Introduction
Lung cancer is the most common cancer as well as the
leading cause of cancer-related mortalities globally (1). Histologically, non-small cell lung
cancer (NSCLC) accounts for ~80% of lung cancer cases. In spite of
the emergence of new cytotoxic drugs and targeted biological
agents, NSCLC remains one of the most clinically challenging cancer
types (2). Thus, providing better
treatment strategies is crucial.
Tumors are composed of an array of cell types,
including cancer and non-cancer cells. The most prominent component
of these non-cancer cells are macrophages, also known as
tumor-associated macrophages (TAMs) (3). There are two well-established
polarized phenotypes, classically activated macrophages (M1) and
alternatively activated macrophages (M2), both of which have been
observed in many tumors (4–6). M1-type macrophages are known to be
induced by granulocyte-macrophage colony-stimulating factor
(GM-CSF), interferon γ (IFN-γ), and/or lipopolysaccharide (LPS) and
have an IL-12high, IL-23high and
IL-10low phenotype (7).
At the opposite extreme, M2-type macrophages are various forms of
macrophages other than the classic M1 including cells exposed to
IL-4, IL-13, immune complexes, IL-10, and glucocorticoids (8). Generally, M1-type macrophages are
regarded as the effector cells that defend the body against
pathogens and tumor cells, while M2-type macrophages suppress
inflammatory responses and adaptive immunity and stimulate
angiogenesis and tumor growth (9,10). The
prognosis of cancer patients is dependent on the ratio of M1 and M2
macrophages (11,12). TAMs play a pivotal role in the
progression of NSCLC. The cytotoxic M1 phenotype explains the
extended survival of patients with NSCLC, suggesting that positive
immunoresponses play a crucial role in the prevention of NSCLC
progression (13–16). Therefore, the molecular mechanisms
underlying TAM polarization to different phenotypes are the focus
of intense investigation.
microRNAs (miRNAs) have been shown to be important
mediators of the macrophage activation process. miRNAs are a small
class of nucleic acids (~20–24 nt) that function in the
transcriptional and post-transcriptional regulation of gene
expression (17). miRNAs play a
vital role in the regulation of most biological and physiological
processes, including development, cell proliferation, cell cycle,
apoptosis, migration, and differentiation, including those
connected to cancer and immunity (18–20).
The role of miRNAs in the regulation of macrophage polarization has
been largely undefined.
In the present study, we examined the role miR-130a
played in the regulation of macrophage activation. The results
showed that M1 macrophage has an elevated expression of miR-130a
compared to M2. miR-130a suppressed the polarization of macrophages
to the M2 phenotype and enhanced M1 polarization. miR-130a
suppressed the expression of PPARγ by directly targeting its 3′UTR.
Additionally, the downregulation of miR-130a was inversely
associated with tumor stage, metastasis, survival and the presence
of the tumor macrophage marker CD163, in nSCLC samples.
Materials and methods
Cell lines
The human acute monocyte THP-1 leukemia cell line
was purchased from the Institute of Biochemistry and Cell Biology,
Chinese Academy of Sciences (Shanghai, China). To generate
macrophage-like differentiated THP-1 cells, THP-1 cells were seeded
in tissue culture flasks at 3×106 cells/flask and
exposed to 320 nM PMA in the culture medium for 48 h as previously
described (21). Following
incubation, the PMA-containing medium was removed and adherent
(differentiated) cells were incubated in fresh culture medium for
subsequent experiments. To generate M1-polarized THP-1 macrophages,
dTHP-1 cells were cultured with IFN-γ (100 U/ml) for another 48 h.
To generate M2-polarized THP-1 macrophages, the dTHP-1 cells were
cultured with M-CSF (100 ng/ml) for an additional 48 h.
Tissue specimens
From December 2010 to november 2014, a total of 75
NSCLC adenocarcinoma specimens and 75 matched normal tissue from
adjacent regions were collected from the patients undergoing
curative resection and diagnosed histopathologically at the
Department of Medical Oncology, Cancer Hospital Institute, Chinese
Academy of Medical Science, Peking Union Medical College (Beijing,
China). The samples were immediately frozen and stored in liquid
nitrogen prior to analysis. None of the patients received
chemotherapy or radiotherapy prior to the surgical excision. All
the patients provided informed consent for the sample collection.
The procedure was approved and supervised by the Institutional
Review Board (IRB) of the Cancer Institute/Hospital of Chinese
Academy of Medical Sciences and Peking Union Medical College.
Quantitative PCR
RNA was isolated using TRIzol (Invitrogen Life
Technologies, Carlsbad, CA, USA), RNeasy (Qiagen, Hilden, Germany),
or miR-Neasy (Qiagen) as per the manufacturer's instructions. The
synthesis of cDnA was performed with 0.5 µg RNA using
PrimeScript® RT Master Mix (Perfect Real-Time) (Takara
Bio Inc., Dalian, China). RT-qPCR analysis was performed in
triplicate on LightCycler® 480 II (Roche Applied Science
Indianapolis, IN, USA) using SYBR® Premix Ex Taq™
(Perfect Real-Time) (Takara Bio Inc.) and the results were
normalized according to the expression levels of gAPDH RnA. Results
were expressed using the 2−ΔΔCT methods. The primers for
the selected genes are shown in Table
I.
 | Table IPrimer sequences used for
RT-qPCR. |
Table I
Primer sequences used for
RT-qPCR.
| Gene | Sense primer | Antisense
primer |
|---|
| IL-1β |
5′-TCCAGGGACAGGATATGG AG-3′ |
5′-TCTTTCAACACGCAGGACAG-3′ |
| TNF-α |
5′-CTGTAGCCCATGTTGTAGCAAAC-3′ |
5′-GCTGGTTATCTCTCAGCTCCAC-3′ |
| iNOS |
5′-GCCAAGCTGAAATTGAATGAGGA-3′ |
5′-TTCTGTGCCGGCAGCTTTAAC-3′ |
| IL-10 |
5′-TTTAAGGGTTACCTGGGTTGC-3′ |
5′-TTGATGTCTGGGTCTTGGTTC-3′ |
| CCL17 |
5′-GGATGCCATCGTTTTTGTAACTG-3′ |
5′-AACTGCATTCTTCACTCTCTTGTTGT-3′ |
| CCL22 |
5′-TGCCGTGATTACGTCCGTTA-3′ |
5′-TCTCCTTATCCCTGAAGGTTAGCA-3′ |
| CD163 |
5′-GCTGCAGTGAATTGCACAGATAT-3′ |
5′-CGGGATGAGCGACCTGTT-3′ |
| PPARγ |
5′CATGGTGCCTTCGCTGAT-3′ |
5′-CAATGGCCATGAGGGAGTTA-3′ |
| GAPDH |
5′-GCACCGTCAAGGCTGAGAAC-3′ |
5′-TGGTGAAGACGCCAGTGGA-3′ |
Western blotting
Western blot analysis was performed as previously
described (22). Briefly, the cells
were lysed in cell lysis buffer [1% NP-40, 20 mM Tris-HCl (pH 7.6),
0.15 M naCl, 3 mM EDTA, 3 mM ethylene glycol tetraacetic acid
(EGTA), 1 mM phenylmethylsulfonyl fluoride, 2 mM sodium vanadate,
20 mg/ml aprotinin and 5 mg/ml leupeptin]. Following treatment, the
lysates were purified by centrifugation and denatured by boiling in
loading buffer. Equal amounts of protein samples were separated on
10% sodium dodecyl sulfate polyacrylamide gel electrophoresis
(SDS-PAgE), and electrophoretically transferred to a nitrocellulose
membrane. Following blocking with 5% non-fat milk at room
temperature for 1.5 h, the membrane was incubated with rabbit
anti-human mono-clonal primary antibody with an appropriate
dilution of antibodies (1:1,000–1:2,000) overnight at 4°C and then
with a horseradish peroxidase-conjugated goat anti-rabbit secondary
antibody at 1:5,000 dilution for 1 h at room temperature, and
detected using the Western Lightning Chemiluminescent detection
reagent (Amersham, Freiburg, Germany).
