Introduction
Pancreatic cancer is a disease with a dismal
outlook. Pancreatic adenocarcinoma is among the leading causes of
cancer-related mortality worldwide (1), and the 1- and 5-year survival rates
are <25 and <6%, respectively. Pancreatic cancer is
characterized by the potential for local invasion, which allows it
to spread during the early developmental stages of the disease.
Therefore, there is an urgent need to discover novel early
diagnostic biomarkers and to identify new therapeutic strategies
(2). However, the molecular
mechanisms underlying the high tumorigenicity of pancreatic cancer
are not well known.
It was recently reported that regulator of G protein
signaling 22 (RGS22) is specifically expressed in the testis during
different stages of development (3). Furthermore, we previously showed that
RGS22 is specifically expressed in adenocarcinoma and squamous
carcinoma. In particular, we observed that RGS22 localized to the
cytoplasm in pancreatic adenocarcinoma tissues. RGS22 also acts as
a tumor suppressor, repressing the metastasis of epithelial cancer
cells (4).
Tumor metastasis involves a series of complex
host-tumor interactions (5). Tumor
cell extravasation into the surrounding tissue is considered a
rate-limiting step in metastasis (6,7) that
involves the active migration of tumor cells across the endothelial
barrier, and penetration through the underlying basement membrane.
Tumor cell interaction with the basement membrane is a 3-step
process: cell attachment, matrix dissolution, and migration
(8). Previously, we found that
tumor cells with high RGS22 expression have decreased metastatic
potential and local invasiveness when compared to the control
cells, but have similar adherence ability to various basement
membrane substrates (4). Therefore,
whether RGS22 is involved in pancreatic adenocarcinoma cell
migration requires investigation.
Several classes of proteins are involved in
metastasis. The heterotrimeric G protein G12 α subunit (GNA12)
signaling promotes the migration of prostate (9) and breast cancer cells (10), and lysophosphatidic acid
(LPA)-induced ovarian cancer cell migration in vitro
(11). GNA12 signaling promotes
metastasis by stimulating cells to become invasive within the
primary tumor. GNA13, has also been shown to stimulate cell
migration in addition to inducing oncogenic transformation
(12). The RGS22 RGS protein (a
diverse family of proteins that regulate heterotrimeric G protein
signaling negatively), interacts with GNA12/13 and GNAq in the
testis (3). Therefore, the Gα
subunit through which RGS22 demonstrates significant ability to
inhibit cancer cell migration, should also be further studied.
Materials and methods
Plasmids and constructs
We constructed a plasmid containing LZRsPBMN-Z
complementary DNA (cDNA) via a retrovirus vector according to the
following protocols. The RGS22 open reading fragment (from 21 to
3783 bp) was PCR-amplified from a pTriplEx2-RGS22 plasmid using a
forward, 5′-GGGGGTCGA CAAGCTTGGCATGCCCGAGAAGACG-3′; containing a
HindIII site and reverse primer, 5′-GGGGCTCGAGCTTCTG
GCTGCTGTGGGC-3′; containing an XhoI site. The RGS22
fragments were purified and digested with HindIII and
XhoI. The LZRsPBMN-Z vector was also digested with
HindIII and XhoI, and ligated with the RGS22
fragments. The RGS22 sequence in the construct was confirmed by DNA
sequencing. Green fluorescent protein (GFP) cDNA was inserted into
the vector as the control.
We purchased a pSilencer 2.1-U6 neo vector for RGS22
RNAi from Ambion (Austin, TX, USA). A 19-mer hairpin with a loop
and 3′ terminal uridine tract efficiently induces RNAi of the
target gene. For each target gene, we designed complementary
55-60-mer oligonucleotides with 5′ single-stranded overhangs for
ligation into the pSilencer neo vectors. The oligonucleotides
encoded 19-mer hairpin sequences specific to the mRNA target, a
loop sequence separating the two complementary domains, and a
polythymidine tract to terminate transcription. Therefore, we
designed the following primers: 484 forward,
5′-GATCCGTCCGGCATGAATTTCAC ATTCAAGAGATGTGAAATTCATGCCGGACTTTTTTGG
AAA-3′ and reverse, 5′-AGCTTTTCCAAAAAAGTCCGGC
ATGAATTTCACATCTCTTGAATGTGAAATTCATGCCG GACG-3′; 3313 forward,
5′-GATCCGGAGTTAGGACCATAT GTATTCAAGAGATACATATGGTCCTAACTCCTTTTTTG
GAAA-3′ and reverse, 5′-AGCTTTTCCAAAAAAGGAGTT
AGGACCATATGTATCTCTTGAATACATATGGTCCTAAC TCCG-3′. The kit also
contained pSilencer-Neg, in which the inserted sequence does not
match human, mouse, or rat genomes, and was used as the negative
control. We annealed the hairpin small interfering RNA (siRNA)
template oligonucleotides, and then ligated the annealed siRNA
templates into the pSilencer vector according to the instruction
manual.
Constructs each containing human GA12 Q231L, GA13
Q226L and GAq Q209L in the pcDNA3.1 vector were purchased from the
University of Missouri-Rolla cDNA Resource Center (Rolla, MO, USA).
The mutations reduce GTPase activity, resulting in a constitutively
active phenotype.
Cell culture and transfection
LZRsPBMN-Z-RGS22 and LZRsPBMN-Z-GFP plasmids were
transfected using a transfection kit (Invitrogen, San Diego, CA,
USA) into BXPC-3 cells purchased from the Institute of Cell,
Chinese Academy of Science (Shanghai, China). Briefly, we seeded
4×105 cells/well in a 6-well plate the day before
transfection and cultured them overnight. Transfection was
performed using Lipofectamine 2000 according to the manufacturer's
protocol (Invitrogen). After transfection, cells were cultured in
RPMI-1640 medium (Invitrogen, Rockville, MD, USA) supplemented with
10% (v/v) fetal calf serum (FCS), 100 μg/ml streptomycin,
and 100 μg/ml penicillin (pH 7.2-7.4) in a humidified
incubator containing 5% CO2 at 37°C, and selected with 1
μg/ml puromycin for one week. We confirmed RGS22 expression
in the cells by western blotting.
We transfected pSilencer-Neg, pSilencer-484 and
pSilencer-3313 into BXPC3-LZR-RGS22 cells using an Invitrogen
transfection kit. We selected the cell lines using 200 μg/ml
G418 for two weeks. We confirmed RGS22 expression in the cells
(LZR-RGS22-Neg, LZR-RGS22-484, LZR-RGS22-3313) by western
blotting.
We transfected constructs, each containing human
Gα12 Q231L, Gα13 Q226L and Gαq Q209L in the pcDNA3.1 vector into
BXPC3-LZR-RGS22 cells using an Invitrogen transfection kit.
