Introduction
Nasal natural killer/T-cell lymphoma (NKTL) is a
rare lymphoma that is more common in East Asia and Central America
than in the West. It accounts for 7–10% of all non-Hodgkin's
lymphomas diagnosed in East Asia and Latin America, but only 1% of
such lymphomas among Caucasians (1–3). Among
the risk factors for NKTL development, Epstein-Barr virus (EBV)
latent infection has been shown to play an important role, and
ENKTL is closely associated with EBV infection (4–7).
EBV is a ubiquitous herpes virus that is linked to
multiple malignancies, including Burkitt's lymphoma, Hodgkin's
disease, gastric, esophageal, cervical and prostate cancer and
nasopharyngeal carcinoma (NPC) (8–12).
Latent membrane protein 1 (LMP1) encoded by EBV functions as an
essential factor in EBV-induced cell transformation and is
expressed in many of the malignancies associated with EBV (13). Clinical studies showed that the
expression of LMP1 was significantly correlated with the prognosis
of patients with ENKTL. Studies of LMP1 in nasopharyngeal carcinoma
indicated that LMP1 can enhance nasopharyngeal carcinoma cell
migration and invasion. Moreover, the human Fab-based
immune-conjugate specific for the LMP1 extracellular domain can
inhibit nasopharyngeal carcinoma growth both in vitro and
in vivo (14,15). These finding suggest that LMP1 may
play an important role in the progression of ENKTL. Therefore,
investigation of the effect and mechanism of action of LMP1 on
ENKTL cells may reveal a potential target for ENKTL treatment.
In the present study, we analyzed the expression of
LMP1 in an NKTL cell line (SNK-6) and aimed to assess the effect of
LMP1 on NKTL progression. After transfection with LMP1 shRNA, cell
proliferation, migration and invasion were analyzed. The underlying
mechanism was also analyzed.
Materials and methods
Antibodies and reagents
Unless stated otherwise, all substances were
purchased from Gibco (Grand Island, NY, USA). RPMI-1640 medium was
purchased from Sigma (St. Louis, MO, USA). Rabbit anti-LMP-1,
eIF4E, NF-κB and IκB monoclonal antibodies were from Santa Cruz
Biotechnology (Santa Cruz, CA, USA). β-actin antibodies were
obtained from Upstate Biotechnology, Inc. (Lake Placid, NY, USA).
HRP-conjugated goat anti mouse and anti-rabbit antibodies were from
Abcam (Cambridge, MA, USA).
Cell lines and cell culture
The SNK-6 cell line was purchased from ScienCell
(San Diego, CA, USA). Cells were cultured in RPMI-1640 media
supplemented with 10% heat-inactivated human plasma, 700 U/ml of
recombinant interleukin-2 (IL-2; Novartis, Surrey, UK), 50 U/ml
penicillin and 50 µg/ml streptomycin. PBMCs from healthy
volunteers were isolated by centrifugation using Ficoll-Hypaque
(Amersham Pharmacia Biotech, Buckinghamshire, UK). Both cell lines
were incubated at 37°C in an atmosphere containing 5%
CO2. Cells used for the experiments were all in the
logarithmic phase.
Infection of SNK-6 cells with lentiviral
LMP1/eIF4E shRNA expression vector
The shRNA-LMP1/eIF4E and its control shRNA-NC
plasmids were designed and synthesized by Shanghai GeneChem
Chemical Technology Co., Ltd. (Shanghai, China). The stable
LMP1/eIF4E knocked down cell line was established via lentiviral
vector transfection. SNK-6 cells in the logarithmic growth phase
were cultured with lentiviral vector solution for 6 h, and
supplemented with lentiviral vector for another 6 h. After 48 h,
cells were selected with hygromycin B until positive cells were
identified.
LMP1 expression vector transfection
The pcDNA3.1-LMP1 plasmid was constructed by
inserting the cDNA fragment retro transcribed from the full-length
cDNA of LMP1 (16). Lipofectamine™
2000 (Invitrogen, Carlsbad, CA, USA) was used for the transfection
of LMP1 plasmid (pcDNA3.1-LMP-1) and empty plasmid (pcDNA3.1)
according to the manufacturer's protocol. Stable expression clones
were selected using G418 (neomycin sulfate, 800 µg/ml). The
cell culture medium was replaced with fresh G418-containing medium
every 2–3 days until resistant colonies were identified. The cells
were collected for further analysis.
MTT assay
Cell viability was monitored via the
2-(4,5-dimethyltriazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
(Sigma) colorimetric assay. Briefly, after adding 20 µl of
MTT (5 mg/ml) to each well, and a 4-h incubation at 37°C, MTT
crystals were dissolved with dimethyl sulfoxide (DMSO) without
discarding the cell supernatants and the absorbance (490 nm) was
measured. All experiments were repeated at least three times with
96-wells per experiment.
Cell cycle and apoptosis analysis by flow
cytometry
For cell cycle analysis, SNK-6 cells were cultured
in 0.01 mmol/l doxorubicin, or not. After 24 h, the cells were
stained in 50 mg/ml propidium iodide, 0.05% Triton X-100, 0.1 mg/ml
RNase A, and 1X PBS at 37°C for 30 min in the dark. The stained
cells were suspended in 500 ml PBS for flow cytometric
analysis.
Cell apoptosis was analyzed using an Annexin V-PI
apoptosis detection kit (Abcam, Cambridge, UK). Briefly,
transfected cells were washed with PBS and resuspended in 500
µl of binding buffer containing Annexin V-fluorescein
isothiocyanate (FITC) and propidium iodide (PI). After incubation
on ice for 10 min, cells were analyzed on a FACSCalibur flow
cytometer (Becton-Dickinson, San Jose, CA, USA). The relative
number of apoptotic cells was calculated.