Luciferase activity assay
To construct pGL3-PPARγ-3′UTR, the full length 3′UTR
of the human PPARγ mRNA was amplified by PCR and cloned into the
pgL3-control vector (Promega, Madison, WI, USA). For the reporter
assays, 293T cells were transiently transfected with reporter
plasmid and miR-130a mimic, using Lipofectamine 2000 (Invitrogen
Life Technologies). Reporter assays were performed 48 h
post-transfection using the dual-luciferase assay system (Promega),
normalized for transfection efficiency by co-transfected
Renilla luciferase.
Flow cytometry
The samples were incubated with PE-CD86 and
FITC-CD206 (BioLegend, San Diego, CA, USA) according to the
manufacturers' instructions. Fluorescent conjugated with Alexa
Fluor 488 (Invitrogen Life Technologies) was used as a secondary
antibody. For each sample at least 1×104 cells were
analyzed.
ELISA
Cytokine concentrations in the culture supernatants
were determined by ELISA kits according to the manufacturer's
instructions (eBioscience, San Diego, CA, USA).
Bioinformatics
Prediction of putative miR-130a targets was
performed by using the online software, TargetScan (http://www.targetscan.org/) in conjunction with
miRanda (http://www.microrna.org/microrna/home.do) and PicTar
(http://pictar.mdc-berlin.de/). MiRanda
was used for the primary screening of miRNA target sites with
cut-off values for free energy (Δg) ≤14 kcal/mole and scores
>70. PicTar and TargetScan is an algorithm for the
identification of miRnA targets.
Statistical analysis
Data are presented as mean ± SD. One-way ANOVA
followed by the Bonferroni test was performed for multiple group
comparisons. The Student's t-test was used for a comparison between
two groups. Pearson's correlation was used to analyze the
relationship between the expression of miR-130a and CD163 and PPARγ
mRNA. P<0.05 was considered to indicate a statistically
significant result.
Results
M1 macrophages demonstrate greater
expression of miR-130a compared to M2 macrophages
We investigated the levels of miR-130a in the
proinflammatory M1 subset and immunosuppressive M2 subset. These
subsets were induced in vitro and characterized based on
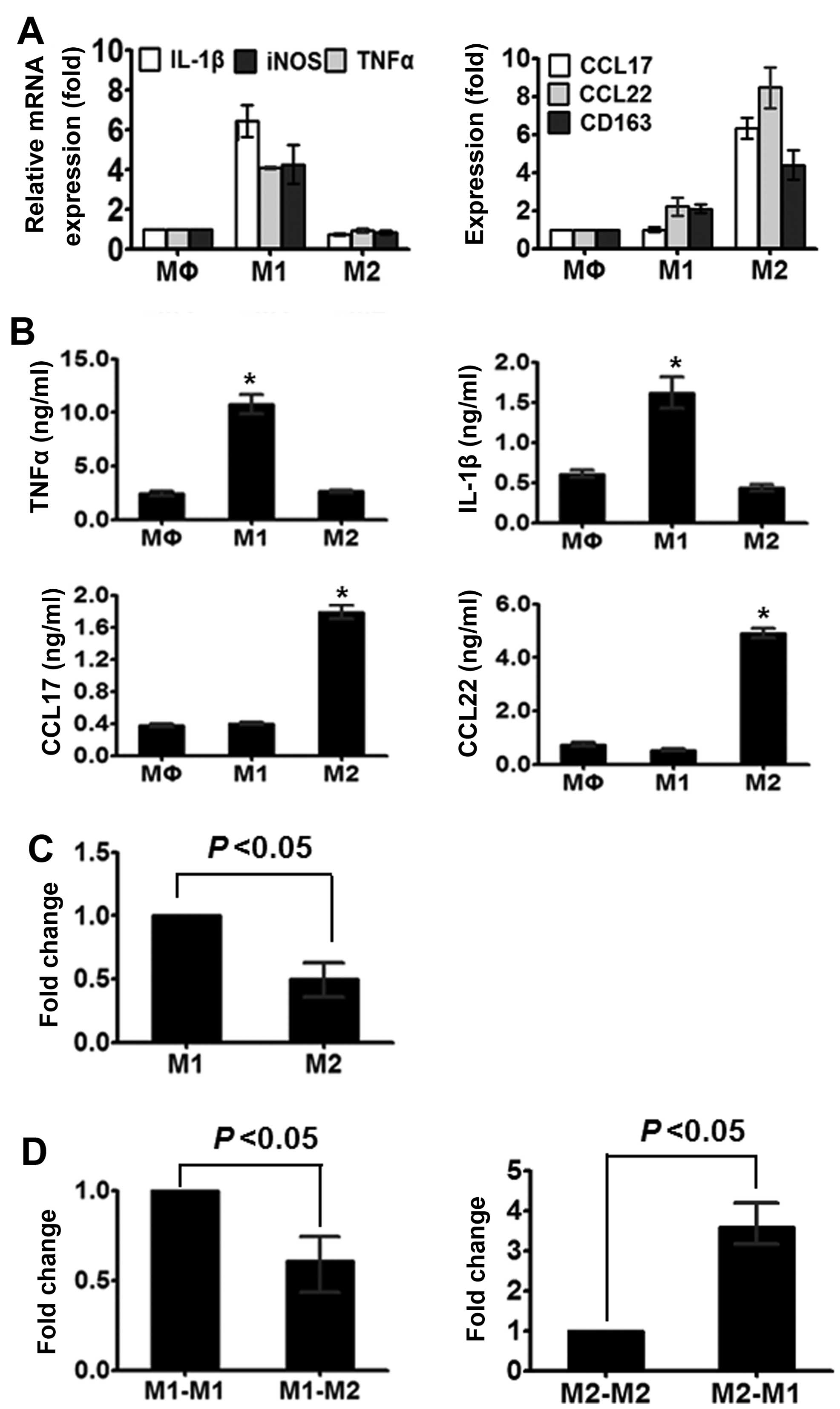
their phenotypic characteristics. As shown in Fig. 1A, the M2 macrophages expressed lower
levels of TNF-α, IL-1β and iNOS compared to the M1 macrophages but
higher levels of CD163, CCL17 and CCL22 mRNA. Furthermore, the
shift in macrophages was detected using ELISA analysis. As shown in
Fig. 1B, M1 enhanced the levels of
TNF-α and IL-1β, whereas M2 reduced the levels of CCL17 and
CCL22.