Western blotting
Lysates of BXPC3-LZR-GFP and BXPC3-LZR-RGS22 cells
or LZR-RGS22-Neg, LZR-RGS22-484 and LZR-RGS22-3313 cells were
subjected to sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and immunoblot analysis. Proteins were
transferred to 0.2-μm nitrocellulose membranes and blotted
with antibody to RGS22 (3) or the
control tubulin (Abcam, Cambridge, UK). We detected the proteins
using secondary antibodies (Beijing Zhongshan Biotechnology,
Beijing, China). Specific proteins were detected using an enhanced
chemiluminescence substrate; bands were analyzed with AlphaImager™
(Amersham, Piscataway, NJ, USA).
Wound-healing assay
Cells were cultured to confluence in 60-mm diameter
dishes and synchronized with serum-free medium for 24 h. A wound
was created by abrasion with a sterile pipette tip, and the cell
layer washed twice to remove non-adherent cells. Cells were
cultured in medium containing 10% FCS for 6 h, and wound healing
was visualized using a Nikon TE2000 microscope (Nikon, Melville,
NY, USA). The cell-free surface area was calculated using Zeiss
AxioVision v4.5 software (Carl Zeiss, Oberkochen, Germany). The
experiments were repeated three times for reproducibility. Data are
presented as the ratio of cell-covered surface area to initial
cell-free surface area.
Cell migration assay
We performed the cell migration assay using a
Transwell chamber with 8-μm pore, 24-mm wide polycarbonate
filters (Coaster, Cambridge, MA, USA). Cells (5×105)
were resuspended in serum-free culture medium, plated onto the
upper chamber, and allowed to migrate for 12 h through the filter
into the lower chamber. The bottom wells were filled with culture
medium supplemented with 10% FCS or 0.1% BSA. Non-migrating cells
were removed from the upper chamber with a cotton swab, the filters
were stained with hematoxylin, and the migrating cells adhering to
the underside of the filter counted in five random areas under a
light microscope. The experiments were repeated three times.
In vitro pull-down assay and
co-immunoprecipitation (Co-IP)
LZRsPBMN-Z-RGS22 BXPC-3 cells were sonicated and
lysed in 20 mM Tris-HCl (pH 7.5) containing 5 mM EDTA, 100 mM NaCl,
5 mM benzamine, 0.2% Triton X-100, and protease inhibitors for 1 h
on ice.
For the pull-down assay (13), the supernatant obtained by
centrifugation was incubated at 48°C for 1 h with recombinant
His6-fusion RGS22 prebound to nickel-Sepharose beads under
GDP−/aberrant lateral root formation
(ALF)4− or
GDP+/ALF4− conditions in
interaction buffer [20 mM Tris-HCl (pH 8.0), 2 mM MgSO4,
6 mM β-mercaptoethanol, 100 mM NaCl, 0.05% Triton X-100, 5%
glycerol]. Bound proteins were separated by 10% SDS-PAGE, and Gα
subunits were visualized using antibodies against GNA12 (S-20),
GNA13 (A-20) and GNA11 (C-19) (all from Santa Cruz Biotechnology)
and G protein subunits (Calbiochem, San Diego, CA, USA).
For Co-IP (13), the
supernatant obtained by centrifugation was incubated with
recombinant His6-fusion RGS22 under
GDP−/ALF4− or
GDP+/ALF4− conditions in
interaction buffer. The supernatant was then divided into two
fractions and immunoprecipitated with specific anti-GNA12,
anti-GNA13 and anti-GNA11 antisera, and the control antibody,
followed by incubation with protein G-Sepharose. The resulting
precipitates were subjected to immunoblot analysis with anti-RGS22
antibody.
Image analysis
BXPC3-LZR-GFP, BXPC3-LZR-RGS22, LZR-RGS22-Neg,
LZR-RGS22-484 or LZR-RGS22-3313 cells were plated at
2×105 cells in RPMI-1640 medium containing 10% FCS in a
6-well chamber slide (Lab-Tek Chambered Coverglass System; Nunc,
Rochester, NY, USA) and incubated for 24 h at 37°C. The medium was
changed to serum-free RPMI-1640 medium, and cells were starved for
17 h; this was set as point 1 (0 min). LPA (final concentration, 1
μmol/l) was warmed for 5 min and added to the wells for 5,
10, 20, 30, 40 and 80 min. Cells without LPA stimulation were used
as the control. Stimulation was terminated by 20-min fixation of
cells with 2% paraformaldehyde. Image capture was carried out using
an Axioskop 2 Plus Microscope (Carl Zeiss). LPA stimulation of
BXPC-3 cells led to a 'shrunken' morphology in a proportion of
cells.
If cell length was less than two times the width,
the cell was defined as having a 'shrunken' morphology (round). We
used the t-test for each of the two groups; P<0.05 was
considered significant. Cells were counted according to round or
stretched morphology in a blinded fashion for genotype and
treatment. Each group was treated in three wells. The experiments
were repeated three times.
Immunofluorescence staining for F-actin,
RGS22, GNA12, GNA13, GNAq and nuclear material
LPA-stimulated BXPC3-LZR-GFP, BXPC3-LZR-RGS22,
LZR-RGS22-Neg, LZR-RGS22-484 or LZR-RGS22-3313 cells (0, 1, 5, 10,
20, 30, 40 and 80 min) were permeabilized with 0.1% Triton X-100
for 1 min prior to staining. For BXPC3-LZR-GFP or BXPC3-LZR-RGS22
cells, Texas Red-X phalloidin (0.5 μM) was added into the
wells and incubated for 20 min at room temperature. After washing
three times with phosphate-buffered saline (PBS), cells were
stained with Hoechst (1 μg/ml; Molecular Probes, Eugene, OR,
USA) for 20 sec at room temperature. LZR-RGS22-Neg, LZR-RGS22-484
or LZR-RGS22-3313 cells were treated with 1:500 anti-RGS22 antibody
at 4°C overnight. After washing, cells were incubated for 1 h with
1:100 fluorescein isothiocyanate (FITC)-labeled goat anti-mouse
immunoglobulin G (IgG) (Beijing Zhongshan Biotechnology). After
washing with PBS, cells were stained with Texas Red-X phalloidin
and Hoechst as above. After washing four times with PBS, cells were
mounted with carbonate buffer solution containing 50% glycerol (pH
9.6).
BXPC3-LZR-RGS22 cells transfected by constructs each
containing human Gα12 Q231L, Gα13 Q226L and Gαq Q209L in the
pcDNA3.1 vector were incubated in RPMI-1640 medium containing 10%
FCS for 24 h at 37°C. The medium was changed to serum-free
RPMI-1640 medium and cells were starved for 17 h; this was set as
point 1 (0 min). LPA (final concentration, 1 μmol/l) was
warmed for 5 min and added to the wells for 5 sec and 5 and 10 min.
Cells without LPA stimulation were used as the control. Stimulation
was terminated by fixing cells with 2% paraformaldehyde for 20 min,
and then permeabilizing cells with 0.1% Triton X-100 for 5 min.
Following 30-min rehydration with 1% BSA at room temperature,
samples were incubated with antibody against GNA12, GNA13 or GNAq
(1:1,000 in PBS containing 1% BSA at a final concentration of 0.2
g/ml) overnight. After washing, samples were incubated for 1 h with
FITC-labeled secondary antibodies (Beijing Zhongshan
Biotechnology), washed twice, incubated with 0.5 μM Texas
Red-X phalloidin and Hoechst DNA stain, and then mounted as
described above. The stained slides were observed under an Axioskop
2 Plus microscope (Carl Zeiss). We used uniform exposure times
during photography to enable optimal comparison of fluorescence
among the test samples.
