Introduction
ADAM12 is a disintegrin and metalloproteinase
family member that plays important roles in embryonic development,
acting on multiple processes including cell adhesion and cell
movement (1). In the majority of
normal adult tissues, the expression of ADAM12 is extremely
low (2), but increases in certain
pathological conditions, including carcinogenesis (3), osteoarthritis (4) and cardiac hypertrophy (5). ADAM12 has been used as a marker
for the diagnosis of breast (6) and
prostate cancer (7). Our previous
study showed that ADAM12 was highly expressed in small cell
lung cancer (SCLC) and can be considered as an effective marker for
diagnosis and prognosis (8), yet
the reasons for the high level of expression of ADAM12 in
SCLC remain unknown.
Studies of the regulation of ADAM12
expression have mainly been focused at the level of transcription.
Transforming growth factor-β (TGFβ) was found to induce the
expression of ADAM12 by activating the PI3K and MAPK
signaling pathways (9).
Z-DNA-binding protein was able to bind a negative element in the
5′-untranslated region of the ADAM12 gene to repress
transcription of ADAM12 in numerous tissues (10). The nuclear factor (NF)-κB signaling
pathway was able to promote the expression of ADAM12 by
inhibiting the expression of miR-29 (11,12).
Our previous research showed that p65 was highly expressed
and regulated by E2F1 in SCLC (13); therefore we speculated that the
transcription factor E2F1 may be a significant factor that
controls the expression of ADAM12 in SCLC. In the present
study, the mechanism by which E2F1 regulates the expression
of ADAM12 was explored, and E2F1 was found to bind to
the promoter and other cis-acting elements to regulate the
expression of ADAM12 in SCLC.
Materials and methods
Reagents and antibodies
RPMI-1640 medium was purchased from HyClone, GE
Healthcare Life Sciences (Logan, UT, USA). Fetal bovine serum
(FBS), siRNA targeting E2F1, scrambled siRNA and
Lipofectamine 2000 were purchased from Invitrogen Co. (Carlsbad,
CA, USA). Penicillin and streptomycin were purchased from Luye
Pharmaceutical Co., Ltd. (Yantai, Shandong, China). Protein A/G
beads, normal mouse IgG and a mouse anti-E2F1 monoclonal
antibody (sc-251) were purchased from Santa Cruz Biotechnology,
Inc. (Dallas, TX, USA). A chromatin immunoprecipitation (ChIP)
assay kit (17–295) and ChIP grade mouse anti-E2F1 monoclonal
antibody (17–10061) were purchased from Merck Millipore (Billerica,
MA, USA). The goat anti-ADAM12 polyclonal antibody (AF1025a)
was purchased from Abgent Co., Ltd. (Suzhou, China). Goat
anti-mouse and rabbit anti-goat secondary antibody with HRP, and
the DAB coloring kit were purchased from Beijing Zhongshan Jinqiao
Biotechnology Co., Ltd. (Beijing, China). A dual-luciferase
analysis kit (E1980) was purchased from Promega Corporation
(Madison, WI, USA). FastDigest enzymes including NheI,
BglII, KpnI and MluI were purchased from
Thermo Fisher Scientific Co. (Waltham, MA, USA). A gel extraction
kit was purchased from Takara Technology Co. (Dalian, China).
Cell culture and tissue samples
Human SCLC cell lines H1688 and H446, and human lung
adenocarcinoma cell line A549 were preserved by our laboratory
(Shandong Province Key Laboratory of Tumor Molecular Biology,
Binzhou Medical University). All cells were cultured at 37°C in
humidified 95% air and 5% CO2 in RPMI-1640 medium
supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml
streptomycin. Forty SCLC tissue samples were obtained from Yantai
Affiliated Hospital of Binzhou Medical University from January to
December 2013. All samples were biopsy samples, and all patients
voluntarily provided informed consent. The present study was
approved by the Medical Ethics Committee of Binzhou Medical
University (no. 2013007). The patient information is listed in
Table I.
 | Table IBasic information of the SCLC
patients. |
Table I
Basic information of the SCLC
patients.