The expression of miR-130a was detected by RT-qPCR.
We found that M1 macrophages exhibited a considerably higher level
of miR-130a compared to the M2 macrophages (Fig. 1C). The initial findings suggested
that miR-130a participates in macrophage polarization. To examine
whether miR-130a contributes to the plasticity of macrophage
polarization, we converted one population into another by culturing
M1 macrophages with M-CSF and M2 macrophages with IFN-γ. As shown
in Fig. 1D, M1 to M2 macrophages
conversion resulted in decreased miR-130a, whereas M2 to M1
conversion led to increased miR-130a expression.
Overexpression of miR-130a reduces
M2-polarized THP-1 macrophages
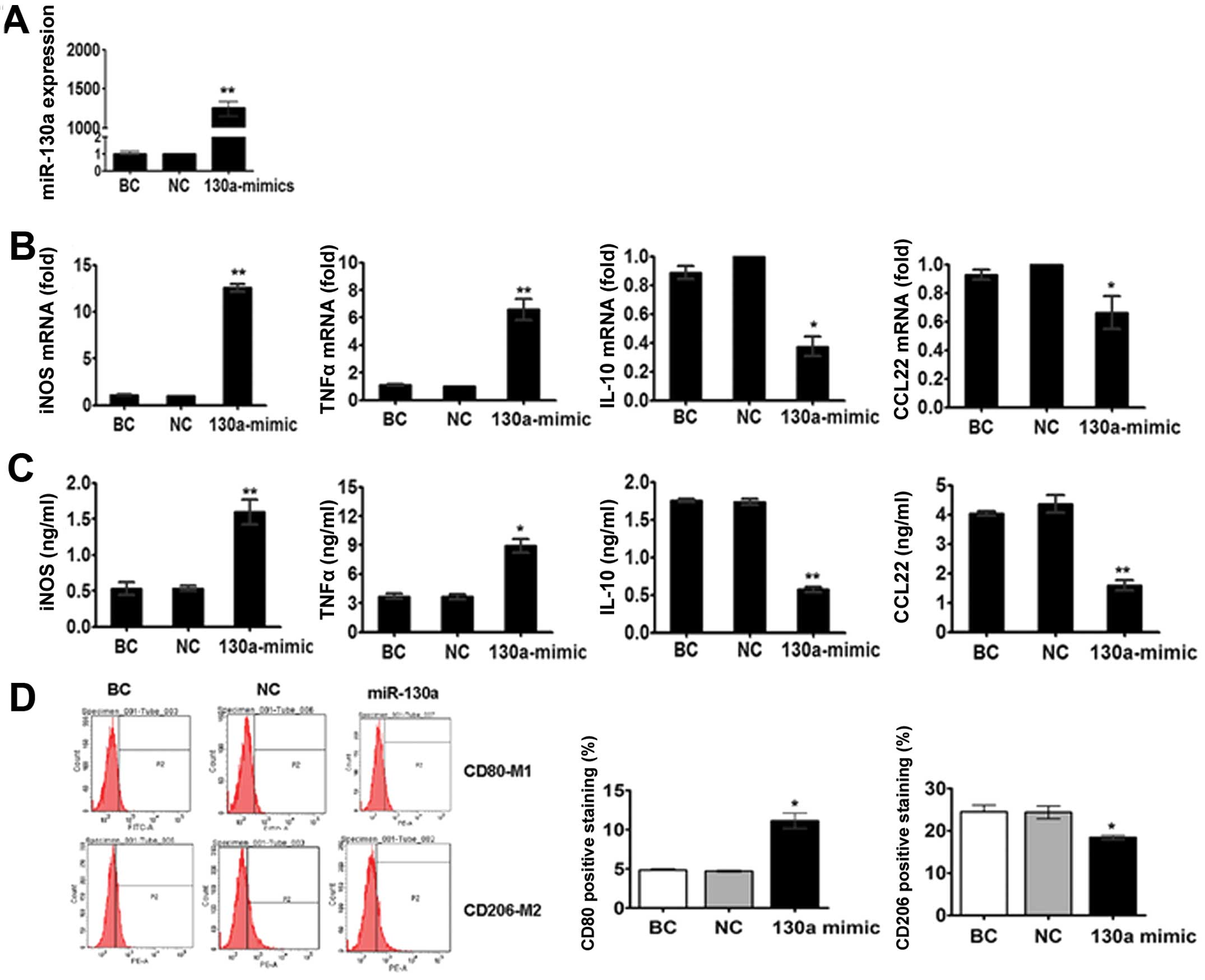
To determine whether miR-130a participates in
macrophage polarization, we transfected M2 macrophage, which have
lower levels of miR-130a compared to M1 macrophages. We found that
the overexpression of miR-130a with miR-130a mimics (Fig. 2A) in M2 macrophages enhanced the
mRNA and protein levels of TNF-α and iNOS, and reduced the
expression of IL-10 and CCL22 (Fig. 2B
and C). Additionally, the flow cytometric analysis revealed
that miR-130a overexpression significantly enhanced CD80 (M1
marker), but inhibited the expression of CD206 (M2 marker) on M2
macrophages (Fig. 2D).