Transwell migration and invasion
assay
The BioCoat Matrigel Invasion Chamber was purchased
from BD Biosciences (Bedford, MA, USA). The membrane has a pore
size of 8 mm and is coated with Matrigel matrix. According to the
manufacturer's instructions, 1×106 SNK-6 cells in 0.5-ml
culture medium were implanted into the upper chamber. The bottom
well contained 0.6 ml of Dulbecco's modified Eagle's medium with
10% fetal bovine serum. After being cultured for 48 h, the cells in
the upper and lower chambers were stained with 0.4% trypan blue and
counted with a hemocytometer. After air-drying, the membrane was
stained with crystal violet. The number of migrating cells was then
counted under a microscope.
An invasion assay was carried out similarly to the
cell migration assay, except that 0.1 ml of Matrigel (50 mg/ml; BD
Biosciences) was added to the membrane surface of the chamber 6 h
before the cells were seeded.
RNA extraction and quantitative real-time
polymerase chain reaction (qRT-PCR)
Cells were harvested after transfection or 48 h
later. Total RNA of the cells was extracted using TRIzol reagent
(Invitrogen). The cDNA was synthesized from 5 µg of the
total RNA using M-MLV reverse transcriptase (Clontech Laboratories,
Palo Alto, CA, USA). The obtained cDNA was then used as a template
for qRT-PCR analysis. First strand cDNA was synthesized from 2
µg of total RNA using the SuperScript II reverse
transcriptase (Invitrogen) with 200 ng of random hexamers. The
qRT-PCR cycling conditions were as follows: 95°C for 2 min for
initial denaturation; 94°C for 15 sec, 58°C for 15 sec, and 72°C
for 20 sec; 2 sec for plate reading for 40 cycles; and melt curve
from 65 to 95°C. The primers used were as follows: LMP-1, 5′-GGT
ACC TAC ATA AGC CTC TCA CAC TG-3′ (forward primer) and 5′-TCT AGA
GAA GGT AAG AGT GCC ATC-3′ (reverse primer); eIF4E, 5′-GGG CCC ATG
GCG ACT GTC GAA CCG GA-3′ (forward primer) and 5′-CTC GAG TTA GTG
GTG GAG CCG CTC TTA-3′ (reverse primer); β-actin, 5′-CTG GGA CGA
CAT GGA GAA AA-3′ (forward primer) and 5′-AAG GAA GGC TGG AAG AGT
GC-3′ (reverse primer).
Western blot analysis
After transfection for 48 h, cells were harvested
after being washed with cold PBS (Invitrogen). Total protein was
extracted using the RIPA lysis buffer system (Santa Cruz
Biotechnology, Dallas, TX, USA). A micro-BCA protein assay kit
(Pierce, Rockford, IL, USA) was used for protein concentration
analysis. Total protein (30 µg/lane) was resolved on a 12%
SDS/PAGE under denatured reducing conditions and transferred onto a
nitrocellulose membrane (Amersham Pharmacia, Freiburg, Germany).