Results
RGS22 overexpression inhibits BXPC-3 cell
migration
Using immunohistochemical studies on a tissue
microarray consisting of 96 cancer tissue types, we previously
found that RGS22 is specifically expressed in adenocarcinoma and
squamous carcinoma (4). In the
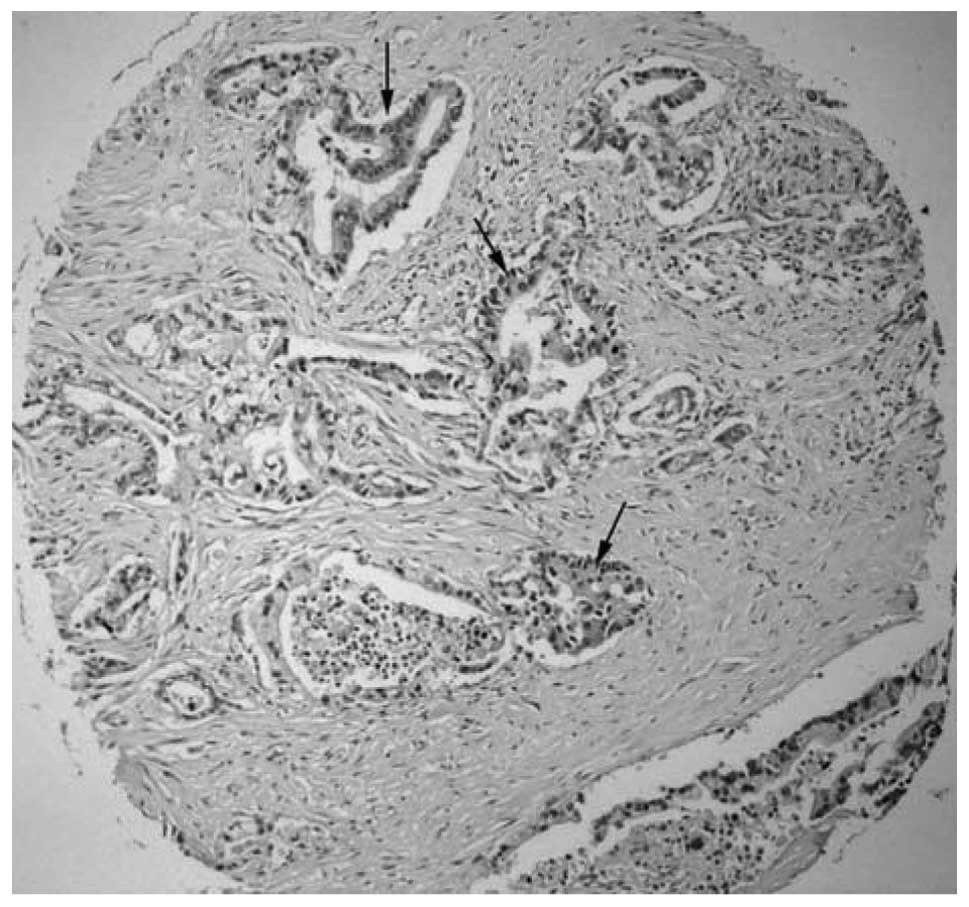
present study, RGS22 localized to the cytoplasm in pancreatic
adenocarcinoma tissues (Fig. 1).
Furthermore, RGS22, a novel cancer/testis (CT) antigen, acts as a
tumor suppressor, repressing epithelial cancer cell invasion and
migration.
To test the effect of RGS22 expression on pancreatic
adenocarcinoma cells, we overexpressed RGS22 in BXPC-3 cells. We
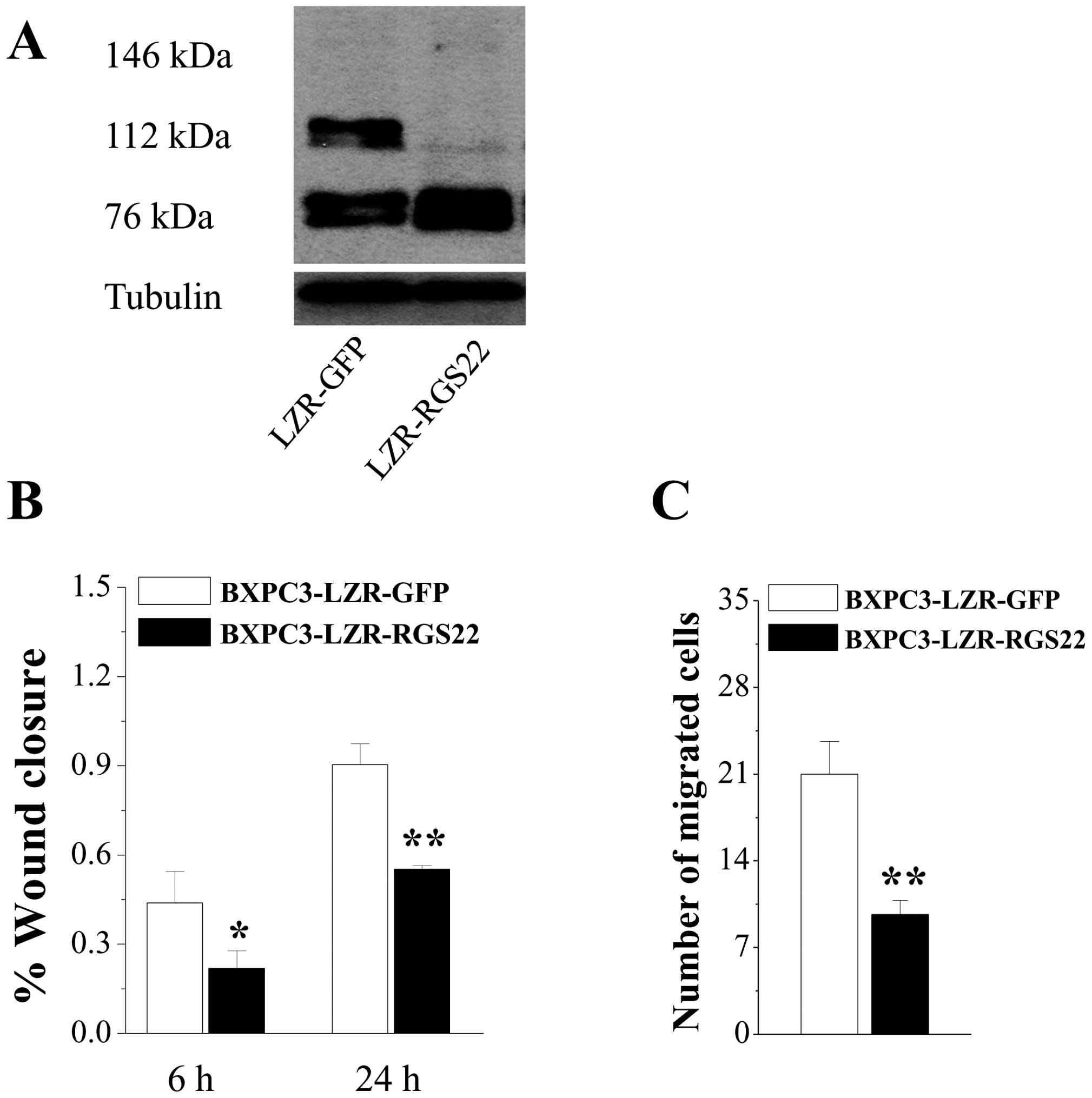
used GFP as the control (BXPC3-LZR-GFP). Western blotting revealed
146- and 76-kDa bands; RGS22-specific antibody was expressed at a
higher level in BXPC3-LZR-RGS22 cells (Fig. 2A). Cells that overexpressed RGS22
had much lower wound-healing rates than the control cells (Fig. 2B). Similarly, RGS22-overexpressing
cells exhibited greatly reduced migration when compared to the
control cells (Fig. 2C). Migration
decreased by >50% in RGS22-overexpressing cells when compared to
BXPC3-LZR-GFP cells. These results suggest that RGS22 expression
suppresses pancreatic adenocarcinoma cell migration.