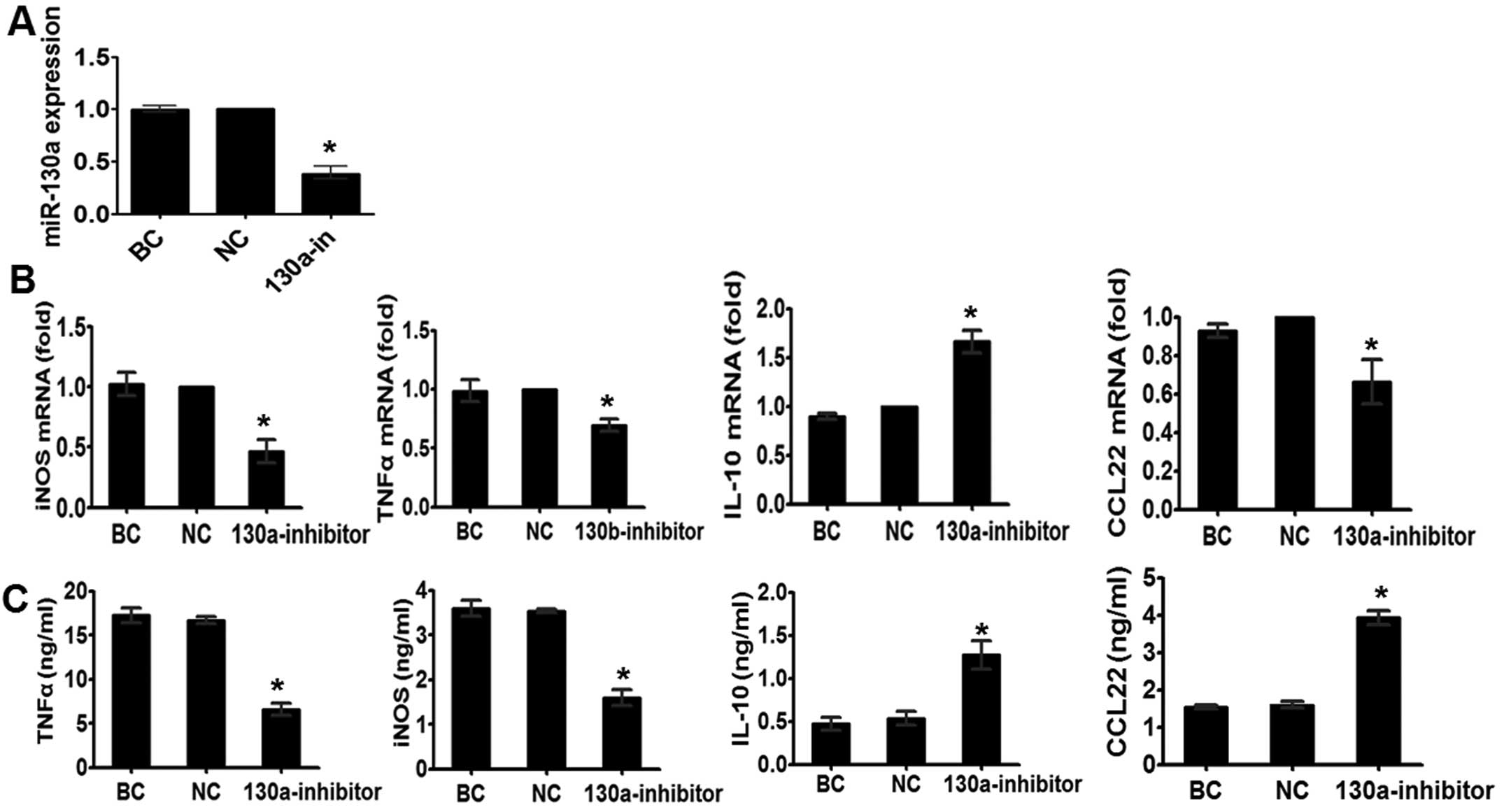
Inhibition of miR-130a results in M2
polarization of macrophages
Earlier, we showed that miR-130a suppresses the
expression of the M2 phenotype. Therefore, we determined whether
miR-130a inhibition in M1 macrophages, which have higher levels of
miR-130a compared to M2 macrophages, demonstrated an effect
opposite to that observed in M2 macrophages transfected with
miR-130a mimics. To verify this, we simulated the THP-1 cells with
IFN-γ for 48 h. As shown in Fig. 3B and
C, miR-130a knockdown decreased the IFn-γ-induced expression of
TNF-α and iNOS, and enhanced the expression of M2-associated genes
IL-10 and CCL22. Given our findings that M2
macrophages with overexpression of miR-130a decreased the
anti-inflammatory response to M-CSF, the results showed that
miR-130a has a suppressive role in M2 macrophage polarization.
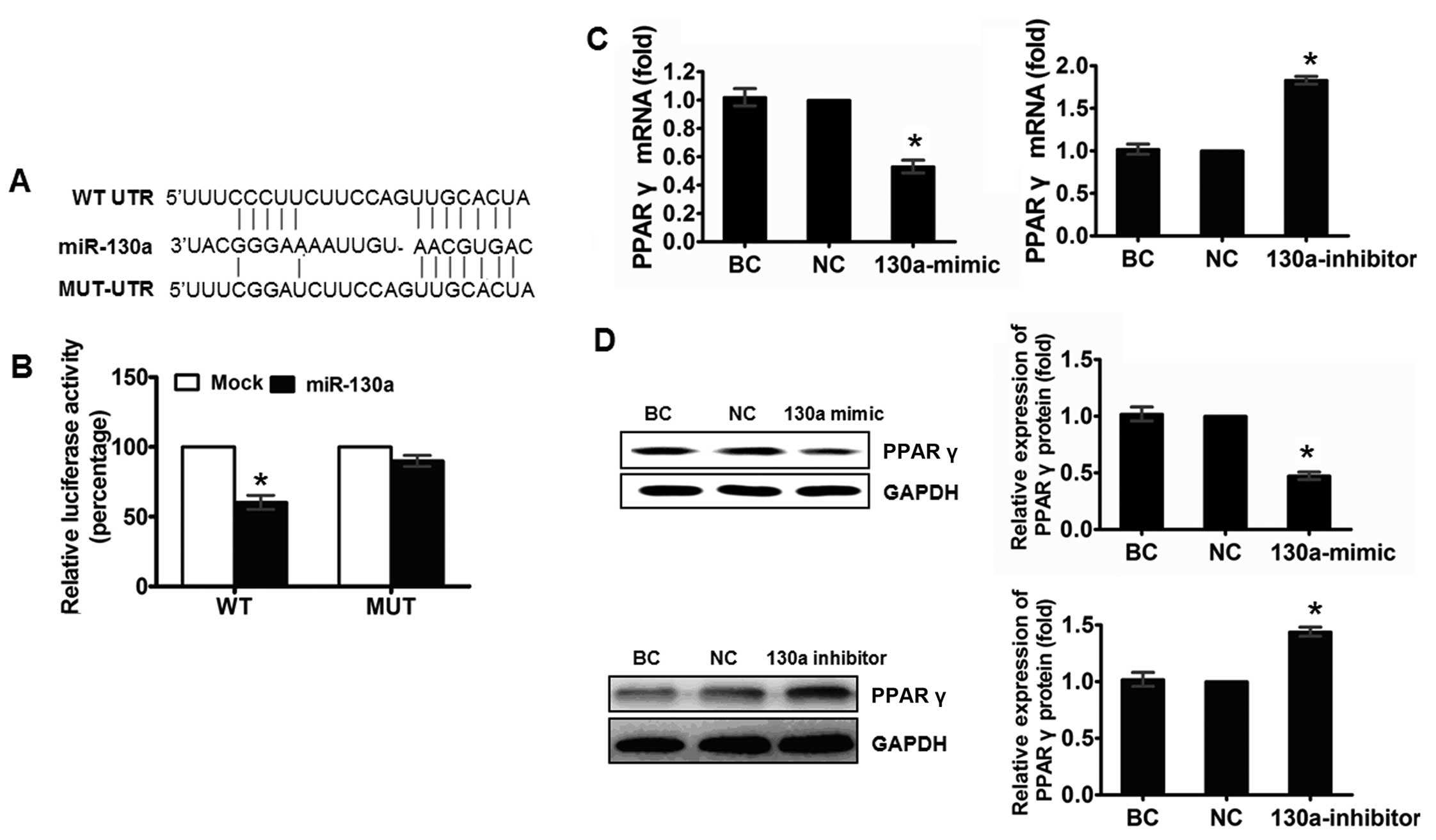
miR-130a inhibits the expression of PPARγ
by targeting its 3′UTR
To elucidate the molecular mechanism by which
miR-130a modulates the macrophage polarization, we predicted the
targets of miR-130a by using bioinformatics tools TargetScan,
PicTar and miRanda. The programs predicted PPARγ as a target of
miR-130a, and one potential miR-130a target sites at thepositions
42 nt in the PPARγ 3′UTR was identified (Fig. 4A). To verify that PPARγ is a
functional target of miR-130a, we cloned a reporter plasmid
containing the wide-type 3′UTR of PPARγ at the 3′ position of the
firefly luciferase reporter gene. In parallel, we constructed
reporter plasmids in which the observed target sequences were
mutated individually or in combination, and transfected 293T cells
with these constructs with miR-130a mimic and NC. Luciferase
activity was markedly reduced in cells transfected with miR-130a
mimics and wild-type PPARγ-3′UTR reporter plasmid-transfected
cells, compared to the cells transfected with NC mimics, but had no
effect on the mutant 3′UTR of PPARγ (Fig. 4B), indicating that miR-130a can
regulate gene expression through the putative binding site in the
3′UTR of PPARγ mRNA.