After 1-h incubation in blocking solution (5% non-fat milk), the
membrane was probed overnight at 4°C with primary antibodies [LMP1
(1:500), eIF4E (1:300), NF-κB (1:400), IκB (1:400), β-actin
(1:500)]. The membranes were washed five times then incubated with
HRP-conjugated secondary antibodies (1:2,000) for 1 h at room
temperature. Chemiluminescent detection was performed using an ECL
kit (Pierce Chemical, Rockford, IL, USA). The gray value of the
bands was analyzed using Image J2x software.
Statistical analysis
Each experiment was repeated at least three times.
The results are presented as the mean ± standard deviation (SD).
Statistical analyses were performed using an independent samples
Student's t-test for direct two-group comparisons and the
Tukey-Kramer test after a significant one-way analysis of variance
(ANOVA) F-test for multiple-group comparisons. A difference was
considered statistically significant at P<0.05.
Results
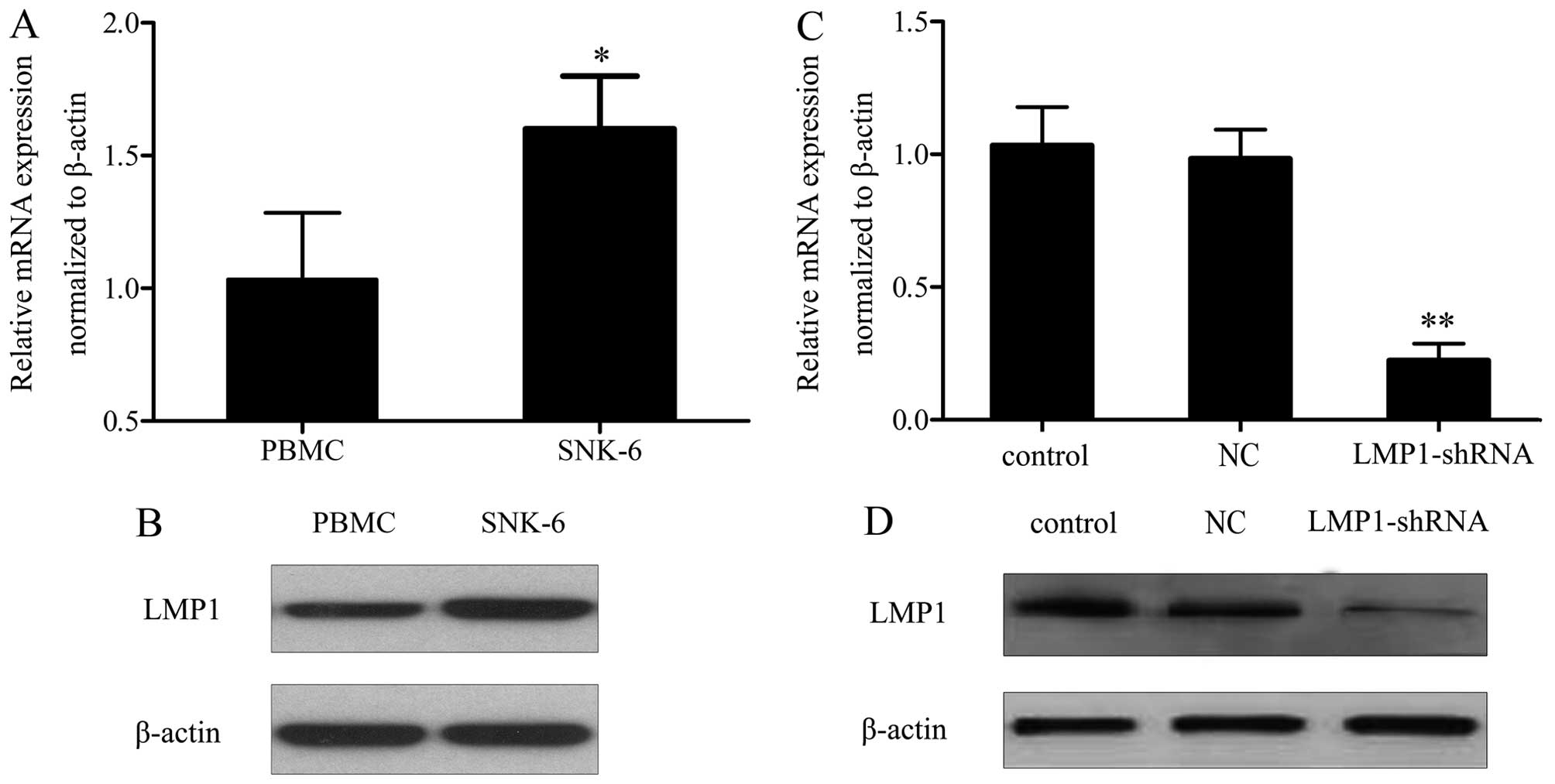
LMP1 is overexpressed in SNK-6 cells
It has been demonstrated that LMP1 is associated
with the development of malignancies (17,18).
To investigate its function in NKTL progression, its levels were
measured. RT-PCR analysis showed that the mRNA levels of LMP1 were
significantly higher in SNK-6 cells than in controls (Fig. 1A). A similar result was observed for
protein levels (Fig. 1B).
LMP1shRNA inhibits SNK-6 cell
proliferation and G0/G1 phase arrest
To evaluate the effect of LMP1 on NKTL development,
LV-LMP1-shRNA transfection was performed. After the selection of
positive cells, LMP1 mRNA and protein expression were detected by
qRT-PCR and western blotting. A statistically significant decrease
in LMP1 levels was observed after LMP1 shRNA transfection, compared
with the NC group (Fig. 1C and D).
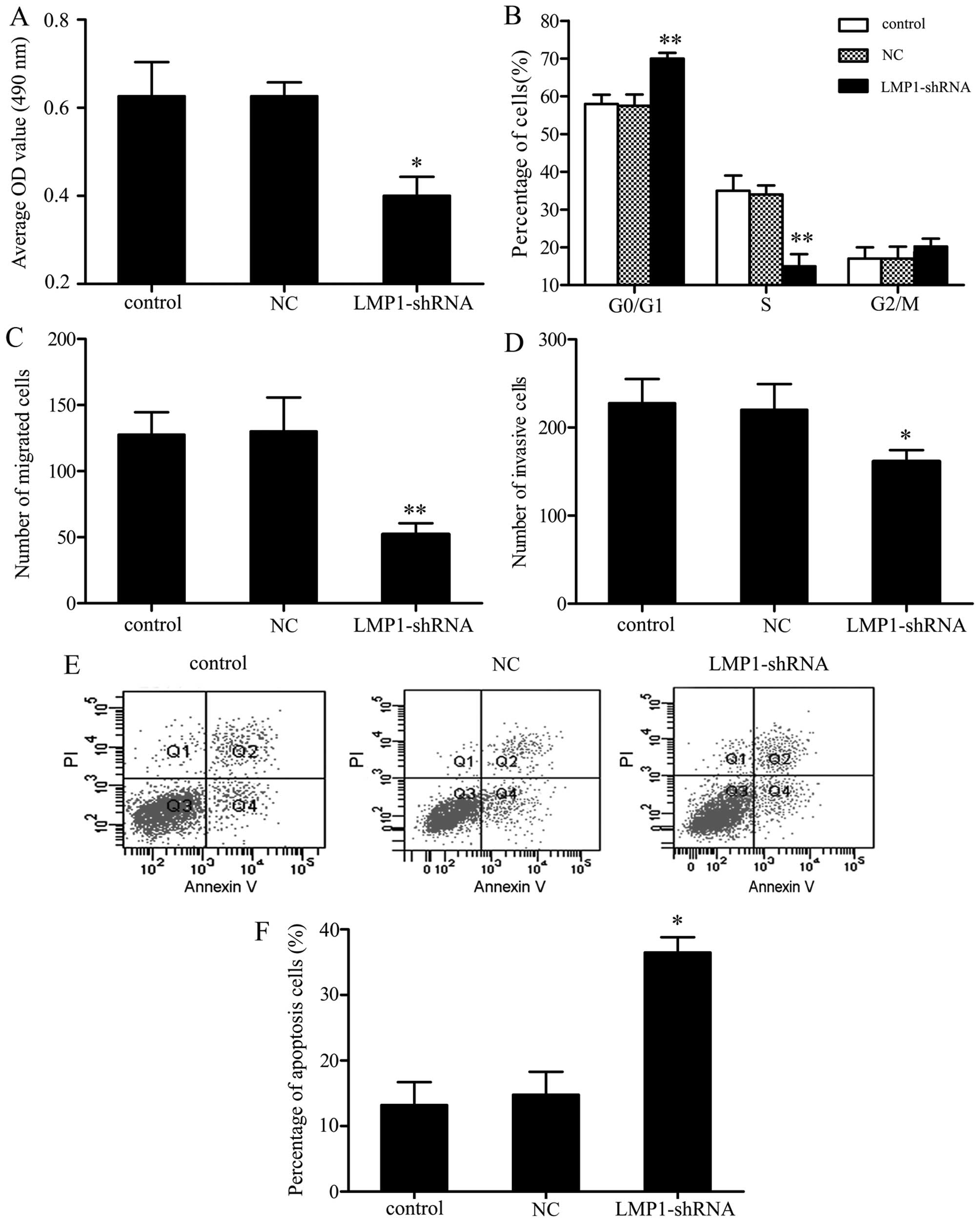
Following transfection with LMP1 shRNA for 48 h, an obvious
decrease in cell proliferation rate was observed, with a 23.0%
inhibition of proliferation compared to control cells (Fig. 2A). Given that LMP1 knockdown
attenuated SNK-6 cell proliferation, we next performed cell cycle
analyses on SNK-6 cells transfected with LMP1 shRNA vs. a control
vector. Further flow cytometry assay showed that in comparison to
the NC group, cells transfected with LMP1 shRNA induced
reproducible and highly significant G0/G1 arrest. The distribution
of transfected cells in the cell cycle increased by 13.4±5.7% in
the G0/G1 phase (P<0.01) and decreased by 21.3±7.2% in the S
phase (P<0.01) compared with controls. These results suggest
that G0/G1 arrest may be the mechanism through which LMP1 regulates
SNK-6 cell growth (Fig. 2B).
