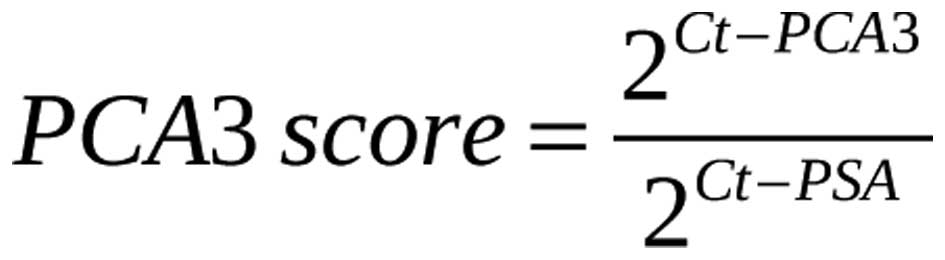Introduction
Prostate cancer remains challenging to clinical
oncologists due to the fact that it has the highest incidence and
second highest mortality rate among all cancer types for males in
developed countries (1–3). In China, the incidence of prostate
cancer is only lower than malignant tumors of the lung, stomach,
esophagus, colon/rectum and liver (4). However, the incidence rate of prostate
cancer is rising significantly. It is expected that the incidence
of prostate cancer in China will reach 40/100,000 males in 2020,
with annual new cases estimated at 350,000.
Prostate cancer is among the few cancers that can be
screened and detected at an early stage (5). Prostate-specific antigen (PSA) has
been widely accepted for prostate cancer screening (6–9). As a
protein secreted only inside the prostate glands (10), its presence in blood/serum is
trivial for healthy subjects. An abnormally high serum PSA level is
evidence of a prostate lesion or a disorder such as prostate
cancer. However, the application of PSA for prostate cancer
diagnosis remains controversial. The debates are mainly focused on
the poor sensitivity and specificity and the resulting
over-diagnosis (11).
An alternative for PSA screening is prostate cancer
antigen 3 (PCA3). PCA3 is a non-coding RNA expressed inside the
prostate gland (12).
Overexpression of PCA3 is common among prostate cancer patients
(13–17). PCA3 is reported to be more
cancer-specific, and thus a promising next-generation marker for
prostate cancer screening (18–21).
With higher specificity than PSA, PCA3 is expected to play more
important roles in clinical practice such as prostate cancer
screening, grading and recurrence monitoring (22). However, the quantification of PCA3
remains difficult due to poor RNA stability and lack of a
purification method from complex matrix.
Nanotechnology has highly influenced biomedical
research and clinical laboratory practice (23–25).
Magnetic beads are nano-sized or micro-sized ferromagnetic iron
oxide particles. Functionalized magnetic beads coated with
antibodies or oligonucleotides have been widely used for the
purification, extraction and detection of biomolecules (26,27).
We herein report a general method for RNA extraction based on
magnetic beads and capturer strands, and its application for
prostate cancer screening and diagnosis. As shown in Fig. 1, magnetic beads covalently
conjugated to poly-T strands were incubated with the capturing
strand (capturer) which involves poly-A and a sequence that is
complementary to part of the target sequence (e.g., PSA or PCA3).
The capturer is then immobilized on the magnetic beads through A-T
pairing. The beads are incubated with the sample, through which the
target sequences are captured by hybridization. Finally the beads
with target immobilization are precipitated by magnetic attraction
and carefully washed so that the interference sequences are removed
with washing buffer. The target strand is eluted by
heat-denaturation at an appropriate temperature.
Patients and methods
Patients and sample acquisition
Prostate cancer cell line 22RV1 (Institute of
Biochemistry and Cell Biology CAS, cat. TCHu100) was cultured in
RPMI-1640 media that contained bovine serum (100 ml/l) (both from
Hyclone) and penicillin (100 U/ml; Sigma-Aldrich) at 37°C and with
50 ml/l CO2. Female urine samples were collected in an
RNAse-free vial. Patients enrolled in the study included 34
pathologically diagnosed primary prostate cancer patients and 18
patients with benign prostatic hyperplasia (BPH). All patients were
enrolled at Peking Union Medical College Hospital. For males, urine
samples of 20–30 ml were collected after prostate massage (28). Urine samples from both male and
female participants were aliquoted; an aliquot of 5 ml urine was
mixed with storage buffer (100 mM Tris-HCl, 500 mM LiCl, 10 mM
EDTA, pH 7.5, 1% lithium dodecyl sulfate, 5 mM dithiothreitol) (all
chemicals from Sigma-Aldrich) in a 1:1 ratio. RNA extraction
occurred within 4 h after sample collection. Urine samples were
stored at 4°C for short term storage or at −80°C for long term
storage.