To confirm that miR-130a represses PPARγ expression
in THP-1 cells, we performed RT-qPCR analysis and found that the
transfection of miR-130a mimic led to a significant decrease in the
PPARγ mRNA level, while the transfection of miR-130a inhibitor led
to a significant increase in the PPARγ mRNA level compared to the
respective controls (Fig. 4C). In
addition, western blot analysis showed that transfection of the
miR-130a mimic led to a significant decrease in the PPARγ protein
level (Fig. 4D) and miR-130a
inhibitor resulted in an increased protein expression of PPARγ.
Taken together, these results provide evidence that miR-130a
inhibits the expression of PPARγ by directly targeting the 3′UTR of
PPARγ.
Expression of miR-130a is inversely
associated with advanced stage and lymph node metastasis of
NSCLC
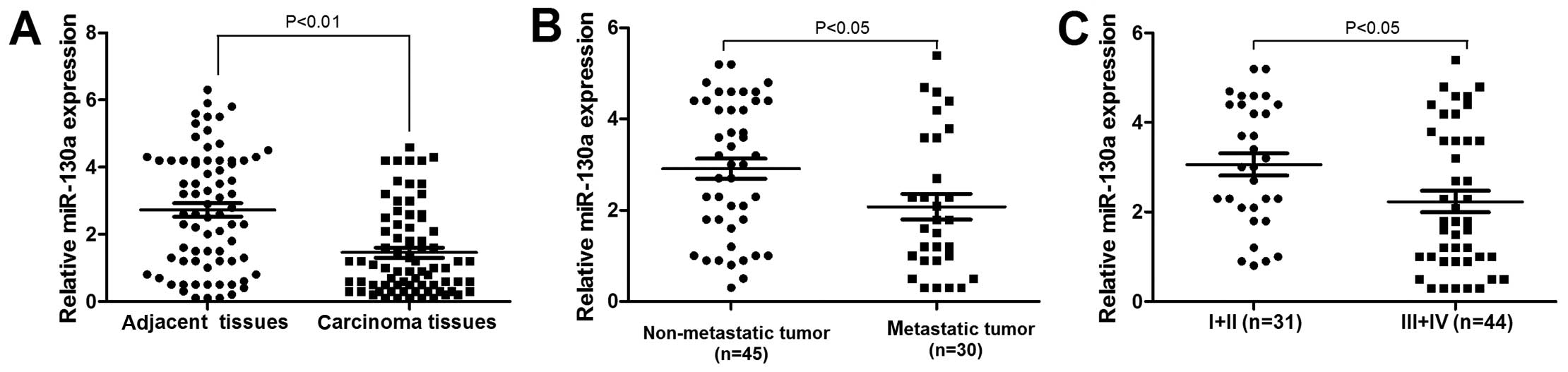
We examined miR-130a expression in 75 NSCLC patients
samples. miR-130a was significantly downregulated in nSCLC tissues,
compared to that in non-tumor tissues (P<0.05, Fig. 5A). We also investigated the
association between miR-130a expression and established
clinicopathological characteristics. As indicated in Table I, miR-130a was significantly
associated with the meta stasis and TNM of NSCLC, and the
expression level of miR-130a in tumor tissues decreased
statistically with the increasing stage of NSCLC (P<0.05)
(Fig. 5B). In addition, miR-130a
expression was significantly reduced in nSCLC, exhibiting lymph
node metastasis compared to NSCLC that did not exhibit lymph node
metastasis (Fig. 5C). no
significant association to age, gender, smoking history or
histological subtype was identified. Therefore, the low miR-130a
expression was closely associated with the progression and
metastasis of NSCLC.