|
Characteristics | No. | Percent (%) |
|---|
| Age (years) | | |
| ≥60 | 34 | 85.0 |
| <60 | 6 | 15.0 |
| Gender | | |
| Male | 29 | 72.5 |
| Female | 11 | 27.5 |
| Smoking
history | | |
| Smoker | 33 | 82.5 |
| Non-smoker | 7 | 17.5 |
| Clinical phage | | |
| LD | 5 | 12.5 |
| ED | 35 | 87.5 |
ChIP-to-sequence
ChIP was conducted according to the manual provided
with the ChIP assay kit (13). In
brief, 5×107 cells were fixed using 1% formaldehyde and
were subsequently incubated in SDS lysis buffer. Ultrasound was
used to fragment the genomic DNA, and the sample was pretreated
with protein A/G beads, and then centrifugation (2,000 rpm, 4°C).
The protein beads were then removed. The resulting sample was
divided into two parts. One part was incubated overnight with the
ChIP grade mouse anti-E2F1 monoclonal antibody (4
µg), and the other with normal mouse IgG (4 µg). On
the following morning, the protein A/G beads were added and
incubated for 2 h at 4°C. The resulting antigen-antibody-protein
bead complexes were reverse crosslinked in the presence of salt at
a high concentration (5 M, NaCl). DNA fragments were purified and
sequenced (BGI Co., www.genomics.cn). The data processing was reported in
our previous study (13).
Immunohistochemistry (IHC)
IHC was carried out according to the protocols of
our laboratory (8,13). In brief, the sections were dewaxed
and rehydrated in a series of alcohols to water. Antigens were
retrieved by heating the sections in citrate buffer (0.01 M,
pH=6.0) for 45 min at 95°C in a boiler. The sections were
subsequently incubated with the primary antibodies overnight at
4°C. The dilution of primary antibodies was 1:50 for E2F1
and 1:200 for ADAM12. On the following day, all the sections
were incubated with a secondary horseradish peroxidase
(HRP)-conjugated antibody, and a brown color reaction was developed
using the DAB kit. Sections were counterstained with hematoxylin,
differentiated, dehydrated, cleared and mounted in neutral gum. The
IHC scores were assessed according to our previous studies
(8,13).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR was performed according to our previous
studies (8,13). The primers for each target gene are
listed in Table II.
 | Table IIPrimers of the target genes for
RT-qPCR. |
Table II
Primers of the target genes for
RT-qPCR.
| Name | | Primer
sequences | Length (bp) | Tm (°C) |
|---|
| Seq0 | F |
ATTCAGGAAGACGGGTGGCT | | |
| R |
TGGTAACCCATCCATTAAGCGG | 70 | 60 |
| Seq1 | F |
GGTGGTCCTAGGTCTGAGCA | | |
| R |
TCAGTTTCCCACAATGCGTG | 81 | 60 |
| Seq2 | F |
GCACTCAGCGTCCTATCTGT | | |
| R |
AAAGTACGCTTGCCAGACCA | 72 | 60 |
| E2F1 | F |
CATCAGTACCTGGCCGAGAG | 118 | 60 |
| R |
TGGTGGTCAGATTCAGTGAGG | | |
| ADAM12 | F |
GCAGTTTCACGGAAACCCAC | | |
| R |
ACACGTGCTGAGACTGACTG | 131 | 60 |
Western blotting
Western blotting was performed according to our
previous studies (8,13). Cells were lysed, and 50 µg of
protein was loaded and separated on 10% polyacrylamide gels (70 V
for 30 min; 140 V for 70 min; 180 V for 10 min). Proteins were
subsequently transferred to NC membranes by wet transfer (300 mA
for 150 min), which were blocked with 5% skimmed milk powder, prior
to incubation with the primary antibodies overnight at 4°C. On the
following morning, the membranes were washed four times (three
times in TBS buffer, one time in TBST buffer), incubated with
HRP-conjugated secondary antibodies (goat anti-mouse for
E2F1 and rabbit anti-goat for ADAM12, 1:5,000), washed four
times and exposed to X-ray film. The dilution of primary antibodies
was as follows: 1:100 for E2F1, 1:200 for ADAM12.
siRNA treatment
siRNAs targeting E2F1 and a scramble- control
siRNA were used to assess the relevance of E2F1 to the
expression of ADAM12 (13).
Cells (1×105) were cultured into a 6-well plate and
transfected with Lipofectamine 2000. The procedure was performed
according to previously described methods (8,13).