RGS22 downregulation in BXPC3-LZR-RGS22
cells recovered cell migration
To confirm the effect of RGS22 expression on
pancreatic adenocarcinoma cells, we downregulated RGS22 expression
in BXPC3-LZR-RGS22 cells by RNA interference (RNAi) (LZR-RGS22-484
and LZR-RGS22-3313). LZR-RGS22-Neg cells were used as the control.
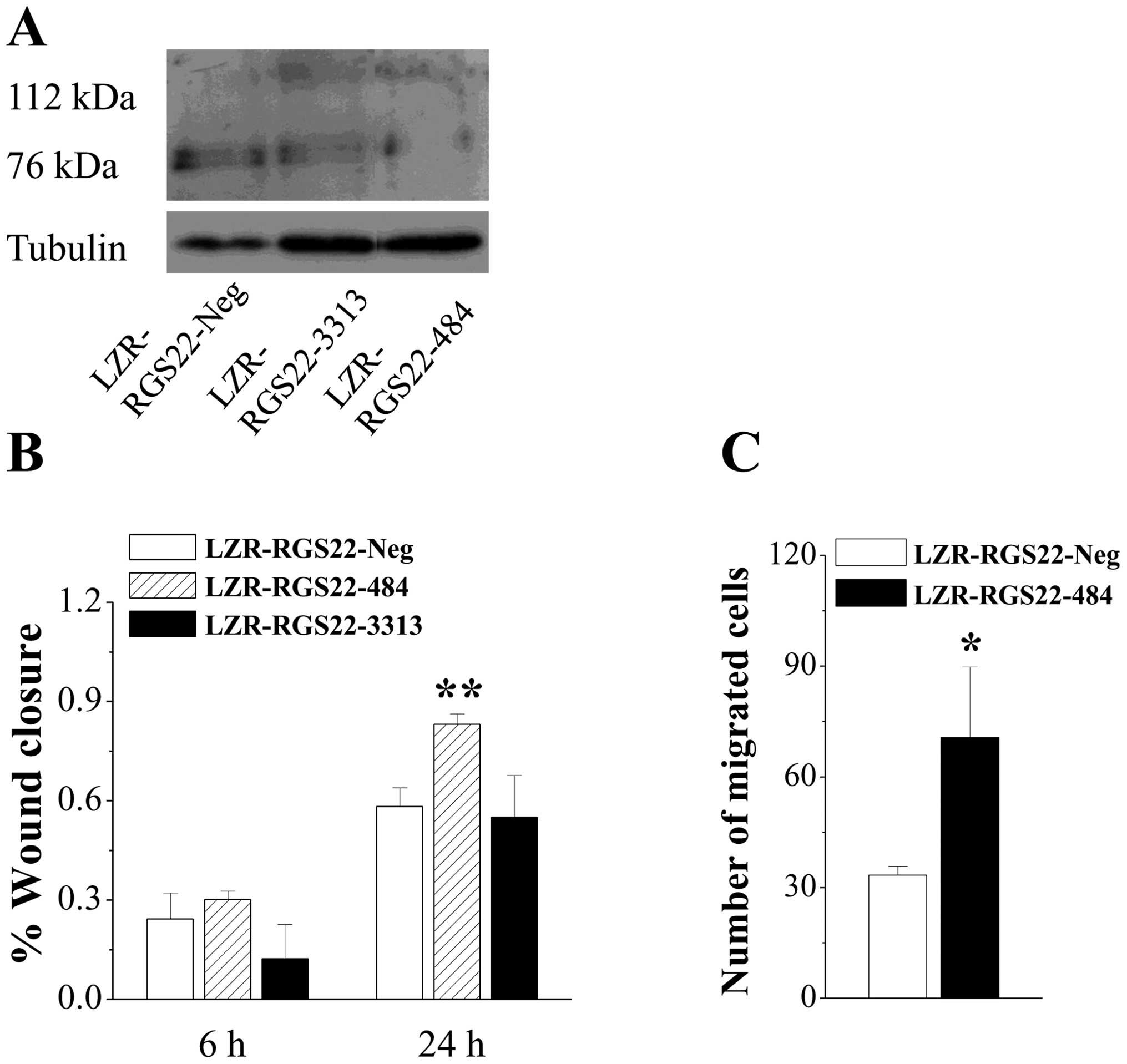
Western blotting revealed 76-kDa bands; RGS22-specific antibody was
expressed at a lower level in BXPC3-LZR-RGS22-3313 cells and at the
lowest level in BXPC3-LZR-RGS22-484 cells (Fig. 3A).
BXPC3-LZR-RGS22-484 cells, in which RGS22 expression
was more greatly downregulated, had higher wound-healing rates at
24 h than the control cells and BXPC3-LZR-RGS22-3313 cells
(Fig. 3B); the BXPC3-LZR-RGS22-3313
cells had similar wound-healing rates to the control cells.
BXPC3-LZR-RGS22-484 cell migration was greatly increased compared
to the control cells (Fig. 3C).
BXPC3-LZR-RGS22-484 cell migration was increased by >100%
compared to that of BXPC3-LZR-RGS22-Neg cells. These results
confirm that RGS22 expression suppresses pancreatic adenocarcinoma
cell migration.