LMP1 shRNA suppresses SNK-6 cell
migration and invasion
We further analyzed whether LMP1 shRNA pretreatment
could influence the chemotaxis of SNK-6 cells. SNK-6 cell migration
and invasion were assessed using a Transwell assay. As shown in
Fig. 2C and D, after transfection
with LMP1 shRNA, the migration and invasive potential of SNK-6
cells was markedly reduced, by 73.8±8.2 and 42.3±7.1% compared with
the controls, respectively.
LMP1 shRNA promotes SNK-6 cell
apoptosis
The apoptosis of differently treated SNK-6 cells was
measured by FCM. The results showed that in parallel with the
control groups, there was a significant increase in apoptosis, with
LMP1 shRNA increasing cell apoptosis by 25.9±6.2% (Fig. 2E). This result indicates that LMP1
may inhibit cellular apoptosis in NKTL.
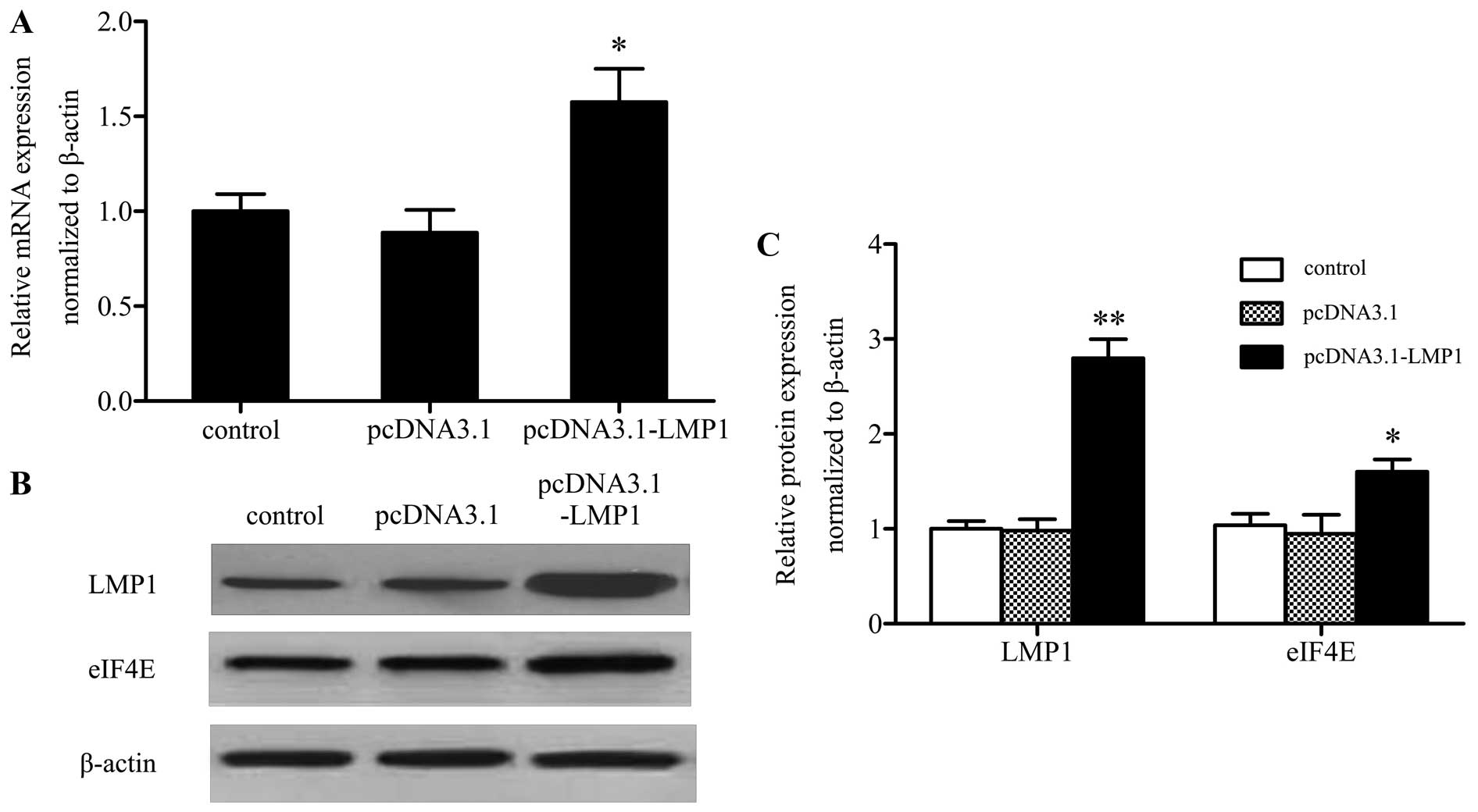
LMP1 promotes the expression of eIF4E in
SNK-6 cells
Increased eIF4E expression is associated with
enhanced invasion and metastasis in many kinds of tumors (ref.?).