The study was approved by the Ethics Committee of
Peking Union Medical College Hospital. All participants provided
their written informed consent to participate in this study.
Phenol-chloroform extraction of RNA
The untreated female urine sample (0.5 ml with the
addition of 104 22RV1 cells) was centrifuged for 5 min
(3,500 rpm, 4°C); the supernatant was discarded. The sample was
mixed with 1 ml TRIzol (Life Technologies) and incubated for 5 min
at ambient temperature. The mixture was added with 200 µl
chloroform (Sigma-Aldrich), vortexed for 15 sec and incubated for 3
min at ambient temperature. The mixture was centrifuged for 15 min
(13,000 rpm, 4°C). The aqueous layer was transferred to an
RNAse-free vial and mixed with isopropanol of the same volume. The
mixture was gently shaken and incubated for 10 min. The mixture was
centrifuged for 15 min (13,000 rpm, 4°C), the supernatant was
discarded, 50 µl of 70% ethanol was added and the vial was
gently shaken. The mixture was centrifuged for another 5 min
(13,000 rpm, 4°C), and the supernatant was discarded. The vial was
dried with a vacuum and the vial was rehydrated with 500 µl
DEPC-treated RNAse-free water. The RNA solution was stored at
−20°C.
RNA extraction by affinity column
(SurePre Urine Exfoliated Cell RNA Purification kit by Thermo
Fisher)
The untreated female urine sample (0.5 ml with the
addition of 104 22RV1 cells) was centrifuged for 5 min
(3,500 rpm, 4°C); the supernatant was discarded. The sample was
mixed with 500 µl TRK buffer, and mixed by pipetting. The
sample was centrifuged for 3 min (≥14,000 rpm, ambient
temperature). An equal volume of anhydrous ethanol was added and
mixed by vortexing for 20 sec. The mixture was transferred to the
affinity column, followed by centrifugation for 1 min (10,000 rpm,
ambient temperature), and the effluent was discarded. RNA wash
buffer I (300 µl) was added, followed by centrifugation for
1 min (10,000 rpm, ambient temperature), and the effluent was
discarded. RNA wash buffer II (500 µl) was added, followed
by centrifugation for 30 sec (10,000 rpm, ambient temperature), and
the effluent was discarded; this step was repeated twice. The
column was dried at an ambient temperature for 2 min. The column
was transferred to a new 1.5 ml Ep tube, and 50 µl of
DEPC-treated RNAse-free water was added to the column to elute RNA
by resting for 2 min and centrifugation for 1 min (10,000 rpm,
ambient temperature). The RNA solution was stored at −20°C.
RNA extraction by Mag-Cap
Poly-T (T25) functionalized magnetic
beads (10–20 mg; NEB) were washed twice with 100 µl
hybridization buffer (20 mM Tris, pH 7.5, 100 mM NaCl). The beads
were incubated with 100 µl hybridization buffer that
contained the capturer strand (1.5 µM, PCA3 capturer: 5′-ATC
TGT TTT CCT GCC CAT CCT TTA AGT TTA (A)30; PSA capturer:
5′-CGA ACT TGC GCA CAC ACG TCA TTG GAT TTA (A)30. The
beads were then mixed with 100 µl hybridization buffer and
100 µl treated urine sample. The vial was shaken to ensure
full mixing and the mixture was heated for 30 min (62±1°C). The
vial was cooled to ambient temperature for 30 min, and the magnetic
beads were collected by magnetic attraction. The beads were washed
for three times with 100 µl elution buffer (10 mM Tris, pH
7.5). The beads were mixed with 50 µl elution buffer, which
were incubated at 70°C for 5 min, and the solution was collected
and subjected to RNA quantification.
RNA extraction by Mag-Bind
The procedure was the same as the Mag-Cap process
only the capturer strand was not contained.