RGS22 interaction with GNA12/13 in BXPC-3
cells
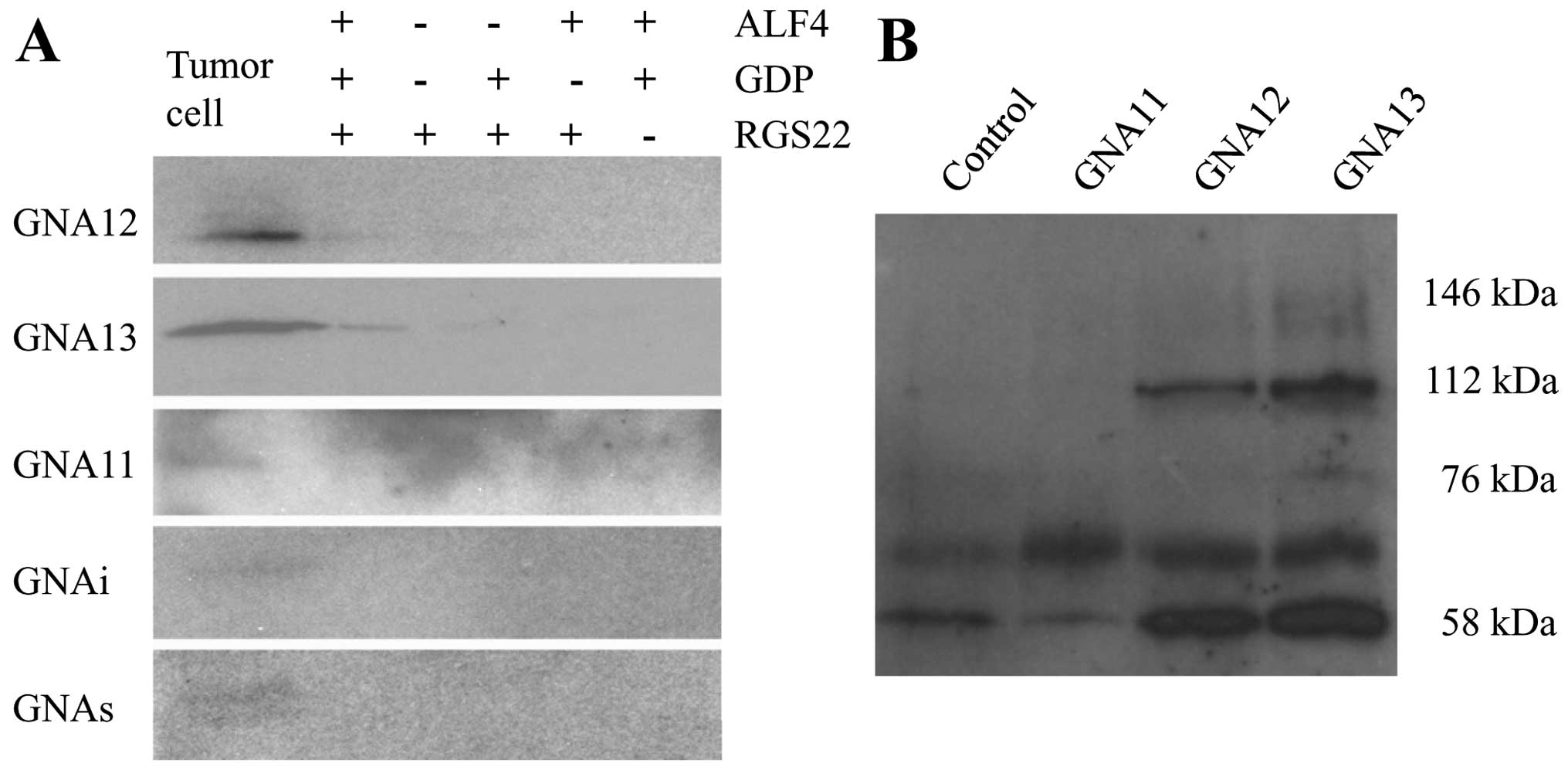
To determine whether RGS22 interacts directly with
Gα subunits in tumor cells, BXPC-3 cell lysates were incubated with
fusion proteins containing His6 immobilized on nickel-Sepharose
beads in the presence or absence of GDP and
ALF4−, which mimics the transition state of
the Gα subunit. RGS22 bound specifically to GNA12 and GNA13 in the
cells under GDP-ALF4− conditions (Fig. 4A).
We confirmed the specificity of RGS22 interaction
with GNA12 and GNA13 by incubating BXPC-3 cell lysates with the
corresponding bead-bound antibodies under the
GDP-ALF4− condition. The 146-, 112- and
76-kDa forms of RGS22 mainly bound to GNA13, while the 112-kDa form
of RGS22 bound primarily to GNA12. RGS22 did not bind to GNAq/11.
The 59-kDa form of RGS22 may have been masked by the heavy chain of
IgG (Fig. 4B). These data indicate
that RGS22 has specific interactions with GNA12 and GNA13 in BXPC-3
cells.
RGS22 inhibits LPA-activated pancreatic
cancer cell morphological changes and F-actin formation
LPA is a small bioactive phospholipid produced by
some cancer cells (14). It is
involved in a variety of physiological and pathophysiological
responses, including wound healing and tumor cell invasion and
metastasis (15). Several G
protein-coupled receptors mediate the biological functions of LPA.
LPA interacts with receptors LPA1-5, which in turn activate the Gi,
Gq, Gs/h and G12/13 proteins to elicit multiple biological
responses.
To characterize the RGS22-induced decrease in BXPC-3
cell migration, we examined the morphology of LPA-stimulated live
cells. We used BXPC3-LZR-RGS22 cells stably transfected with
full-length human RGS22 or GFP-tagged BXPC3-LZR-GFP cells. The
control BXPC3-LZR-GFP cells began to appear rounded 5 min after LPA
stimulation, while BXPC3-LZR-RGS22 cells began to shrink 20 min
after LPA stimulation. The images obtained showed that the control
cells achieved the maximum rate of cell deformation 20 min after
LPA stimulation. However, the BXPC3-LZR-RGS22 cells achieved the
maximum rate of cell deformation only after 30 min (Fig. 5A and B). Downregulating RGS22
expression reversed its inhibitory effect on BXPC3-LZR-RGS22-484
cells compared to the BXPC3-LZR-RGS22-Neg cells. The reaction began
at 5 min, and the time taken to achieve the maximum deformation
cell ratio was 20 min (Fig. 5A and
C). So, the higher the RGS22 expression, the later the cell
deformation after LPA stimulation.