Assembly of luciferase reporter
constructs
Genomic DNA was extracted from the H1688 cells, and
the fragments (Chr10: 128010444–128011026, located in the intron of
ADAM12, named seq0; Chr10: 128076927–128078127, located in
the promoter of ADAM12, named seq1; Chr10:
128086195–128086876, located in the upstream 20 kb from
transcription start site of ADAM12, named seq2) pulled down
by E2F1 were amplified by PCR using primers with the
sequences shown in Table III.
These PCR fragments and the pGL3-basic promoter-less vector were
digested with FastDigest NheI, BglII for seq1,
KpnI and MluI for seq0 and seq2. The digested
fragments were extracted using a gel extraction kit and ligated
using T4 DNA ligase to generate three luciferase reporter
constructs. The luciferase reporter vector containing a mutant
E2F1 binding site was constructed by overlap PCR using the
primers listed in Table III.
These three luciferase reporter vectors were digested with
FastDigest KpnI and MluI, and the seq0 and seq2
fragments were ligated into the digested seq1 vector using T4 DNA
ligase to generate the fusion luciferase reporter constructs
containing seq0-seq1 and seq2-seq1. All the constructs were
verified by sequencing (BioSune Co., Jinan, China).
 | Table IIIPrimers of the target fragments for
luciferase reporter constructs. |
Table III
Primers of the target fragments for
luciferase reporter constructs.
| Fragment | | Primers | E |
|---|
| Seq0 | F | ACTGGTACCAGTATGTACAAATGAAGTGTCATG | KpnI |
| R | AATACGCGTAGACCATGCGGTTCCCA | MluI |
| Seq1 | F | ACTGCTAGCGTGCTCCGTCAGGAATCGGT | NheI |
| R | ACTAGATCTTTCTGGCACAAGCCAGCCTT | BglII |
| Mut-Seq1 | P1 |
TCTTATTAaaaaGGAAC | |
| P2 |
GTTCCttttTAATAAGA | |
| Seq2 | F | AATGGTACCGGGCAGTTGGCTCTGTTA | KpnI |
| R | AATACGCGTAACCCAAATAGCCCTGCC | MluI |
Luciferase reporter analysis
H1688, H446 and A549 cells were transfected with 0.5
µg luciferase reporter vector, 0.3 µg E2F1
expression vector or pcDNA3.1 and 0.02 µg pRL-TK