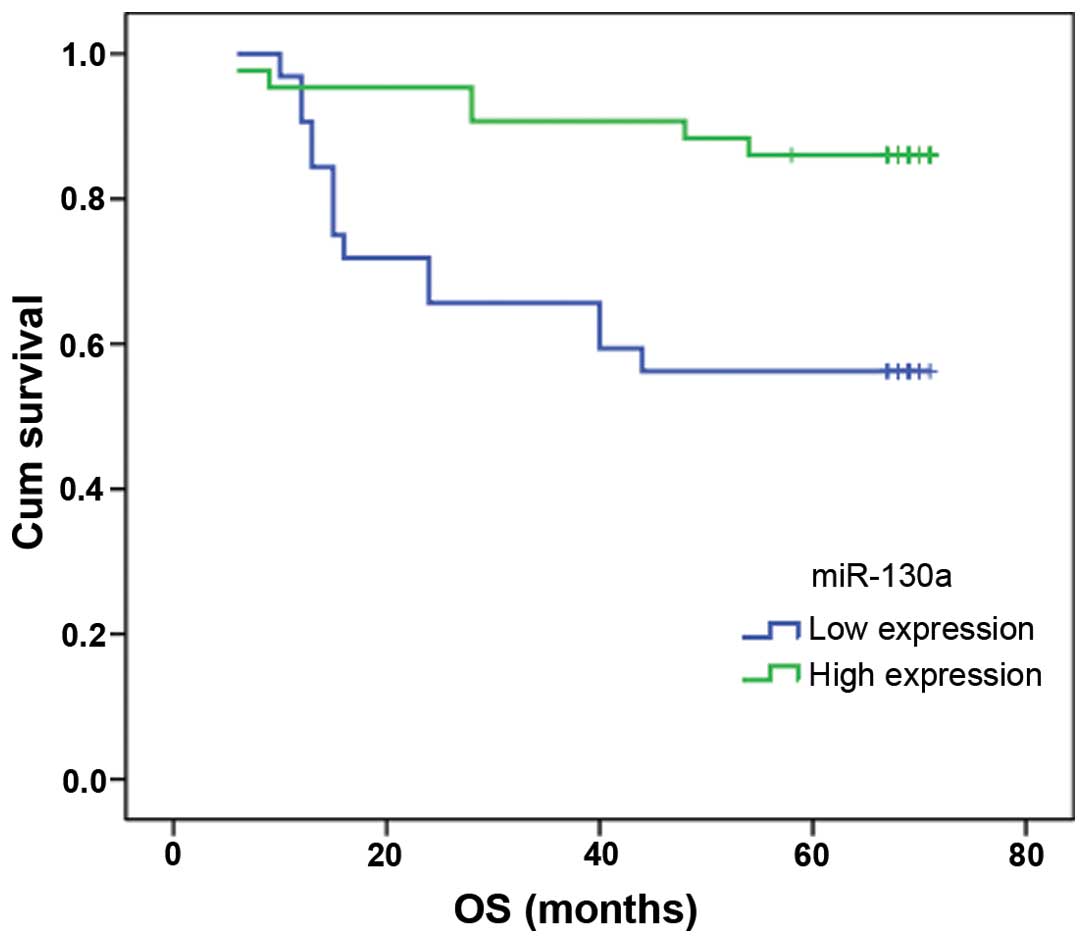
miR-130a downregulation predicts poor
overall survival in NSCLC patients
To evaluate the potential clinical relevance of the
downregulated miR-130a with regard to prognosis, the Kaplan-Meier
survival analysis was performed using overall survival. The results
indicated that miR-130a was significantly associated with patient
survival (Fig. 6). Patients with a
high fold change of miR-130a survived longer (n=43; median survival
of 65 months) than the patients with a low fold change of miR-130a
(n=32; median survival of 49 months) (P=0.003).
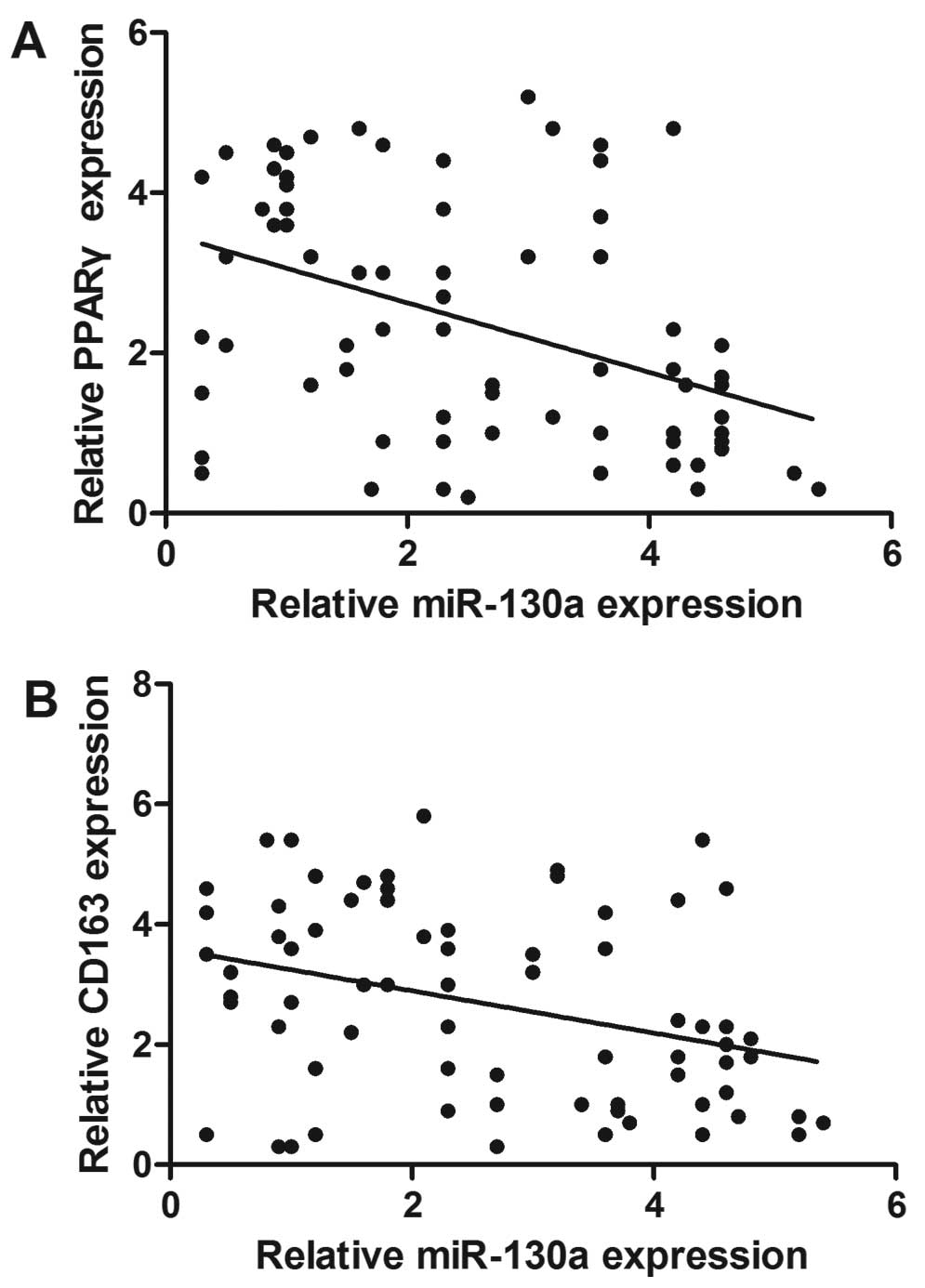
miR-130a expression is associated with
CD163 and PPARγ expression in NSCLC tissues
We investigated the association between miR-130a and
CD163, an M2 macrophage marker. As shown in Fig. 7, the Spearman's correlation analysis
revealed a direct correlation between CD163 and miR-130a. The lower
CD163 mRnA expression tumors had a substantially higher miR-130a
content compared to the reduction in miR-130a levels with
increasing CD163 macrophage contents indicating an inverse
association between miR-130a expression and macrophages. In
addition, the inverse association was observed between PPARγ mRNA
expression and miR-130a level.
Discussion
One of the hallmarks of malignancy is the
polarization of TAMs from a pro-immune (M1-like) phenotype to an
immun-0suppressive (M2-like) phenotype. The two distinct subsets,
which coexist in tumors, adapt to the changing tumor
microenvironment, and can be re-educated by immunoregulatory cues
(23,24). This event has primed interest in
developing therapies, with the aim of skewing TAMs to an M1-like
phenotype (25). Nonetheless, only
a few molecules have been identified to orchestrate this process
thus far. Evidence has shown that miRNAs are relevant in macrophage
activation and function. For example, miR-155, -146, -147, -9 and
-21 are induced by TLR ligands (26,27).