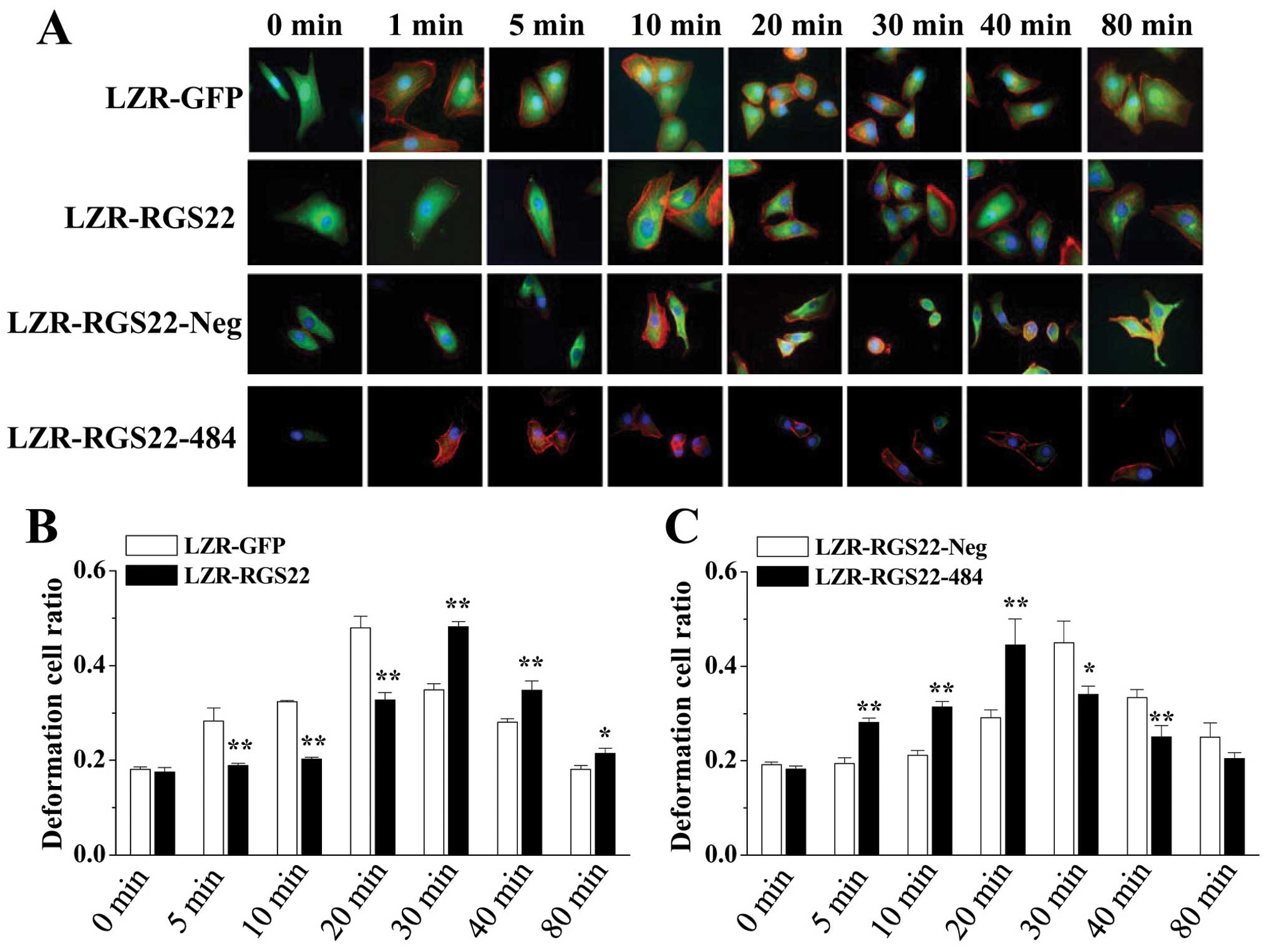 | Figure 5RGS22 inhibits LPA-activated
pancreatic cancer cell morphological changes and F-actin formation.
(A) RGS22 delayed LPA-stimulated F-actin formation and cell
deformation. LPA-activated F-actin formation in BXPC3-LZR-GFP
cells. F-actin (red) began to form thick fibers 1 min after LPA
stimulus; GFP (green) was expressed throughout the cell; Hoechst
stained the nuclei blue (magnification, ×1,000). Cells start to be
round after 5 min LPA stimulation. While, the F-actin (red)
reaction began 5 min after LPA stimulus in RGS22 (green, anti-RGS22
antibody) high expressed BXPC3-LZR-RGS22 cells and cells begin to
shrink 20 min after stimulation. F-actin (red) began to form thick
fibers 10 min after LPA stimulus in LZR-RGS22-Neg cells. RGS22
(green) is expressed in the cytoplasm; Hoechst stained the nuclei
blue (magnification, ×1,000). Cells start to be round after 20 min
LPA stimulation While, F-actin (red) began to form thick fibers in
1 min, in LZR-RGS22-484 cells after LPA stimulus and cells begin to
shrink 5 min after stimulation. (B) RGS22 inhibition of
LPA-activated pancreatic cancer cell morphological changes. y-axis,
ratio of deformation cells. RGS22 delayed LPA-stimulated cell
deformation. The reaction began 5 min after LPA stimulus in
BXPC3-LZR-GFP cells and 20 min in BXPC3-LZR-RGS22 cells. Maximum
cell deformation rates after LPA stimulus occurred after 20 min in
BXPC3-LZR-GFP cells and 30 min in BXPC3-LZR-RGS22 cells. Each of
the two groups was analyzed by t-test. *P<0.05 and
**P<0.01. (C) Downregulated RGS22 accelerates
LPA-activated pancreatic cancer cell morphological changes. y-axis,
ratio of deformation cells. Cell deformation reaction began 20 min
after LPA stimulus in LZR-RGS22-Neg cells and 5 min in RGS22
downregulated LZR-RGS22-484 cells. Maximum cell deformation rates
after LPA stimulus occurred after 30 min in LZR-RGS22-Neg cells and
20 min in LZR-RGS22-484 cells. Each of the two groups was analyzed
by t-test. *P<0.05 and **P<0.01. |
The actin cytoskeleton has been implicated in the
management of cell shape and migration activity (16). To determine whether RGS22 involves
LPA-mediated F-actin polymerization, we triple-stained cells with
Hoechst (stains nuclei blue), RGS22-specific antibody (control
cells were tagged with GFP, green), and Texas Red-X phalloidin
(stains F-actin red). LPA induced the formation of a dense and
organized network of thick, parallel F-actin stress fibers starting
at 1 min in BXPC3-LZR-GFP cells (Fig.
5A). In BXPC3-LZR-RGS22 cells, in which RGS22 expression was
higher, LPA-induced F-actin stress fiber formation was delayed to
10 min (Fig. 5A). Downregulating
RGS22 expression recovered its inhibitory effect on
BXPC3-LZR-RGS22-484 cells compared to BXPC3-LZR-RGS22-Neg cells
(Fig. 5A). LPA-induced F-actin
stress fiber formation was delayed to 10 min in BXPC3-LZR-RGS22-484
cells. Therefore, our studies demonstrate that higher RGS22
expression delays LPA-induced F-actin formation.
Effect of constitutive activation of Gα
subunits by RGS22 on LPA-activated morphological changes and
F-actin formation
We used LZR-RGS22 BXPC-3 cells transiently
transfected with constructs containing human Gω12 Q231L, Gα13 Q226L
or Gαq Q209L to test whether RGS22 inhibits F-actin formation
through the Gα12, Gα13 or Gαq subunit. These mutations reduce
guanosine triphosphatase (GTPase) activity, resulting in a
constitutively active phenotype with or without RGS proteins. That
is, RGS proteins bind to constitutively active Gα subunits without
accelerating GTP hydrolysis to GDP. We did not investigate the
function of RGS proteins as negative regulators of heterotrimeric G
protein signaling.