Renilla reniformis luciferase. Luciferase activity was
analyzed with dual-luciferase assay kits according to the
instruction manual.
Statistical analysis
All the data were analyzed using SPSS 17.0 software
(SPSS, Inc., Chicago, IL, USA) and paired t-tests were used to
assess the significance of the differences in expression among the
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
ADAM12 is highly expressed in SCLC
samples in which E2F1 is strongly positive
Our previous results found that ADAM12 and
E2F1 were highly expressed in SCLC tissue samples,
respectively (8,13). NF-κB induced the expression of
ADAM12 (11,12) and p65 was highly expressed and
regulated by E2F1 in the SCLC samples (13), which indicated that E2F1 may
be a significant factor for promoting the expression of
ADAM12. Since the tissues used for detection in our previous
studies were from differential hospitals (8,13), it
was unconvincing that E2F1 may regulate the expression of
ADAM12. In order to solve this issue, an additional 40 SCLC
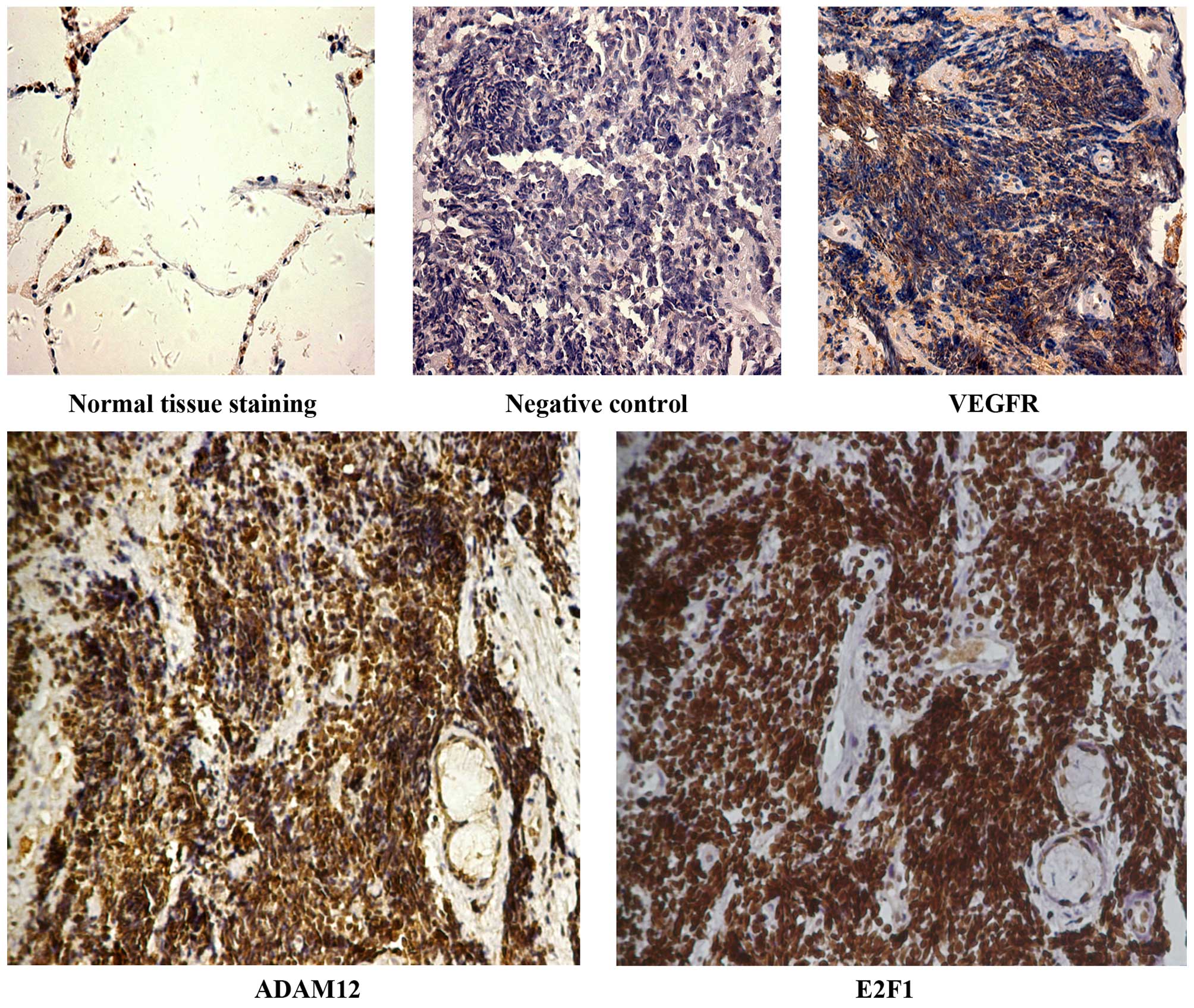
tissue samples were obtained. IHC results revealed that
negative/weak (<10%), moderate (10–60%) and strong (>60%)
positive expression was noted in 2, 3, 12 and 22 cases for
E2F1; and in 5, 4, 11 and 19 cases for ADAM12
(Table IV). These results were
consistent with our previous studies (8,13). The
positive expression (including moderate and strong staining) of
E2F1 and ADAM12 was 85 and 75%, respectively. In the
same tissue samples, ADAM12 was highly expressed in the
tissue samples for which anti-E2F1 staining was
strong-positive (Fig. 1). The
section incubated with phosphate-buffered saline (PBS) instead of
the primary antibody was considered as the negative control, and
the expression of vascular endothelial growth factor receptor
(VEGFR) in SCLC has been reported and was considered as the
positive control (14), and the
expression levels of E2F1 and ADAM12 were negative in
normal alveolar epithelial cells (Fig.