RNA quantification
Purified RNA was converted to cDNA through
reverse-transcription using a commercial kit (Fermentas). The
quantification of cDNA was realized by qPCR. The primers and probe
sequences (Life Technologies) are listed as following: primers for
PCA3: forward, 5′-CCAGGAAGATCTGCATGGTGGG-3′ and reverse,
5′-GATGACCCAAGATGGCGGC-3′; probe for PCA3:
FAM-5′-GCACAGAGATCCCTGGGAGAAATGCC-TAMRA; primers for PSA:
forward, 5′-CCT GCT CGG GTG ATT CTG-3′ and reverse, 5′-GCC ACG ATG
GTG TCC TTG AT-3′; probe for PSA: FAM-5′-GGG CCC ACT TGT CTG
TAA TGG TGT GC-TAMRA. The PCR mixture was prepared using 2
µl 10X PCR buffer (Roche), 4 µl MgCl2 (25
mM; Sigma-Aldrich), 4 µl dNTP solution (2.5 mM; Fermentas),
0.5 µl primer solutions (10 µM), 0.2 µl probe
solution (10 µM), 0.5 µl Taq polymerase
(Roche), 2 µl cDNA solution and water to make the mixture 20
µl. One PCR cycle consisted of 95°C for 15 sec and 60°C for
30 sec. Forty-five cycles were run for each PCR reaction. PCA3
scores were calculated using the following equation:
where Ct-PCA3 is the Ct value during the amplification of the cDNA
of PCA3; Ct-PSA is the Ct value during the amplification of the
cDNA of PSA.
Statistical analysis
We used significance testing to compare the
performance of Mag-Cap with the other methods. The power of PCA3
score for prostate cancer detection was evaluated using receiver
operating characteristic (ROC) analysis. All statistical analysis
was realized on SPSS (version 13.0).
Results
Comparison of Mag-Cap with other
methods
We first evaluated Mag-Cap using artificial samples
(prostate cancer cell line 22RV1 in 10 urine samples from healthy
female subjects enrolled at Peking Union Medical College Hospital).
The mRNA of PSA was extracted by phenol-chloroform method, affinity
column (SurePre Urine Exfoliated Cell RNA Purification kit; Thermo
Fisher) or Mag-Cap. The RNA molecules were quantified by
quantitative PCR (qPCR) after reverse transcription, and Ct values
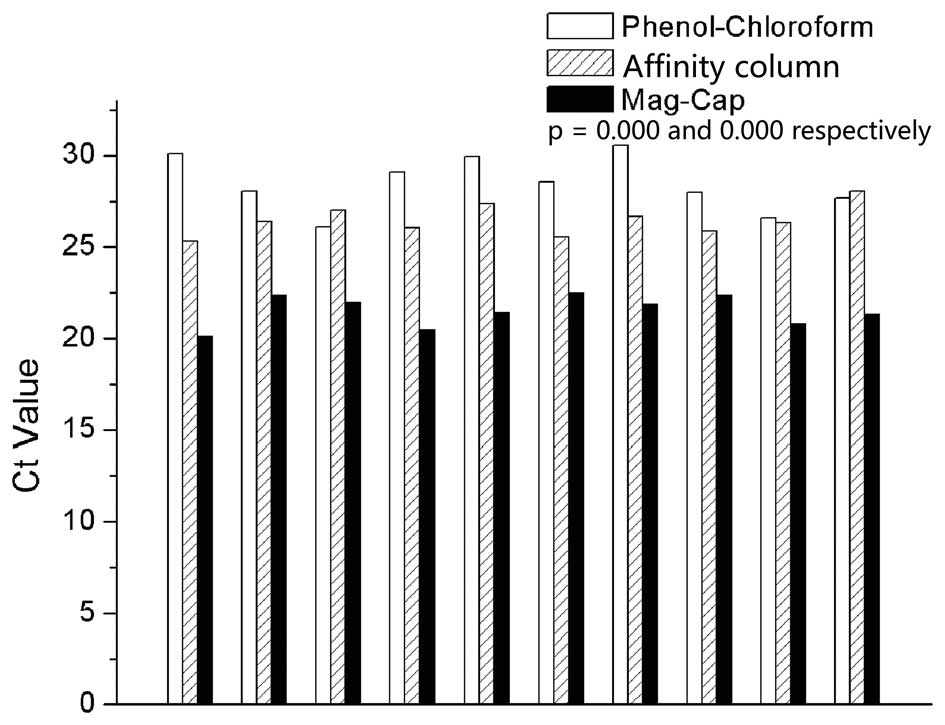
are presented in Fig. 2. The
medians of Ct values for phenol-chloroform extraction, affinity
column and Mag-Cap were 28.3, 26.4 and 21.6, respectively. The
difference between Mag-Cap and the other two extraction methods
achieved statistical significance.