To clarify the underlying mechanism involved in LMP1 regulated
SNK-6 cell proliferation, migration, invasion and apoptosis, the
expression of eIF4E was analyzed. As expected, after transfection
with pcDNA3.0 or pcDNA3.0-LMP1 for 48 h, qRT-PCR analysis showed
that compared with the control group, pcDNA3.0-LMP1 transfection
increased the mRNA level of eIF4E by ~1.5-fold (Fig. 3A). Further analysis of LMP1 and
eIF4E protein expression by western blotting showed that along with
the increased protein level of LMP1, eIF4E expression was also
upregulated compared with controls, suggesting that LMP1 tended to
increase eIF4E expression (Fig. 3B and
C).
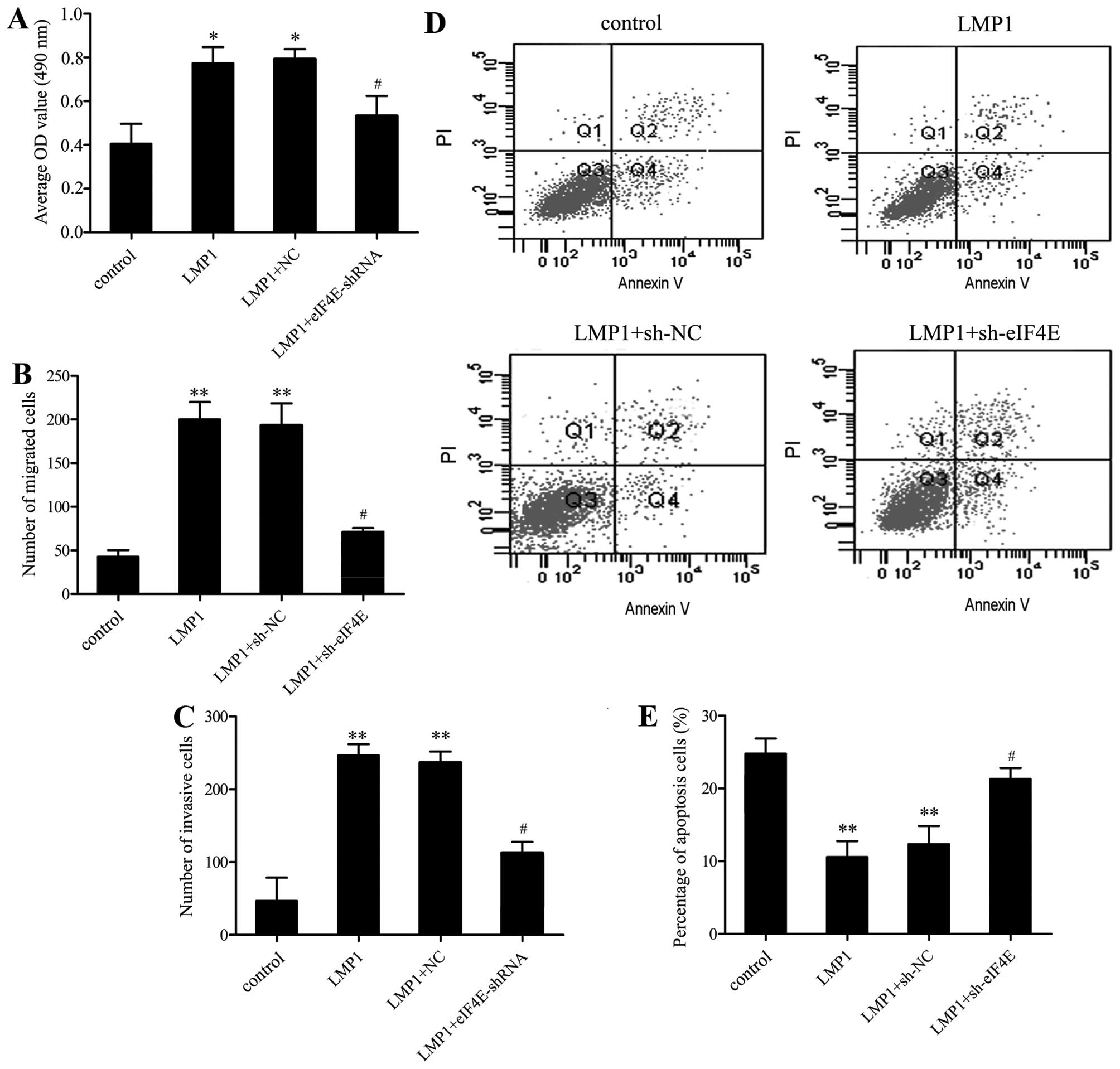
eIF4E is responsible for LMP1-induced
SNK-6 cell proliferation, migration, invasion and apoptosis
To further analyze whether LMP1 regulated SNK-6 cell
function via eIF4E expression, we performed a series of functional
restoration assays. Following pretreatment with eIF4EshRNA, cells
were then transfected with pcDNA3.0-LMP1, and the corresponding
effect on cell proliferation, migration, invasion and apoptosis was
assessed as before. The results showed that eIF4E silencing
decreased the proliferation (Fig.
4A), migration (Fig. 4B), and
invasion (Fig. 4C) of SNK-6 cells
induced by LMP1 upregulation. The inhibitory effect on apoptosis
triggered by LMP1 overexpression was also ameliorated in the
eIF4E-silenced groups (Fig. 4D and
E). These results suggested that LMP1 may trigger the
proliferation, migration, invasion and apoptosis of SNK-6 cells by
increasing eIF4E expression.