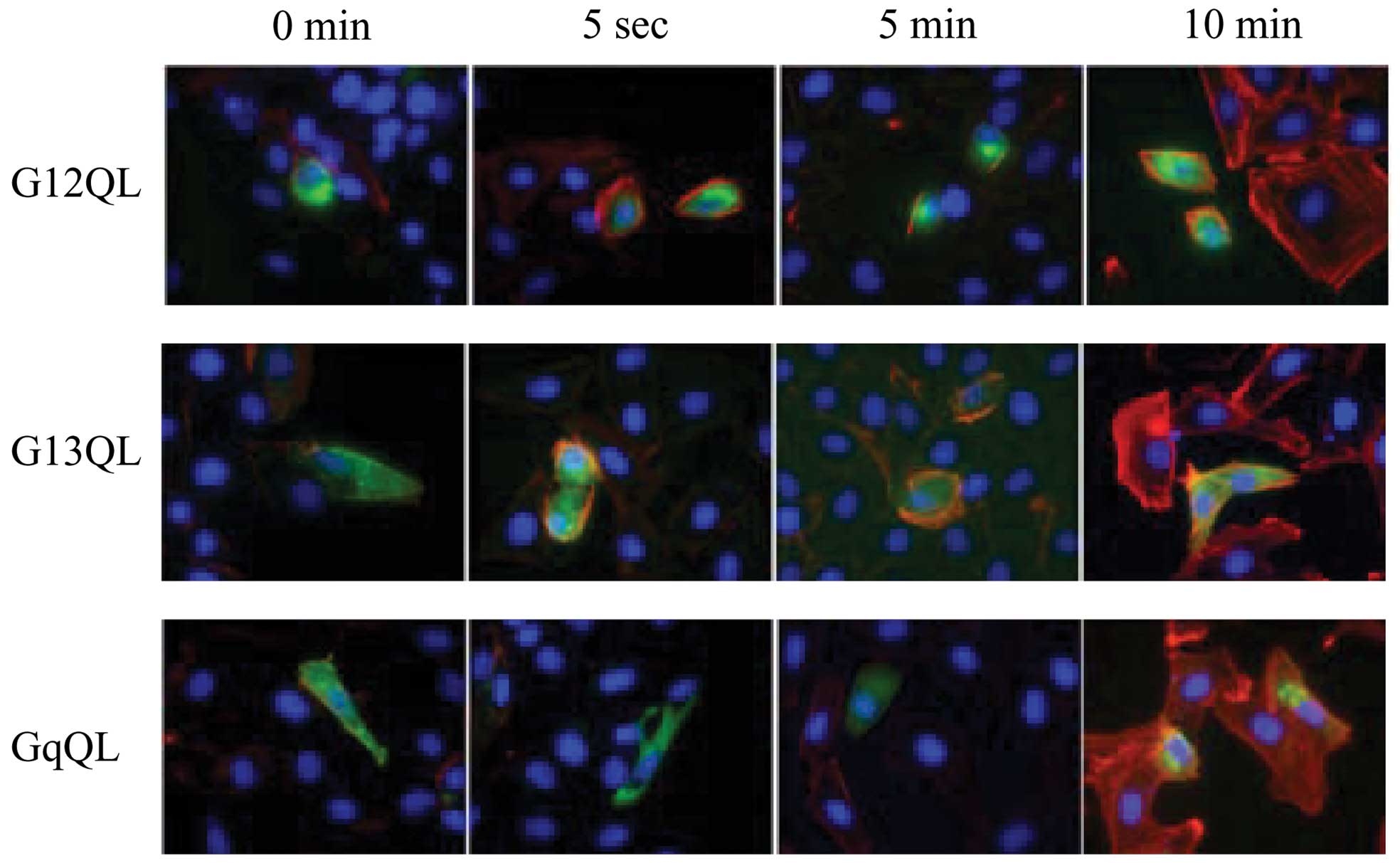
Cells were triple stained with Hoechst (blue), Gα
subunit-specific antibody (green), and Texas Red-X phalloidin.
Cells transfected with Gα12 Q231L and Gα13 Q226L began to appear
rounded and F-actin stress fibers began to form 5 sec after LPA
stimulation. F-actin stress fiber formation in untransfected cells
and Gαq Q209L-transfected cells (Fig.
6) began 10 min after LPA stimulation.
Discussion
Several features of pancreatic cancer are
responsible for the high mortality rate; it is difficult to detect
precursor lesions or notice symptoms until the disease is in the
advanced stages (17). Pancreatic
cancer often undergoes micrometastasis, which is responsible for
its poor prognosis. In a previous study, we showed that RGS22
localized to the cytoplasm in pancreatic adenocarcinoma tissues,
indicating its association with pancreatic adenocarcinoma. We also
found that RGS22 expression is higher in cancers without lymph node
metastasis than in cancers with lymph node metastasis. RGS22 can
inhibit tumor metastasis. To confirm this, we used the human
pancreatic cancer cell line BXPC-3 to test the association of RGS22
with pancreatic cancer cell migration. BXPC3-LZR-RGS22 cells, with
higher RGS22 expression, had lower wound-healing rates and
migration than BXPC3-LZR-GFP control cells. Conversely,
downregulating RGS22 in BXPC3-LZR-RGS22-484 cells resulted in
higher wound-healing rates and migration than in
BXPC3-LZR-RGS22-Neg control cells. Therefore, RGS22 is inhibitory
toward human pancreatic cancer cell migration.
Several classes of proteins are involved in cell
migration and metastasis. GNA12 signaling promotes cell migration
in prostate (9) and breast cancer
(10), and in LPA-induced ovarian
cancer cells in vitro (11).
In these studies, Rho activation appeared critical to the effect of
GNA12 in promoting invasion. GNA12 signaling promoting metastasis
takes place via the stimulation of cells to become invasive within
the primary tumor. GNA13 also stimulates cell migration in addition
to inducing oncogenic transformation (12). Via pull-down and Co-IP assays, we
found that RGS22 interacted with GNA12/13.
Increased cell migration is an acquired feature of
metastatic cancer cells. Cell migration is an important component
in the spread of pancreatic cancer (18). In the early stages of the disease,
cancer spread is believed to occur after tumor cells infiltrate the
peritoneal cavity and gain access to the blood vessels. Thus,
studying the migratory abilities of pancreatic cancer cells is
necessary for providing insight into the biological processes that
mediate metastasis. One of the simplest natural phospholipids, LPA
is a lipid mediator that evokes hormone- and growth factor-like
responses in almost every cell type. LPA is an efficacious
chemoattractant of human pancreatic carcinoma cells (19). In the malignant ascites from
pancreatic cancer patients, LPA is an active component that
stimulates pancreatic cancer cell migration and invasion. We used
LPA as a stimulator to investigate the possible mechanism of RGS22
in pancreatic cancer cell migration. LPA stimulation led to altered
cell morphology; BXPC3-LZR-RGS22 cells began to appear rounded and
achieved the maximum rate of cell deformation more quickly after
stimulus than control cells. Furthermore, the downregulated RGS22
expression in BXPC3-LZR-RGS22-484 cells recovered the inhibitory
effect of RGS22 compared to BXPC3-LZR-RGS22-Neg cells.