Comparison with a commercial kit
PCA3 in the 10 urine samples mentioned in the
previous section was extracted by both Mag-Cap and a commercial
magnetic bead-based RNA extraction kit (Mag-Bind mRNA kit; Omega
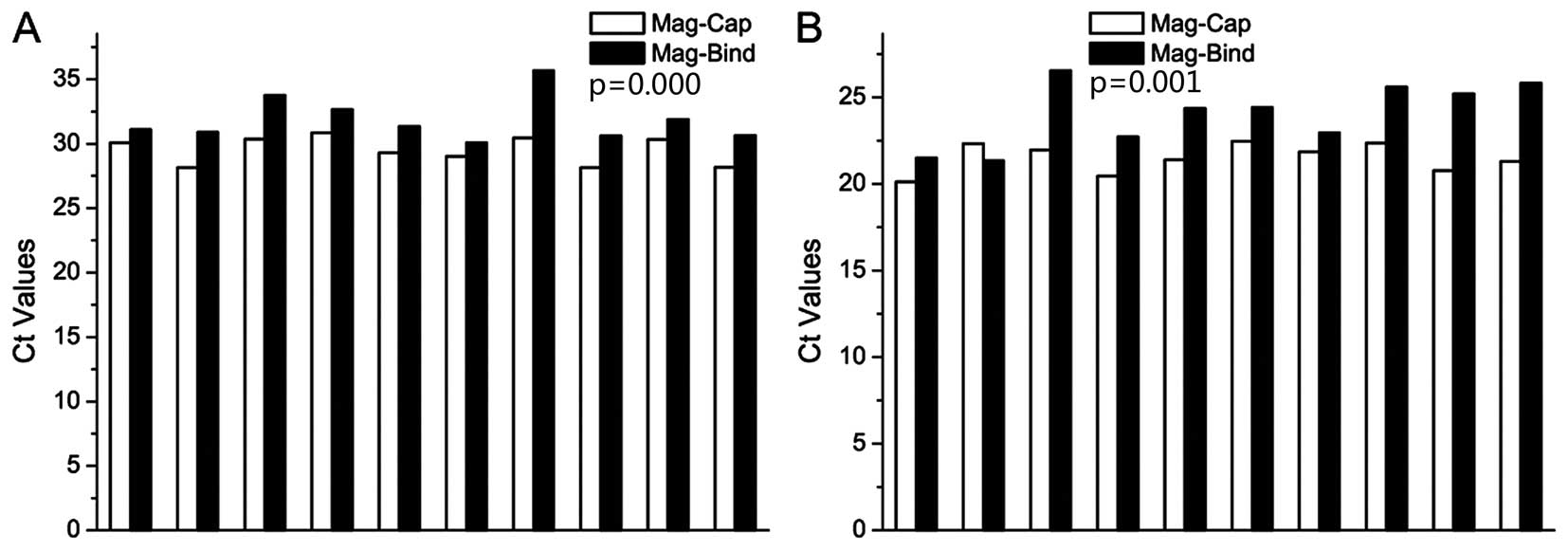
Bio-Tek), and then quantified by qPCR. The results are shown in
Fig. 3A. Medians of the Ct values
of Mag-Cap and the Mag-Bind kit were 29.7 and 31.2, respectively.
We also extracted and quantified PSA mRNA using the Mag-Bind kit.
Median of the Ct values of 10 samples was 24.4, compared with 21.6
when mRNA was extracted using Mag-Cap (Fig. 3B). The extraction yields of the two
methods achieved significant difference for PCA3 and PSA
extraction, respectively.
Clinical applications
We assessed the values of our method for prostate
cancer prediction and diagnosis retrospectively. We enrolled 52
male patients including 34 with diagnosed prostate cancer and 18
with BPH. Their urine samples were collected after prostate
massage; PCA3 and PSA mRNA were extracted and quantified using
Mag-Cap and real-time PCR, respectively. PCA3 scores were
calculated as 1,000-fold of the ratio of PCA3 to PSA mRNA
expression. The clinicopathological information and PCA3 scores of
the patients enrolled are summarized in Tables I and II.
 | Table IClinicopathological information and
PCA3 scores of the prostate cancer patients. |
Table I
Clinicopathological information and
PCA3 scores of the prostate cancer patients.