Effect of LMP1 on eIF4E expression is
mediated by the NF-κB pathway
To further explore the underlying mechanism involved
in LMP1-induced eIF4E expression, the NF-κB pathway was
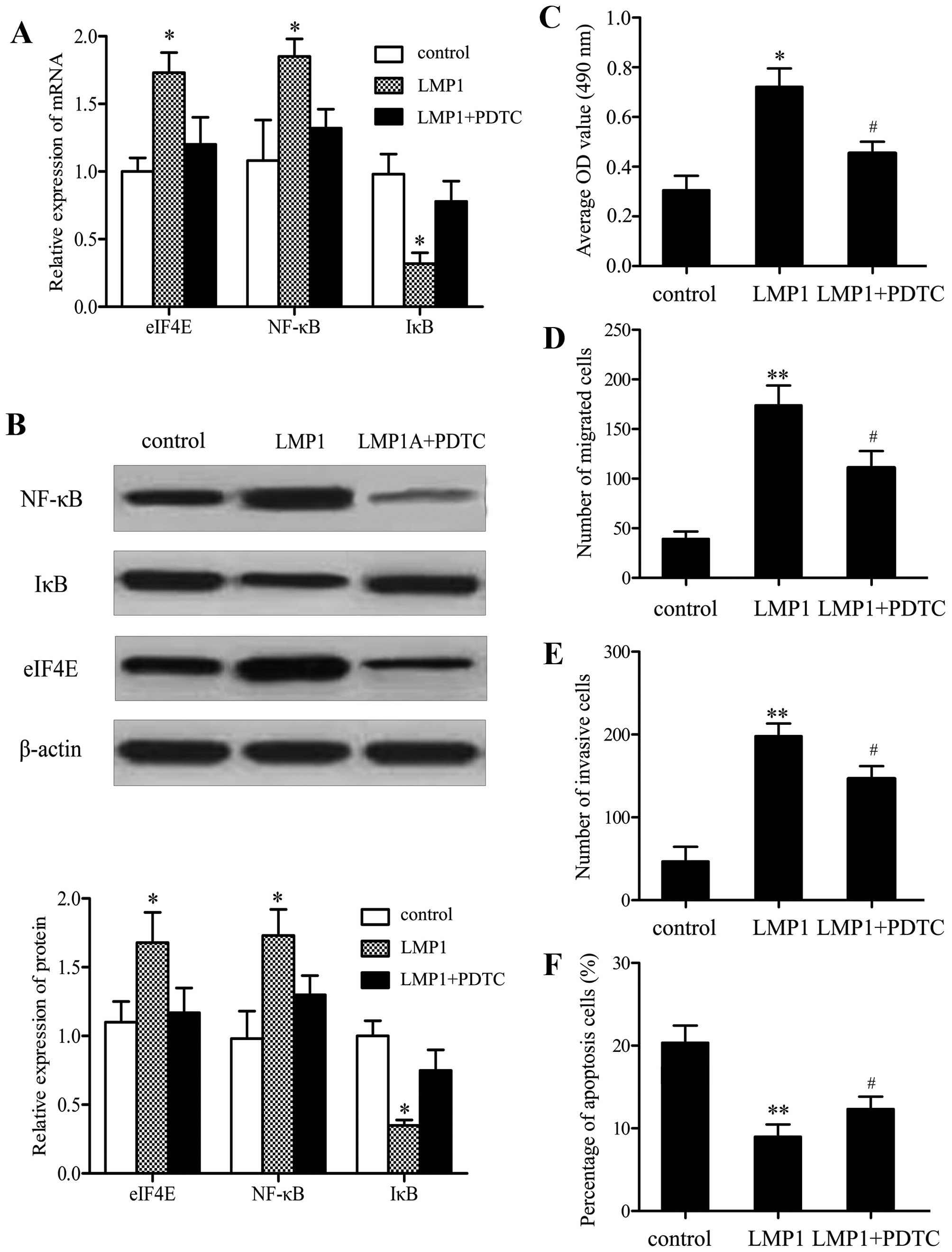
investigated. As expected, LMP1 overexpression clearly induced
activation of the NF-κB pathway (Fig.
5A). However, following pretreatment with
pyrrolidinedithiocarbamate (PDTC; Sigma), an inhibitor of the
pathway, the expression of eIF4E induced by LMP1 was dramatically
downregulated. Similar effects on eIF4E protein levels were
observed, indicating that LMP1 induces eIF4E expression by
activating the NF-κB pathway (Fig.
5B). SNK-6 cell proliferation, migration, invasion, and
apoptosis were also assessed in order to certify whether the effect
of LMP1 on SNK-6 cell function was mediated by the NF-κB pathway.
MTT analysis showed that after pretreatment with PDTC, the
proliferation induced by LMP1 was effectively attenuated (Fig. 5C). Moreover, the results of the
transwell assay indicated that the changes in cell migration
(Fig. 5D) and invasion (Fig. 5E) were consistent with
proliferation. PDTC pretreatment enhanced cell apoptosis
significantly compared with the LMP1 group (Fig. 5F). The results demonstrated that
eIF4E is responsible for LMP1-induced SNK-6 cell proliferation,
migration, invasion and apoptosis via the NF-κB signaling
pathway.
Discussion
NKTL is an uncommon disease, usually showing a
highly aggressive clinical course (19). Improved understanding of this deadly
cancer at the basic molecular level is greatly needed. The present
study provides insight into a potential regulatory mechanism
involved in NKTL.
NKTL is universally associated with EBV infection,
while LMP1 is known as a major viral oncogene in the EBV
carcinogenic process (20).
However, the effect of LMP1 on NKTL is not clear. Our data
confirmed that LMP1 is highly expressed in the NKTL cell line SNK-6
compared to PBMCs. The findings indicate that LMP1 might play an
important role in NKTL progression. Further functional analysis
revealed that transfecting LMP1 shRNA1 into SNK-6 cells induces
slower proliferation, migration, and invasion with cell cycle
arrest at the G0/G1 phase, and silencing of LMP1 also promoted
apoptosis in SNK-6 cells. The opposite effect was seen, when cells
were transfected with pcDNA3.0-LMP1, as expected. Those findings
suggest that LMP1 may act as an oncogene in NKTL progression.