1).
 | Table IVExpression levels of E2F1 and
ADAM12 in 40 SCLC tissue samples. |
Table IV
Expression levels of E2F1 and
ADAM12 in 40 SCLC tissue samples.
| E2F1 | ADAM12 |
|---|
|
|---|
Negative
| Positive
| Negative
| Positive
|
|---|
| Negative | Weak | Moderate | Strong | Negative | Weak | Moderate | Strong |
| 2 | 3 | 12 | 22 | 5 | 4 | 11 | 19 |
E2F1 knockdown significantly inhibits the
expression of ADAM12
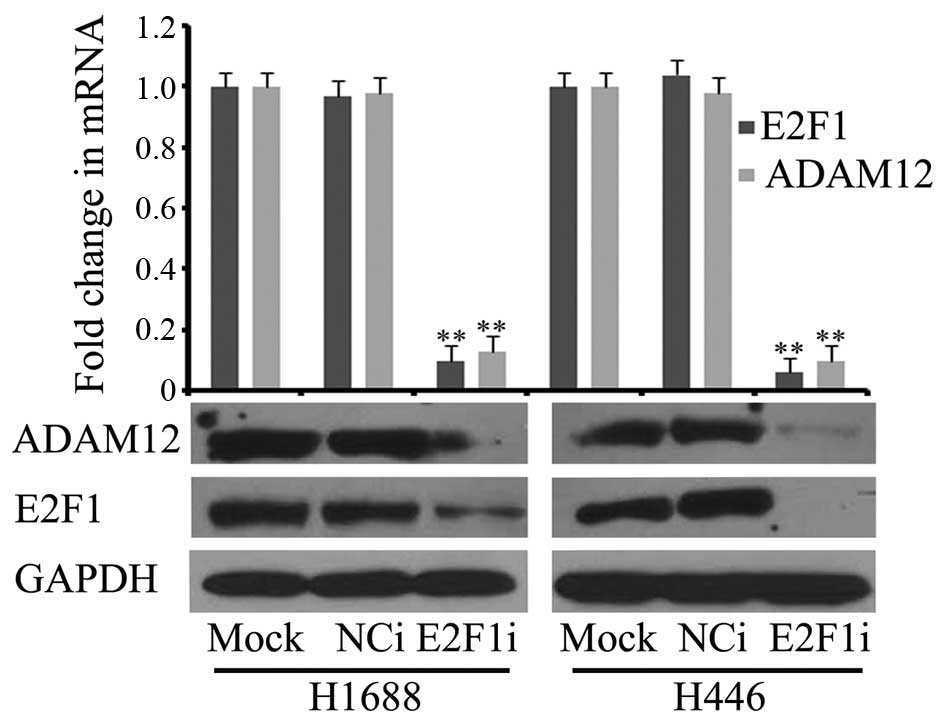
As shown in Fig. 1,
E2F1 may regulate the expression of ADAM12. siRNA targeting
E2F1 was transfected into H1688 and H446 cells, and
E2F1 expression was significantly decreased at the
transcription and translation levels (Fig. 2). Meanwhile, ADAM12
expression was also significantly reduced (Fig. 2). This result showed that
E2F1 knockdown significantly inhibited the expression of
ADAM12 at the mRNA and protein levels, thus indicating that
E2F1 was able to regulate the expression of ADAM12 at
the transcription level.
ChIP-to-seq analysis indicates the