| Patient ID | Age (years) | Gleason score | T-PSAa (ng/ml) | PCA3 score |
|---|
| C01 | 61 | 4+3 | 8.8 | 41 |
| C02 | 76 | 3+4 | 37.5 | 79 |
| C03 | 75 | 3+4 | 4.4 | 33 |
| C04 | 69 | 3+2 | 4.8 | 39 |
| C05 | 74 | NA | 4.1 | 59 |
| C06 | 71 | 5+4 | 31.4 | 103 |
| C07 | 57 | 4+3 | 4.7 | 66 |
| C08 | 73 | 3+3 | 11.7 | 35 |
| C09 | 64 | 3+3 | 7.7 | 23 |
| C10 | 69 | 3+3 | 6.2 | 70 |
| C11 | 73 | 4+4 | 5.0 | 85 |
| C12 | 68 | NA | 5.7 | 19 |
| C13 | 75 | 4+5 | 10.0 | 226 |
| C14 | 76 | 3+3 | 3.2 | 137 |
| C15 | 70 | 4+4 | 206.8 | 48 |
| C16 | 64 | 4+3 | 33.3 | 71 |
| C17 | 75 | NA | 6.4 | 64 |
| C18 | 78 | 3+4 | 0.2 | 33 |
| C19 | 70 | 3+3 | 14.8 | 29 |
| C20 | 59 | 3+3 | 12.8 | 88 |
| C21 | 75 | 4+5 | 27.3 | 91 |
| C22 | 64 | 4+3 | 33.3 | 48 |
| C23 | 59 | 3+3 | 12.8 | 0 |
| C24 | 70 | 3+3 | 14.8 | 59 |
| C25 | 64 | 3+3 | 7.7 | 33 |
| C26 | 58 | NA | 361.6 | 68 |
| C27 | 75 | 4+5 | 27.3 | 69 |
| C28 | 75 | 3+4 | 4.4 | 83 |
| C29 | 76 | 3+4 | 37.5 | 73 |
| C30 | 73 | 3+3 | 11.7 | 0 |
| C31 | 57 | 4+3 | 4.7 | 33 |
| C32 | 66 | NA | 8.2 | 36 |
| C33 | 71 | 3+4 | 24.3 | 0 |
| C34 | 70 | 3+3 | 12.9 | 24 |
 | Table IIClinicopathological information and
PCA3 scores of the BPH patients. |
Table II
Clinicopathological information and
PCA3 scores of the BPH patients.
| Patient ID | Age (years) | Pathological
diagnosis | T-PSA (ng/ml) | PCA3 score |
|---|
| B01 | 71 | BPH | 7.1 | 20 |
| B02 | 72 | BPH | 1.9 | 18 |
| B03 | 72 | BPH | 3.1 | 28 |
| B04 | 58 | BPH | 0.6 | 13 |
| B05 | 74 | BPH | 2.1 | 9 |
| B06 | 78 | BPH | 11.2 | 18 |
| B07 | 62 | BPH | 7.4 | 8 |
| B08 | 60 | BPH | 25.5 | 45 |
| B09 | 89 | BPH | 19.7 | 11 |
| B10 | 69 | BPH | 10.6 | 51 |
| B11 | 74 | BPH | 10.1 | 16 |
| B12 | 68 | BPH | 6.0 | 3 |
| B13 | 71 | BPH | 9.8 | 0 |
| B14 | 74 | BPH | 10.1 | 23 |
| B15 | 69 | BPH | 10.6 | 11 |
| B16 | 40 | BPH | 1.2 | 2 |
| B17 | 74 | BPH | 3.4 | 32 |
| B18 | 71 | BPH | 7.1 | 61 |
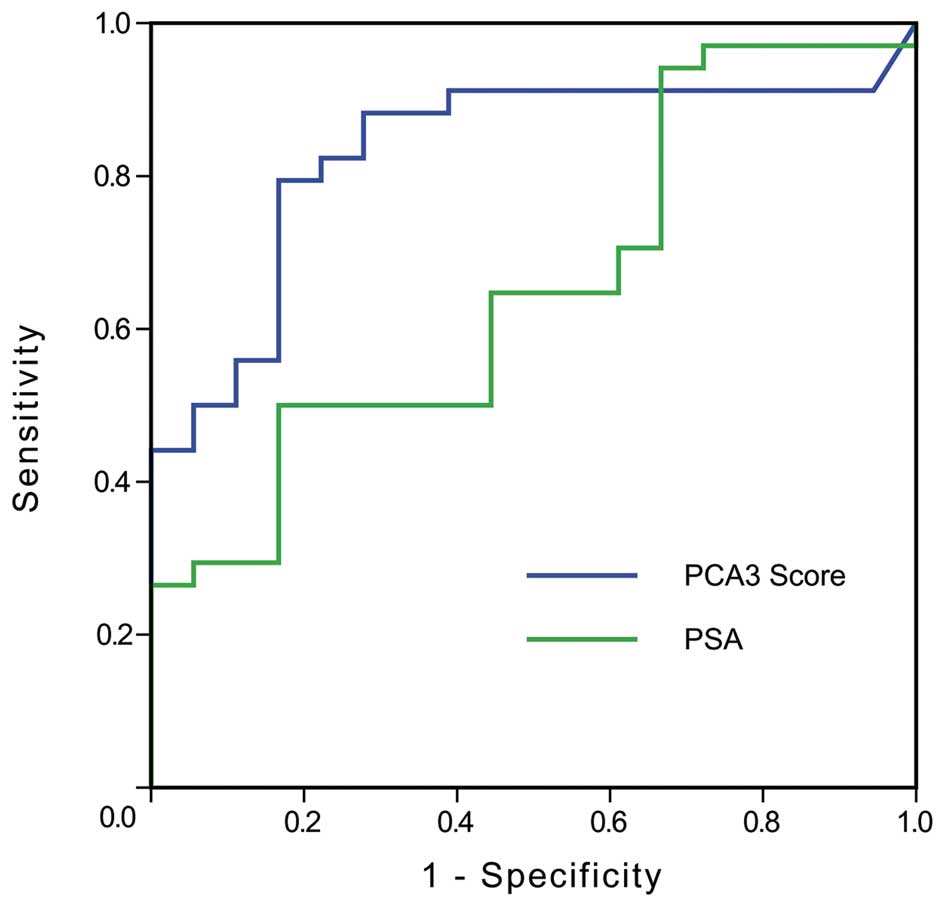
Median values of the PCA3 scores for patients with
prostate cancer and BPH were 53.5 and 17, respectively. ROC
analysis was applied to assess the performance of the PCA3 score as
a prostate-specific cancer marker. The curve was plotted in
Fig. 4 (blue trace). For
comparison, we also plotted the ROC curve of total PSA in Fig. 4 (green trace). It was noted that the
area under the curve (AUC) values for the PCA3 score and PSA were
0.831 and 0.655, respectively. A sensitivity of 82.4% and a
specificity of 77.8% were obtained when the cut-off value for the
PCA3 score was 28.5.
Discussion
Researchers have focused on improving methods with
higher specificity for prostate cancer diagnosis, and thus reduce
unnecessary prostate biopsy. Plasma PSA has been widely used as a
