colon carcinoma cells: A late event in
cancer progression associated with tumor dedifferentiation and poor
prognosis. Int J Cancer. 130:278–287. 2012. View Article : Google Scholar
|
|
19
|
Carney DN: Lung cancer - time to move on
from chemotherapy. N Engl J Med. 346:126–128. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen WQ, Zhang SW, Zou XN and Zhao P: An
analysis of lung cancer mortality in China, 2004–2005. Zhonghua Yu
Fang Yi Xue Za Zhi. 44:378–382. 2010.In Chinese. PubMed/NCBI
|
|
22
|
Schütz A, Härtig W, Wobus M, Grosche J,
Wittekind Ch and Aust G: Expression of ADAM15 in lung carcinomas.
Virchows Arch. 446:421–429. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rosell R, Bivona TG and Karachaliou N:
Genetics and biomarkers in personalisation of lung cancer
treatment. Lancet. 382:720–731. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou H, Chen JH, Hu J, Luo YZ, Li F, Xiao
L and Zhong MZ: High expression of Toll-like receptor 5 correlates
with better prognosis in non-small-cell lung cancer: An anti-tumor
effect of TLR5 signaling in non-small cell lung cancer. J Cancer
Res Clin Oncol. 140:633–643. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pang X, Yi Z, Zhang X, Sung B, Qu W, Lian
X, Aggarwal BB and Liu M: Acetyl-11-keto-beta-boswellic acid
inhibits prostate tumor growth by suppressing vascular endothelial
growth factor receptor 2-mediated angiogenesis. Cancer Res.
69:5893–5900. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cox G, Jones JL and O'Byrne KJ: Matrix
metalloproteinase 9 and the epidermal growth factor signal pathway
in operable non-small cell lung cancer. Clin Cancer Res.
6:2349–2355. 2000.PubMed/NCBI
|
|
27
|
Cheng CY, Hsieh HL, Hsiao LD and Yang CM:
PI3-K/Akt/JNK/NF-κB is essential for MMP9 expression and outgrowth
in human limbal epithelial cells on intact amniotic membrane. Stem
Cell Res. 9:9–23. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Deryugina EI and Quigley JP: Matrix
metalloproteinases and tumor metastasis. Cancer Metastasis Rev.
25:9–34. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Albrechtsen R, Kveiborg M, Stautz D, et
al: ADAM12 redistributes and activates MMP-14, resulting in gelatin
degradation, reduced apoptosis and increased tumor growth. J Cell
Sci. 126:4707–4720. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiao LJ, Lin P, Lin F, et al: ADAM17
targets MMP-2 and MMP-9 via EGFR-MEK-ERK pathway activation to
promote prostate cancer cell invasion. Int J Oncol. 40:1714–1724.
2012.
|
|
31
|
Sato H, Kida Y, Mai M, Endo Y, Sasaki T,
Tanaka J and Seiki M: Expression of genes encoding type IV
collagen-degrading metalloproteinases and tissue inhibitors of
metalloproteinases in various human tumor cells. Oncogene. 7:77–83.
1992.PubMed/NCBI
|
|
32
|
Kim S, Choi JH, Lim HI, Lee SK, Kim WW,
Cho S, Kim JS, Kim JH, Choe JH, Nam SJ, et al: EGF-induced MMP9
expression is mediated by the JAK3/ERK pathway, but not by the
JAK3/STAT-3 pathway in a SKBR3 breast cancer cell line. Cell
Signal. 21:892–898. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
O-Charoenrat P, Rhys-Evans P, Modjtahedi
H, Court W, Box G and Eccles S: Overexpression of epidermal growth
factor receptor in human head and neck squamous carcinoma cell
lines correlates with matrix metalloproteinase-9 expression and in
vitro invasion. Int J Cancer. 86:307–317. 2000. View Article : Google Scholar : PubMed/NCBI
|