However, the potential of miRNAs to alter macrophage phenotype and
function has been rarely studied. Our investigation provides
insight into the role of miR-130a in the control of macrophage
polarization.
The biological role of miR-130a in the macrophage
has yet to be reported. In the present study, we demonstrate that
miR-130a is at a higher level (M1 macrophages) comapred to M2
macrophages. The transfection of macrophages with miR-130a mimic
resulted in the downregulation of markers and cytokines associated
with the phenotype of the classically activated (M2) macrophages
CD206, IL-10 and CCL22, whereas cytokines and markers associated
with the phenotype of alternatively activated (M1) regulatory
macrophages CD80, inOS and TnF-α, were upregulated. These results
suggest that miR-130a skews their polarization from an M2 towards
an M1 phenotype. Our results therefore reveal a critical role of
miR-130a in the induction of pathogenic M1 macrophage activation
and the transition between the pro-and anti-inflammatory
phenotypes, which is believed to provide novel insight into the
molecular regulation of the functional shaping of macrophages and
associated inflammatory disorders.
PPARγ is a ligand-activated transcription factor
belonging to the nuclear receptor superfamily. It plays a pivotal
role in the control of lipid metabolism and maintenance of
energetic homeostasis. PPARγ has been known to inhibit
pro-inflammatory gene expression through several mechanisms,
including the transrepression of NF-κB (28). For macrophage programming towards M2
polarization, the activation of PPARγ is considered to be critical
(29). It has been demonstrated
that IL-4 and IL-13 induce the expression and activation of PPARγ
(30,31). The present study provides evidence
that miR-130a regulates inflammatory cytokine production via PPARγ
targeting. The results of four sets of experiments from the present
study support this conclusion. First, the bioinformatics analysis
reveal that PPARγ is a potential target of miR-130a. Second, the
results from the luciferase reporter assay demonstrate that
miR-130a may regulate PPARγ protein expression through the
conserved miR-130a binding site in the 3′UTR of the PPARγ mRNA.
Third, the level of PPARγ protein is downregulated by the ectopic
expression of miR-130a, but is upregulated by the inhibition of
endogenous miR-130a with the synthetic inhibitor. Fourth, an
inverse association was observed between PPARγ mRNA expression and
the miR-130a level in NSCLC tissues. These results clearly indicate
that PPARγ is a target for miR-130a and that miR-130a controls
cytokine production in THP-1 cells by releasing its translational
inhibition of PPARγ.
TAMs are abundant components of NSCLC and play a key
role in the progression of nSCLC (32). Aberrant miRnAs expressions have been
observed in different types of cancer and their expression
signatures can be extremely informative for the diagnosis of cancer
(33–35). The above results show that miR-130a
is a key factor in M1/M2 modulation, raising the question of
whether the evaluation of miR-130a expression has a prognostic role
in NSCLC patients. Therefore, we examined the role of miR-130a in
NSCLC. To the best of our knowledge, we report for the first time
that miR-130a expression was downregulated in NSCLC samples
compared with the adjacent tissues. Tumors with low miR-130a levels
were associated with high tumor stage and poor recurrence-free
survival suggesting that miR-130a is a potential marker for tumor
progression. Therefore, miR-130a may be a novel tumor-suppressor
miRNA, and its downregulation may contribute to lung cancer
progression and metastasis. Few reports have shown the involvement
of miR-130a in tumorigenesis. Pan et al (36) have demonstrated that mRNA-130a
inhibits cell proliferation, invasion and migration in human breast
cancer by targeting the RAB5A. Chen et al (37) have reported that miR-130a can
predict response to temozolomide in patients with glioblastoma
multiforme. Acunzo et al (38) showed that miR-130a targets MET and
induces TRAIL sensitivity in NSCLC by downregulating miR-221 and
−222. In addition to the link to tumor prognosis, miR-130a was
strongly and inversely correlated with CD163 expression. CD163, a
marker of M2 macrophages, has been studied in several aggressive
tumors, and the increased expression of CD163 was significantly
associated with a poor overall survival in various types of cancer
(39–41). Our results are in accordance with
those obtained from THP-1 cells, suggesting that miR-130a are
important factors in macrophage polarization.
In conclusion, in the present study, we have
identified an unknown role for miR-130a in macrophages, providing
further insight into the complexities of macrophage plasticity,
suggesting that targeting miR-130a may have unforeseen effects on
macrophage function. Additionally, miR-130a is frequently
downregulated in NSCLC and correlates with tumor stage and poorer
patients' prognosis. These results suggest that miR-130a functions
as a tumor suppressor in NSCLC and is a potential molecular target
for NSCLC therapy.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Byers LA and Rudin CM: Small cell lung
cancer: Where do we go from here? Cancer. 121:664–672. 2015.
View Article : Google Scholar
|
|
3
|
Murray PJ, Allen JE, Biswas SK, Fisher EA,
Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence
T, et al: Macrophage activation and polarization: Nomenclature and
experimental guidelines. Immunity. 41:14–20. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cook J and Hagemann T: Tumour-associated
macrophages and cancer. Curr Opin Pharmacol. 13:595–601. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gordon S: Alternative activation of
macrophages. Nat Rev Immunol. 3:23–35. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ruffell B and Coussens LM: Macrophages and
therapeutic resistance in cancer. Cancer Cell. 27:462–472. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Krausgruber T, Blazek K, Smallie T,
Alzabin S, Lockstone H, Sahgal N, Hussell T, Feldmann M and Udalova
IA: IRF5 promotes inflammatory macrophage polarization and TH1-TH17
responses. Nat Immunol. 12:231–238. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Allavena P, Sica A, Solinas G, Porta C and
Mantovani A: The inflammatory microenvironment in tumor
progression: The role of tumor-associated macrophages. Crit Rev
Oncol Hematol. 66:1–9. 2008. View Article : Google Scholar
|
|
9
|
Sica A and Mantovani A: Macrophage
plasticity and polarization: In vivo veritas. J Clin Invest.
122:787–795. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Santoni M, Massari F, Amantini C, Nabissi
M, Maines F, Burattini L, Berardi R, Santoni G, Montironi R,
Tortora G, et al: Emerging role of tumor-associated macrophages as
therapeutic targets in patients with metastatic renal cell
carcinoma. Cancer Immunol Immunother. 62:1757–1768. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ruhrberg C and De Palma M: A double agent
in cancer: Deciphering macrophage roles in human tumors. Nat Med.
16:861–862. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen P and Bonaldo P: Role of macrophage
polarization in tumor angiogenesis and vessel normalization:
Implications for new anticancer therapies. Int Rev Cell Mol Biol.