diagnostic marker for prostate cancer, but also shares certain
deficiencies. PSA is prostate tissue-specific rather than
cancer-specific, thus conditions such as prostatitis, benign
prostatic hyperplasia, acute urinary retention, digital rectal
examination, and cystoscopy can interfere. In addition, prostate
cancer patients are not always characterized by abnormal plasma
PSA. Although PSA derivatives have been developed to increase the
specificity and sensitivity, such as the fPSA/tPSA ratio, PSA
density, and PSA velocity, the outcome is still unsatisfactory.
PCA3, located in 9q21-22, was originally named
differential display code 3 (DD3) in 1999. PCA3 is a non-coding RNA
expressed inside the prostate gland. The PCA3 gene is overexpressed
specifically in human prostate cancer cells, and is thus considered
to be an independent and stable marker for prostate cancer. The
quantification of urine PCA3 after prostate massage is a
non-invasive detection method, which can significantly improve the
sensitivity, specificity and accuracy of prostate cancer diagnosis,
and reduce unnecessary prostate biopsy. However, the quantification
of PCA3 remains difficult due to poor RNA stability and the lack of
a purification method from complex matrix.
In the present study, we present a general method
for RNA extraction (Mag-Cap). A key step for the extraction of
nucleic acids using magnetic beads is to immobilize a capturing
strand which is complimentary to the target strand on the beads. A
straight forward way is covalent conjugation, which is time- and
labor-consuming and not general. Each target strand requires unique
functionalized magnetic beads, which significantly limits its
clinical applications. Our method avoided the covalent
immobilization of various capturing strands, which were instead
immobilized through DNA hybridization. This avoided the conjugation
of the magnetic beads with various strands and is general for DNA,
RNA and even artificial nucleic acids such as PNA and LNA.
We compared Mag-Cap with phenol-chloroform
extraction, affinity column and a commercial magnetic bead-based
nucleic acid extraction kit using controlled samples. The
extraction yield was quantified by real-time PCR after reverse
transcription. Ct values of the Mag-Cap group were 3–6 cycles less
than those extracted by phenol-chloroform extraction and affinity
column, which indicated that the yield of Mag-Cap was at least 8
(23) times higher than the other
two theoretically. We also found that Mag-Cap was more efficient
than the commercial Mag-Bind kit. All RNA sequences were
non-specifically extracted. Those unwanted nucleic acids might
induce interference for further analysis.