Eukaryotic translation initiation factor 4E (eIF4E)
is a recently discovered oncogene (21). As a key factor in the translation
initiation complex, eIF4E plays a rate limiting role in the
initiation of translation of many mRNAs of oncogenes and growth
factor genes (22,23). Several clinical studies have
reported that the expression of eIF4E is increased in lung, breast
and esophageal cancer, among others, and is closely associated with
invasion and metastasis in these tumors (22,24,25).
Silencing of eIF4E expression by siRNA or antisense polynucleotide
reduces proliferation and changes the cell cycle of
laryngocarcinoma, esophageal carcinoma, and breast carcinoma
(26–28). To explore whether eIF4E is
associated with LMP1-induced cell function changes or plays a role
in NKTL progression, we transfected SNK-6 cells with LMP1, and
found that LPM1 significantly activated the transcriptional
activity of eIF4E in SNK-6 cells, indicating that LMP1 might affect
NKTL progression by activating the transcription of eIF4E. We then
transfected SNK-6 cells with LV-eIF4E-shRNA and found that the
expression of eIF4E decreased. Further mechanistic analysis showed
that blocking eIF4E expression significantly reduced SNK-6 cell
proliferation, migration, and invasion induced by LMP1
overexpression, and increased cell apoptosis following eIF4E
silencing. The results confirmed that LMP1 may regulate the
biological function of SNK-6 cells by enhancing eIF4E expression.
However, the mechanism by which LMP1 enhances the expression of
eIF4E in NKTL remains unclear.
A previous study proved that LMP1 increased the
development of lymphoma in LMP1 transgenic mice through the NF-κB
pathway (29). In addition, eIF4E
is a direct transcriptional target of NF-κB and is aberrantly
regulated in acute myeloid leukemia (30). Thus, the role of NF-κB in the
induction of eIF4E expression by LMP1 was evaluated in the present
study. As expected, LMP1 overexpression induced activation of the
NF-κB pathway. Furthermore, eIF4E expression induced by LMP1
overexpression was inhibited by PDTC, an inhibitor of NF-κB,
indicating that LMP1 may increase eIF4E expression via the NF-κB
signaling pathway. Further functional analysis demonstrated that
pretreatment with PDTC, ameliorated the effect on cell
proliferation, invasion and migration triggered by LMP1, and was
accompanied by increased cell apoptosis. Together, these results
suggest that LMP1 may act as an oncogene in NKTL by regulating
eIF4E via activating the NF-κB pathway.
In summary, we have shown that LMP1 is overexpressed
in NKTL and that silencing it inhibits cell proliferation,
cell-cycle progression, migration, invasion and promotes apoptosis.
Furthermore, we suggest that LMP1 regulated the development of NKTL
by regulating eIF4E expression via the NF-κB signaling pathway.
This might be a major mechanism for NKTL and other EBV-associated
tumors. Therefore, the present study provides insight into the
underlying mechanism by which LMP1 regulates the progression of
NKTL and provides a new target for treatment of NKTL.
Abbreviations:
|
NKTL
|
nasal natural killer T-cell
lymphoma
|
|
EBV
|
Epstein-Barr virus
|
|
LMP1
|
Latent membrane protein1
|
|
eIF4E
|
eukaryotic translation initiation
factor 4E
|
|
MTT
|
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium
bromide
|
|
PI
|
propidium iodide, NPC, nasopharyngeal
carcinoma
|
|
DMSO
|
dimethyl sulfoxide
|
|
FITC
|
fluorescein isothiocyanate
|
References
|
1
|
Vose J, Armitage J and Weisenburger D;
International T-Cell Lymphoma Project: International peripheral
T-cell and natural killer/T-cell lymphoma study: Pathology findings
and clinical outcomes. J Clin Oncol. 26:4124–4130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Au W-Y, Ma S-Y, Chim C-S, Choy C, Loong F,
Lie AK, Lam CC, Leung AY, Tse E, Yau CC, et al: Clinicopathologic
features and treatment outcome of mature T-cell and natural
killer-cell lymphomas diagnosed according to the World Health
Organization classification scheme: A single center experience of
10 years. Ann Oncol. 16:206–214. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
No authors listed. A clinical evaluation
of the International Lymphoma Study Group classification of
non-Hodgkin's lymphoma. The Non-Hodgkin's Lymphoma Classification
Project. Blood. 89:3909–3918. 1997.PubMed/NCBI
|
|
4
|
Takahashi E, Ohshima K, Kimura H, Hara K,
Suzuki R, Kawa K, Eimoto T and Nakamura S; NK-cell Tumor Study
Group: Clinicopathological analysis of the age-related differences
in patients with Epstein-Barr virus (EBV)-associated extranasal
natural killer (NK)/T-cell lymphoma with reference to the
relationship with aggressive NK cell leukaemia and chronic active
EBV infection-associated lymphoproliferative disorders.
Histopathology. 59:660–671. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carbone A, Gloghini A and Dotti G:
EBV-associated lymphoproliferative disorders: Classification and
treatment. Oncologist. 13:577–585. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cohen JI, Bollard CM, Khanna R and
Pittaluga S: Current understanding of the role of Epstein-Barr
virus in lymphomagenesis and therapeutic approaches to
EBV-associated lymphomas. Leuk Lymphoma. 49(Suppl 1): 27–34. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kohrt H and Advani R: Extranodal natural
killer/T-cell lymphoma: Current concepts in biology and treatment.