301:1–35. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ohri CM, Shikotra A, Green RH, Waller DA
and Bradding P: Macrophages within NSCLC tumour islets are
predominantly of a cytotoxic M1 phenotype associated with extended
survival. Eur Respir J. 33:118–126. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang J, Cao J, Ma S, Dong R, Meng W, Ying
M, Weng Q, Chen Z, Ma J, Fang Q, et al: Tumor hypoxia enhances
non-small cell lung cancer metastasis by selectively promoting
macrophage M2 polarization through the activation of ERK signaling.
Oncotarget. 5:9664–9677. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun J, Mao Y, Zhang YQ, Guo YD, Mu CY, Fu
FQ and Zhang XG: Clinical significance of the induction of
macrophage differentiation by the costimulatory molecule B7-H3 in
human non-small cell lung cancer. Oncol Lett. 6:1253–1260.
2013.PubMed/NCBI
|
|
16
|
Domagala-Kulawik J: The role of the immune
system in non-small cell lung carcinoma and potential for
therapeutic intervention. Transl Lung Cancer Res. 4:177–190.
2015.PubMed/NCBI
|
|
17
|
Bartel DP: MicroRnAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hammond SM: An overview of microRnAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lan H, Lu H, Wang X and Jin H: MicroRNAs
as potential biomarkers in cancer: opportunities and challenges.
Biomed Res Int. 2015:1250942015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jasinski-Bergner S, Mandelboim O and
Seliger B: The role of microRNAs in the control of innate immune
response in cancer. J Natl Cancer Inst. 106:dju2572014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Park EK, Jung HS, Yang HI, Yoo MC, Kim C
and Kim KS: Optimized THP-1 differentiation is required for the
detection of responses to weak stimuli. Inflamm Res. 56:45–50.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang XF, Wang HS, Wang H, Zhang F, Wang
KF, Guo Q, Zhang G, Cai SH and Du J: The role of indoleamine
2,3-dioxygenase (IDO) in immune tolerance: Focus on macrophage
polarization of THP-1 cells. Cell Immunol. 289:42–48. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Movahedi K, Laoui D, Gysemans C, Baeten M,
Stangé G, Van den Bossche J, Mack M, Pipeleers D, In't Veld P, De
Baetselier P, et al: Different tumor microenvironments contain
functionally distinct subsets of macrophages derived from
Ly6C(high) monocytes. Cancer Res. 70:5728–5739. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pucci F, Venneri MA, Biziato D, Nonis A,
Moi D, Sica A, Di Serio C, Naldini L and De Palma M: A
distinguishing gene signature shared by tumor-infiltrating
Tie2-expressing monocytes, blood 'resident' monocytes, and
embryonic macrophages suggests common functions and developmental
relationships. Blood. 114:901–914. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rolny C, Mazzone M, Tugues S, Laoui D,
Johansson I, Coulon C, Squadrito ML, Segura I, Li X, Knevels E, et
al: HRG inhibits tumor growth and metastasis by inducing macrophage
polarization and vessel normalization through downregulation of
PlGF. Cancer Cell. 19:31–44. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alam MM and O'Neill LA: MicroRNAs and the
resolution phase of inflammation in macrophages. Eur J Immunol.
41:2482–2485. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He M, Xu Z, Ding T, Kuang DM and Zheng L:
MicroRnA-155 regulates inflammatory cytokine production in
tumor-associated macrophages via targeting C/EBPbeta. Cell Mol
Immunol. 6:343–352. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ricote M, Li AC, Willson TM, Kelly CJ and
Glass CK: The peroxisome proliferator-activated receptor-gamma is a
negative regulator of macrophage activation. Nature. 391:79–82.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bouhlel MA, Derudas B, Rigamonti E,
Dièvart R, Brozek J, Haulon S, Zawadzki C, Jude B, Torpier G, Marx
N, et al: PPARgamma activation primes human monocytes into
alternative M2 macrophages with anti-inflammatory properties. Cell
Metab. 6:137–143. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang JT, Welch JS, Ricote M, Binder CJ,
Willson TM, Kelly C, Witztum JL, Funk CD, Conrad D and Glass CK:
Interleukin-4-dependent production of PPAR-gamma ligands in
macrophages by 12/15-lipoxygenase. Nature. 400:378–382. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Berry A, Balard P, Coste A, Olagnier D,
Lagane C, Authier H, Benoit-Vical F, Lepert JC, Séguéla JP,
Magnaval JF, et al: IL-13 induces expression of CD36 in human
monocytes through PPARgamma activation. Eur J Immunol.
37:1642–1652. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Becker M, Müller CB, De Bastiani MA and
Klamt F: The prognostic impact of tumor-associated macrophages and
intratumoral apoptosis in non-small cell lung cancer. Histol
Histopathol. 29:21–31. 2014.
|
|
33
|
Xue Z, Wen J, Chu X and Xue X: A microRNA
gene signature for identification of lung cancer. Surg Oncol.
23:126–131. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wali RK, Hensing TA, Ray DW, Dela Cruz M,
Tiwari AK, Radosevich A, Jepeal L, Fernando HC, Litle VR, Charlot
M, et al: Buccal microRnA dysregulation in lung field
carcinogenesis: gender-specific implications. Int J Oncol.
45:1209–1215. 2014.PubMed/NCBI
|
|
35
|
Piva R, Spandidos DA and Gambari R: From
microRnA functions to microRNA therapeutics: Novel targets and
novel drugs in breast cancer research and treatment (Review). Int J
Oncol. 43:985–994. 2013.PubMed/NCBI
|
|
36
|
Pan Y, Wang R, Zhang F, Chen Y, Lv Q, Long
G and Yang K: MicroRNA-130a inhibits cell proliferation, invasion
and migration in human breast cancer by targeting the RAB5A. Int J
Clin Exp Pathol. 8:384–393. 2015.PubMed/NCBI
|
|
37
|
Chen H, Li X, Li W and Zheng H: miR-130a
can predict response to temozolomide in patients with glioblastoma
multiforme, independently of O6-methylguanine-DnA
methyltransferase. J Transl Med. 13:692015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Acunzo M, Visone R, Romano G, Veronese A,
Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli
G, et al: miR-130a targets MET and induces TRAIL-sensitivity in
NSCLC by downregulating miR-221 and 222. Oncogene. 31:634–642.
2012.
|
|
39
|
Maniecki MB, Etzerodt A, Ulhøi BP,
Steiniche T, Borre M, Dyrskjøt L, Orntoft TF, Moestrup SK and
Møller HJ: Tumor-promoting macrophages induce the expression of the
macrophage-specific receptor CD163 in malignant cells. Int J
Cancer. 131:2320–2331. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
He KF, Zhang L, Huang CF, Ma SR, Wang YF,
Wang WM, Zhao ZL, Liu B, Zhao YF, Zhang WF, et al: CD163
tumor-associated macrophages correlated with poor prognosis and
cancer stem cells in oral squamous cell carcinoma. Biomed Res Int.
2014:8386322014. View Article : Google Scholar
|
|
41
|
De Palma M and Lewis CE: Macrophage
regulation of tumor responses to anticancer therapies. Cancer Cell.
23:277–286. 2013. View Article : Google Scholar : PubMed/NCBI
|