Migrating cells are crucial for accurate
organization of the actin cytoskeleton and the migratory response
of pancreatic carcinoma cells (20). To determine whether RGS22 is
involved in LPA-mediated F-actin polymerization, we triple-stained
the cells. In the high-RGS22 expression BXPC3-LZR-RGS22 cells,
F-actin stress fiber formation was delayed compared to
BXPC3-LZR-GFP cells. The inhibitory effect of RGS22 was recovered
by downregulating RGS22 expression in BXPC3-LZR-RGS22-484 cells
compared to BXPC3-LZR-RGS22-Neg cells. Therefore, our studies
demonstrate that higher RGS22 expression delays LPA-induced F-actin
formation.
The G12/13 G protein family has been implicated in
various cellular processes, such as Rho-mediated organization of
the cytoskeleton, and subsequently, cell shape (21). G12/13-mediated stress fiber
formation is important for cell motility. The complex interplay of
actin polymerization orchestrates cell movement. We inhibited RGS22
signaling by expressing constitutively active GNA12 and GNA13 (but
not GNAq) in BXPC3-LZR-RGS22 cells. These results further support
the notion of RGS22 signaling, whereby RGS22 couples to GNA12/13,
which in turn inhibits stress fiber formation.
In conclusion, our data demonstrate that RGS22 acts
as a tumor suppressor, repressing human pancreatic adenocarcinoma
cell migration by coupling to GNA12/13, which in turn leads to the
inhibition of stress fiber formation. RGS22 may be a promising
therapeutic target for tumor metastasis pathway intervention.
Acknowledgments
This study was supported by grants from the Six
Talent Peaks Project of Jiangsu Province of China (no.
2013-WSN-021), the 333 Project of Jiangsu Province of China, the
Key Medical Talents of Yangzhou City, the National Natural Science
Foundation of China (NSFC) (no. 81200127), and the Natural Science
Foundation of Jiangsu Province of China (no. BK20131230).
References
|
1
|
Bardeesy N, Sharpless NE, DePinho RA and
Merlino G: The genetics of pancreatic adenocarcinoma: A roadmap for
a mouse model. Semin Cancer Biol. 11:201–218. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang XJ, Ye H, Zeng CW, He B, Zhang H and
Chen YQ: Dysregulation of miR-15a and miR-214 in human pancreatic
cancer. J Hematol Oncol. 3:462010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hu Y, Xing J, Chen L, Guo X, Du Y, Zhao C,
Zhu Y, Lin M, Zhou Z and Sha J: RGS22, a novel testis-specific
regulator of G-protein signaling involved in human and mouse
spermiogenesis along with GNA12/13 subunits. Biol Reprod.
79:1021–1029. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hu Y, Xing J, Wang L, Huang M, Guo X, Chen
L, Lin M, Zhou Y, Liu Z, Zhou Z, et al: RGS22, a novel
cancer/testis antigen, inhibits epithelial cell invasion and
metastasis. Clin Exp Metastasis. 28:541–549. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fokas E, Engenhart-Cabillic R, Daniilidis
K, Rose F and An HX: Metastasis: The seed and soil theory gains
identity. Cancer Metastasis Rev. 26:705–715. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liotta LA, Steeg PS and Stetler-Stevenson
WG: Cancer metastasis and angiogenesis: An imbalance of positive
and negative regulation. Cell. 64:327–336. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Orr FW, Wang HH, Lafrenie RM, Scherbarth S
and Nance DM: Interactions between cancer cells and the endothelium
in metastasis. J Pathol. 190:310–329. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu J and Verkman AS: Increased migration
and metastatic potential of tumor cells expressing aquaporin water
channels. FASEB J. 20:1892–1894. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kelly P, Stemmle LN, Madden JF, Fields TA,
Daaka Y and Casey PJ: A role for the G12 family of heterotrimeric G
proteins in prostate cancer invasion. J Biol Chem. 281:26483–26490.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kelly P, Moeller BJ, Juneja J, Booden MA,
Der CJ, Daaka Y, Dewhirst MW, Fields TA and Casey PJ: The G12
family of heterotrimeric G proteins promotes breast cancer invasion
and metastasis. Proc Natl Acad Sci USA. 103:8173–8178. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bian D, Mahanivong C, Yu J, Frisch SM, Pan
ZK, Ye RD and Huang S: The G12/13-RhoA signaling pathway
contributes to efficient lysophosphatidic acid-stimulated cell
migration. Oncogene. 25:2234–2244. 2006. View Article : Google Scholar
|
|
12
|
Radhika V, Onesime D, Ha JH and
Dhanasekaran N: Galpha13 stimulates cell migration through
cortactin-interacting protein Hax-1. J Biol Chem. 279:49406–49413.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mao H, Zhao Q, Daigle M, Ghahremani MH,
Chidiac P and Albert PR: RGS17/RGSZ2, a novel regulator of Gi/o,
Gz, and Gq signaling. J Biol Chem. 279:26314–26322. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Choi KU, Yun JS, Lee IH, Heo SC, Shin SH,
Jeon ES, Choi YJ, Suh DS, Yoon MS and Kim JH: Lysophosphatidic
acid-induced expression of periostin in stromal cells: Prognoistic
relevance of periostin expression in epithelial ovarian cancer. Int
J Cancer. 128:332–342. 2011. View Article : Google Scholar
|
|
15
|
Yatomi Y, Igarashi Y, Yang L, Hisano N, Qi
R, Asazuma N, Satoh K, Ozaki Y and Kume S: Sphingosine 1-phosphate,
a bioactive sphingolipid abundantly stored in platelets, is a
normal constituent of human plasma and serum. J Biochem.
121:969–973. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Roffers-Agarwal J, Xanthos JB and Miller
JR: Regulation of actin cytoskeleton architecture by Eps8 and Abi1.
BMC Cell Biol. 6:362005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yadav D and Lowenfels AB: The epidemiology
of pancreatitis and pancreatic cancer. Gastroenterology.
144:1252–1261. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Doi Y, Yashiro M, Yamada N, Amano R, Noda
S and Hirakawa K: VEGF-A/VEGFR-2 signaling plays an important role
for the motility of pancreas cancer cells. Ann Surg Oncol.
19:2733–2743. 2012. View Article : Google Scholar
|
|
19
|
Yamada T, Sato K, Komachi M, Malchinkhuu
E, Tobo M, Kimura T, Kuwabara A, Yanagita Y, Ikeya T, Tanahashi Y,
et al: Lysophosphatidic acid (LPA) in malignant ascites stimulates
motility of human pancreatic cancer cells through LPA1. J Biol
Chem. 279:6595–6605. 2004. View Article : Google Scholar
|
|
20
|
Hage B, Meinel K, Baum I, Giehl K and
Menke A: Rac1 activation inhibits E-cadherin-mediated adherens
junctions via binding to IQGAP1 in pancreatic carcinoma cells. Cell
Commun Signal. 7:232009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Offermanns S: G-proteins as transducers in
transmembrane signalling. Prog Biophys Mol Biol. 83:101–130. 2003.
View Article : Google Scholar : PubMed/NCBI
|