Leuk Lymphoma. 50:1773–1784. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Raab-Traub N: Epstein-Barr virus
transforming proteins: biologic properties and contribution to
oncogenesis. DNA Tumor Viruses. Springer; pp. 259–284. 2009,
View Article : Google Scholar
|
|
9
|
Strong MJ, Xu G, Coco J, Baribault C,
Vinay DS, Lacey MR, Strong AL, Lehman TA, Seddon MB, Lin Z, et al:
Differences in gastric carcinoma microenvironment stratify
according to EBV infection intensity: Implications for possible
immune adjuvant therapy. PLoS Pathog. 9:e10033412013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Beek J, Zur Hausen A, Klein Kranenbarg
E, van de Velde CJ, Middeldorp JM, van den Brule AJ, Meijer CJ and
Bloemena E: EBV-positive gastric adenocarcinomas: A distinct
clinicopathologic entity with a low frequency of lymph node
involvement. J Clin Oncol. 22:664–670. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang YY, Koh LW, Tsai JH, Tsai CH, Wong
EF, Lin SJ and Yang CC: Correlation of viral factors with cervical
cancer in Taiwan. J Microbiol Immunol Infect. 37:282–287.
2004.PubMed/NCBI
|
|
12
|
Whitaker NJ, Glenn WK, Sahrudin A, Orde
MM, Delprado W and Lawson JS: Human papillomavirus and Epstein-Barr
virus in prostate cancer: Koilocytes indicate potential oncogenic
influences of human papillomavirus in prostate cancer. Prostate.
73:236–241. 2013. View Article : Google Scholar
|
|
13
|
Cao Y: Epstein-Barr virus encoded LMP1
regulates cyclin D1 promoter activity by nuclear EGFR and STAT3 in
CNE1 cells. J Exp Clin Cancer Res. 14:192013.
|
|
14
|
Ho CH, Chen CL, Li WY and Chen CJ: Decoy
receptor 3, upregulated by Epstein-Barr virus latent membrane
protein 1, enhances nasopharyngeal carcinoma cell migration and
invasion. Carcinogenesis. 30:1443–1451. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen R, Zhang D, Mao Y, Zhu J, Ming H, Wen
J, Ma J, Cao Q, Lin H, Tang Q, et al: A human Fab-based
immunoconjugate specific for the LMP1 extracellular domain inhibits
nasopharyngeal carcinoma growth in vitro and in vivo. Mol Cancer
Ther. 11:594–603. 2012. View Article : Google Scholar
|
|
16
|
Devergne O, Cahir McFarland ED, Mosialos
G, Izumi KM, Ware CF and Kieff E: Role of the TRAF binding site and
NF-kappaB activation in Epstein-Barr virus latent membrane protein
1-induced cell gene expression. J Virol. 72:7900–7908.
1998.PubMed/NCBI
|
|
17
|
Shair KH, Bendt KM, Edwards RH, Nielsen
JN, Moore DT and Raab-Traub N: Epstein-Barr virus-encoded latent
membrane protein 1 (LMP1) and LMP2A function cooperatively to
promote carcinoma development in a mouse carcinogenesis model. J
Virol. 86:5352–5365. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang CF, Peng LX, Huang TJ, Yang GD, Chu
QQ, Liang YY, Cao X, Xie P, Zheng LS, Huang HB, et al: Cancer
stem-like cell characteristics induced by EB virus-encoded LMP1
contribute to radioresistance in nasopharyngeal carcinoma by
suppressing the p53-mediated apoptosis pathway. Cancer Lett.
344:260–271. 2014. View Article : Google Scholar
|
|
19
|
Aozasa K and Zaki MA: Epidemiology and
pathogenesis of nasal NK/T-cell lymphoma: A mini-review. Sci World
J. 11:422–428. 2011. View Article : Google Scholar
|
|
20
|
Zheng H, Li LL, Hu DS, Deng XY and Cao Y:
Role of Epstein-Barr virus encoded latent membrane protein 1 in the
carcinogenesis of nasopharyngeal carcinoma. Cell Mol Immunol.
4:185–196. 2007.PubMed/NCBI
|
|
21
|
Shatsky IN, Dmitriev SE, Andreev DE and
Terenin IM: Transcriptome-wide studies uncover the diversity of
modes of mRNA recruitment to eukaryotic ribosomes. Crit Rev Biochem
Mol Biol. 49:164–177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fischer PM: Cap in hand: Targeting eIF4E.
Cell Cycle. 8:2535–2541. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Zeng J, Zhou M, Li B, Zhang Y,
Huang T, Wang L, Jia J and Chen C: The tumor suppressive role of
miRNA-370 by targeting FoxM1 in acute myeloid leukemia. Mol Cancer.
11:562012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
De Benedetti A and Harris AL: eIF4E
expression in tumors: Its possible role in progression of
malignancies. Int J Biochem Cell Biol. 31:59–72. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
De Benedetti A and Graff JR: eIF-4E
expression and its role in malignancies and metastases. Oncogene.
23:3189–3199. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nasr Z, Robert F, Porco JA Jr, Muller WJ
and Pelletier J: eIF4F suppression in breast cancer affects
maintenance and progression. Oncogene. 32:861–871. 2013. View Article : Google Scholar
|
|
27
|
Oridate N, Kim HJ, Xu X and Lotan R:
Growth inhibition of head and neck squamous carcinoma cells by
small interfering RNAs targeting eIF4E or cyclin D1 alone or
combined with cisplatin. Cancer Biol Ther. 4:318–323. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
DeFatta RJ, Nathan CO and De Benedetti A:
Antisense RNA to eIF4E suppresses oncogenic properties of a head
and neck squamous cell carcinoma cell line. Laryngoscope.
110:928–933. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thornburg NJ, Kulwichit W, Edwards RH,
Shair KH, Bendt KM and Raab-Traub N: LMP1 signaling and activation
of NF-kappaB in LMP1 transgenic mice. Oncogene. 25:288–297.
2006.
|
|
30
|
Hariri F, Arguello M, Volpon L,
Culjkovic-Kraljacic B, Nielsen TH, Hiscott J, Mann KK and Borden
KL: The eukaryotic translation initiation factor eIF4E is a direct
transcriptional target of NF-kappaB and is aberrantly regulated in
acute myeloid leukemia. Leukemia. 27:2047–2055. 2013. View Article : Google Scholar : PubMed/NCBI
|