binding of E2F1 to ADAM12
As ADAM12 may be regulated by E2F1,
ChIP-to-seq was employed to discover the E2F1 binding sites
to explore the detailed mechanism. A total of 4,700 genes regulated
by E2F1 were identified by ChIP-to-seq (data not shown;
these data will be reported in a subsequent study). There were
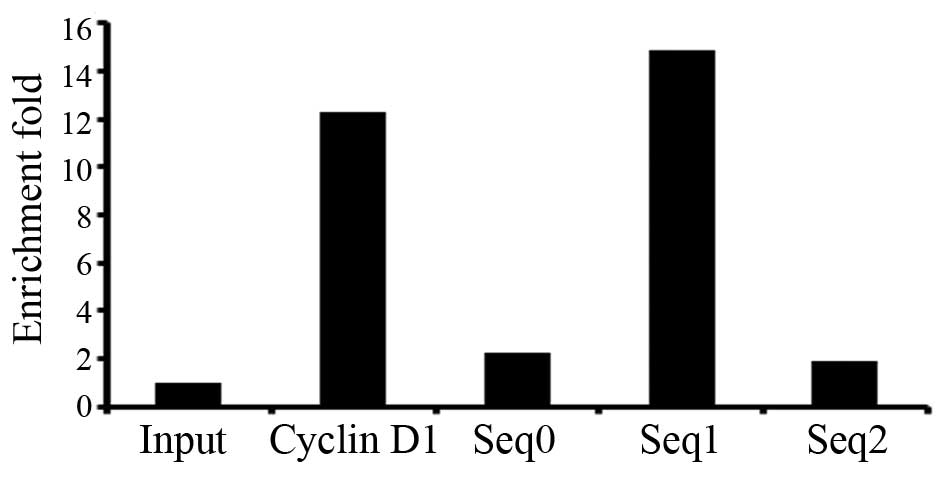
three E2F1 binding sites in the ADAM12 gene (Table V). They were the following: Chr10:
128010444–128011026, located in the intron of ADAM12, named
seq0; Chr10: 128076927–128078127, located in the promoter of
ADAM12, named seq1; Chr10: 128086195–128086876, located in
the upstream 20 kb from the transcription start site of
ADAM12, named seq2. Since there were three E2F1
binding sites, the enrichment fold was calculated. The results
showed that the enrichment fold of seq0, seq1 and seq2 by
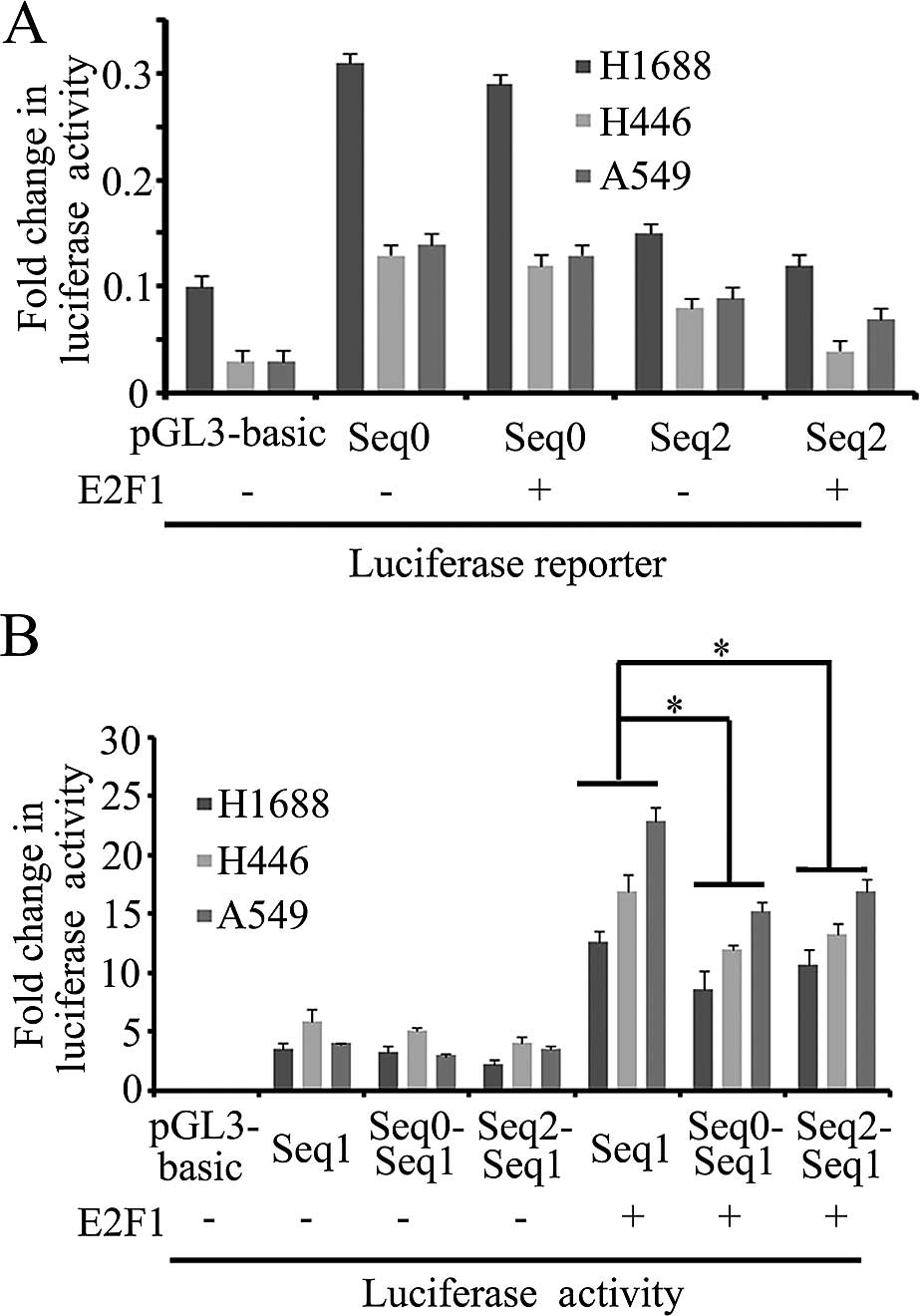
E2F1 was 2.3, 14.9 and 1.9 as determined by qPCR (Fig. 3), indicating that the ability of
E2F1 binding seq1 was stronger than seq0 and seq2.