We finally applied our method for PCA3 score
measurement of clinical samples. The PCA3 score has been widely
accepted as a more specific marker for prostate cancer screening.
Irreproducibility and poor RNA extraction and detection limit the
clinical applications of the PCA3 score (29–31).
We applied our method for PCA3 score testing retrospectively and
assessed its value for prostate cancer diagnosis by ROC analysis.
The AUC value was over 0.83, while AUC of the serum total PSA level
was 0.66, which is consistent with previous studies (32).
In this research, 2 cases of prostate cancer
patients with serum tPSA <4 µg/l were both diagnosed with
prostate cancer using the urinary PCA3 score when the cutoff value
was 28.5, and 16 cases of prostate cancer patients with serum tPSA
4–10 µg/l. This indicates that the urinary PCA3 score
contributed to the diagnosis of prostate cancer with normal serum
tPSA or patients in the gray area. Cases (5 of 7) of BPH patients
with serum tPSA >10 µg/l were excluded as prostate cancer
using the urinary PCA3 score method, which indicates that urinary
PCA3 scores contribute to exclude prostate cancer in patients with
elevated serum tPSA, thus reducing unnecessary prostate biopsy.
In conclusion, we present a novel RNA extraction
technique based on magnetic beads while avoiding tedious covalent
functionalization of the beads with nucleic acids. We found that
the RNA extraction yields were higher than traditional
phenol-chloroform extraction, affinity column and a commercial RNA
purification kit based on magnetic beads. We also report its
application for PCA3 RNA detection. At the optimized condition, the
sensitivity and specificity were 82.4 and 77.8%, respectively.
However, this result was limited due to the small sample.
Fortunately, this research is still ongoing, and further
investigation is warranted for the application of the urinary PCA3
score in the diagnosis of prostate cancer.
Acknowledgments
The authors thank Xiangyun Qu and Yong Qin for the
helpful discussions.
References
|
1
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Surveillance E; End Results (SEER)
Program: (www.seer.cancer.gov) SEER*Stat
Database: Mortality - All COD, Aggregated With State, Total U.S.
(1969–2010) <Katrina/Rita Population Adjustment>, National
Cancer Institute, DCCPS, Surveillance Research Program,
Surveillance Systems Branch, released April 2013. Underlying
mortality data provided by NCHS (www.cdc.gov/nchs)
|
|
4
|
Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu
L and He J: The incidences and mortalities of major cancers in
China, 2009. Chin J Cancer. 32:106–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smith RA, Cokkinides V and Eyre HJ: Cancer
screening in the United States, 2007: A review of current
guidelines, practices, and prospects. CA Cancer J Clin. 57:90–104.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Slawin KM, Ohori M, Dillioglugil O and
Scardino PT: Screening for prostate cancer: An analysis of the
early experience. CA Cancer J Clin. 45:134–147. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Catalona WJ, Richie JP, Ahmann FR, Hudson
MA, Scardino PT, Flanigan RC, deKernion JB, Ratliff TL, Kavoussi
LR, Dalkin BL, et al: Comparison of digital rectal examination and
serum prostate specific antigen in the early detection of prostate
cancer: Results of a multicenter clinical trial of 6,630 men. J
Urol. 151:1283–1290. 1994.PubMed/NCBI
|
|
8
|
Cher ML and Carroll PR: Screening for
prostate cancer. West J Med. 162:235–242. 1995.PubMed/NCBI
|
|
9
|
Schmidt JD: Clinical diagnosis of prostate
cancer. Cancer. 70(Suppl 1): 221–224. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lilja H: Biology of prostate-specific
antigen. Urology. 62(Suppl 1): 27–33. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yates DR and Catto JW: Distinct patterns
and behaviour of urothelial carcinoma with respect to anatomical
location: How molecular biomarkers can augment clinicopathological
predictors in upper urinary tract tumours. World J Urol. 31:21–29.