 | Table VFeatures of the E2F1 binding
DNA fragments in the ADAM12 gene from ChIP-to-seq. |
Table V
Features of the E2F1 binding
DNA fragments in the ADAM12 gene from ChIP-to-seq.
| Name | Size
(kb) | Ch | Sites |
|---|
| ADAM12 | 0.582 | 10 |
128010444–128011026 |
| 1.2 | 10 |
128076927–128078127 |
| 0.681 | 10 |
128086195–128086876 |
E2F1 regulates ADAM12 expression via an
E2F1 binding motif in the promoter
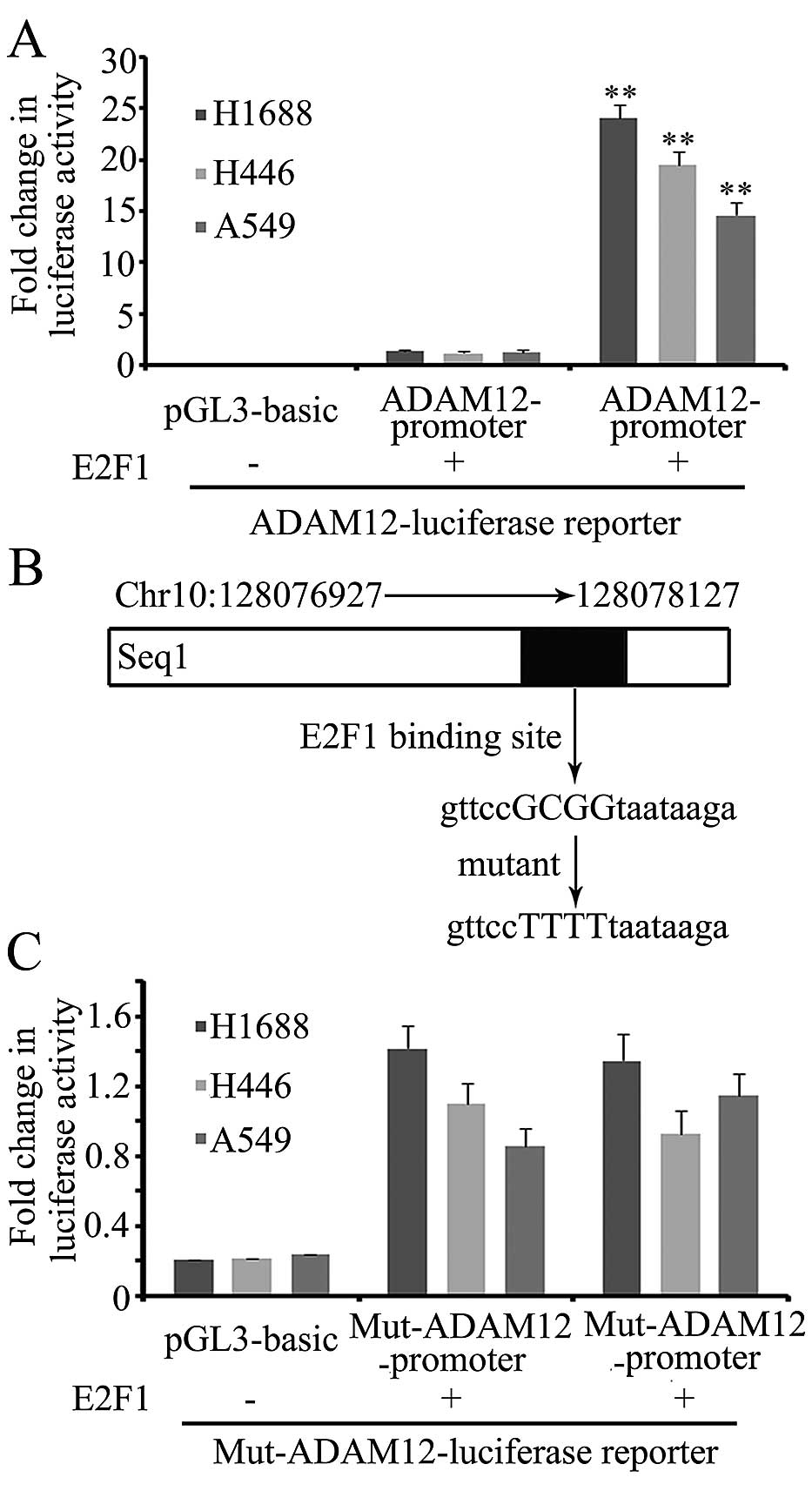
After considering the above-mentioned results, we
speculated that E2F1 could directly regulate ADAM12
expression and tested this using dual-luciferase reporter
constructs. The seq1 fragment was cloned and incorporated into the
pGL3 basic vector (known as the wild-type ADAM12 promoter).
The wild-type ADAM12 promoter was transfected into H1688,
H446 and A549 cells, and this signifi-cantly promoted the
luciferase activity. When co-transfected with the E2F1
expression vector, the luciferase activity was significantly
increased (Fig. 4A). One putative
E2F1 binding site (gttccGCGGtaataaga) was identified by
MatInspector software (http://www.genomatix.de/) in the seq1 fragment
(Fig. 4B). An E2F1 binding
site mutant luciferase reporter (known as the mut-ADAM12
promoter) was constructed. Following transfection into H1688, H446
and A549 cells, overexpression of E2F1 did not significantly
promote the activity of the mut-ADAM12 promoter, indicating
that E2F1 stimulated the expression of ADAM12 via the
E2F1 binding site in the ADAM12 promoter region
(Fig. 4C).
Cis-acting elements seq0 and seq2 inhibit
the activity of the ADAM12 promoter
ChIP-to-seq results showed that there were three
E2F1 binding sites in the ADAM12 gene (Table V), in which seq1 as a promoter could
recruit E2F1 to drive ADAM12 expression. Seq0 and