2013. View Article : Google Scholar
|
|
12
|
Clarke RA, Zhao Z, Guo AY, Roper K, Teng
L, Fang ZM, Samaratunga H, Lavin MF and Gardiner RA: New genomic
structure for prostate cancer specific gene PCA3 within BMCC1:
Implications for prostate cancer detection and progression. PLoS
One. 4:e49952009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Roddam AW, Duffy MJ, Hamdy FC, Ward AM,
Patnick J, Price CP, Rimmer J, Sturgeon C, White P and Allen NE;
NHS Prostate Cancer Risk Management: Use of prostate-specific
antigen (PSA) isoforms for the detection of prostate cancer in men
with a PSA level of 2–10 ng/ml: Systematic review and
meta-analysis. Eur Urol. 48:386–399. 2005. View Article : Google Scholar
|
|
14
|
Bussemakers MJ, van Bokhoven A, Verhaegh
GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N and Isaacs
WB: DD3: A new prostate-specific gene, highly overexpressed in
prostate cancer. Cancer Res. 59:5975–5979. 1999.PubMed/NCBI
|
|
15
|
Hessels D, Klein Gunnewiek JM, van Oort I,
Karthaus HF, van Leenders GJ, van Balken B, Kiemeney LA, Witjes JA
and Schalken JA: DD3(PCA3)-based molecular urine analysis for the
diagnosis of prostate cancer. Eur Urol. 44:8–16. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schalken JA, Hessels D and Verhaegh G: New
targets for therapy in prostate cancer: Differential display code 3
(DD3(PCA3)), a highly prostate cancer-specific gene. Urology.
62(Suppl 1): 34–43. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
de Kok JB, Verhaegh GW, Roelofs RW,
Hessels D, Kiemeney LA, Aalders TW, Swinkels DW and Schalken JA:
DD3(PCA3), a very sensitive and specific marker to detect prostate
tumors. Cancer Res. 62:2695–2698. 2002.PubMed/NCBI
|
|
18
|
Day JR, Jost M, Reynolds MA, Groskopf J
and Rittenhouse H: PCA3: From basic molecular science to the
clinical lab. Cancer Lett. 301:1–6. 2011. View Article : Google Scholar
|
|
19
|
Ploussard G and de la Taille A: Urine
biomarkers in prostate cancer. Nat Rev Urol. 7:101–109. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Roobol MJ: Contemporary role of prostate
cancer gene 3 in the management of prostate cancer. Curr Opin Urol.
21:225–229. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Roobol MJ, Haese A and Bjartell A: Tumour
markers in prostate cancer III: Biomarkers in urine. Acta Oncol.
50(Suppl 1): 85–89. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marks LS, Fradet Y, Deras IL, Blase A,
Mathis J, Aubin SM, Cancio AT, Desaulniers M, Ellis WJ, Rittenhouse
H, et al: PCA3 molecular urine assay for prostate cancer in men
undergoing repeat biopsy. Urology. 69:532–535. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Janegitz BC, Cancino J and Zucolotto V:
Disposable biosensors for clinical diagnosis. J Nanosci
Nanotechnol. 14:378–389. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McCarroll J, Teo J, Boyer C, Goldstein D,
Kavallaris M and Phillips PA: Potential applications of
nanotechnology for the diagnosis and treatment of pancreatic
cancer. Front Physiol. 5:22014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thakor AS and Gambhir SS: Nanooncology:
The future of cancer diagnosis and therapy. CA Cancer J Clin.
63:395–418. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chan CP, Cheung YC, Renneberg R and
Seydack M: New trends in immunoassays. Adv Biochem Eng Biotechnol.
109:123–154. 2008.
|
|
27
|
Sandhu A, Handa H and Abe M: Synthesis and
applications of magnetic nanoparticles for biorecognition and point
of care medical diagnostics. Nanotechnology. 21:4420012010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Casadio V, Calistri D, Salvi S, Gunelli R,
Carretta E, Amadori D, Silvestrini R and Zoli W: Urine cell-free
DNA integrity as a marker for early prostate cancer diagnosis: A
pilot study. Biomed Res Int. 2013:2704572013. View Article : Google Scholar
|
|
29
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ahyai SA, Graefen M, Steuber T, Haese A,
Schlomm T, Walz J, Köllermann J, Briganti A, Zacharias M, Friedrich
MG, et al: Contemporary prostate cancer prevalence among T1c
biopsy-referred men with a prostate-specific antigen level ≤4.0 ng
per milliliter. Eur Urol. 53:750–757. 2008. View Article : Google Scholar
|
|
31
|
Thompson IM, Pauler DK, Goodman PJ, Tangen
CM, Lucia MS, Parnes HL, Minasian LM, Ford LG, Lippman SM, Crawford
ED, et al: Prevalence of prostate cancer among men with a
prostate-specific antigen level ≤4.0 ng per milliliter. N Engl J
Med. 350:2239–2246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Groskopf J, Aubin SM, Deras IL, Blase A,
Bodrug S, Clark C, Brentano S, Mathis J, Pham J, Meyer T, et al:
APTIMA PCA3 molecular urine test: development of a method to aid in
the diagnosis of prostate cancer. Clin Chem. 52:1089–1095. 2006.
View Article : Google Scholar : PubMed/NCBI
|