seq2 also bind E2F1, but the function of seq0 and seq2 in
the regulation of ADAM12 is unknown. Seq0 and seq2 fragments
were cloned into the pGL3 basic vector. After transfection into
H1688, H446 and A549 cells, there was no luciferase activity,
indicating that seq0 and seq2 fragments could not promote the
expression of luciferase (Fig. 5A).
We next ascertained whether seq0 or seq2 has enhancer function.
Subsequently, the seq0-seq1 and seq2-seq1 fusion luciferase
expression constructs were analyzed. Compared with the seq1
fragments, the luciferase activity of fusion fragment vectors was
on average decreased, but there was no statistically significant
difference. When co-transfected with E2F1, the luciferase
activity of the fusion fragments was significantly decreased,
indicating that seq1 and seq2 repressed the ADAM12 promoter
(Fig. 5B).
Discussion
ADAM12 exhibits a wide range of expression
levels in various tissues and cells (15), but the distribution of ADAM12
is strictly regulated (16). During
embryonic development of mice, ADAM12 is expressed in
stromal cells and results in bone and muscle development. It is
absent in adult rat muscle cells, but is expressed again during the
process of muscle regeneration (16–19).
In addition, ADAM12 is also expressed in osteoclasts
(20,21), macrophages (22), trophoblast cells during embryonic
development (23), adipocytes
(24), chon-drocytes (25) and liver stellate cells (26). These results demonstrate that
ADAM12 expression has strict temporal and spatial
specificity. Additionally, ADAM12 is highly expressed in
certain types of tumors, such as breast (6), prostate (7) and small cell lung cancer (SCLC)
(8), but there are limited studies
reporting the regulation of ADAM12 expression in tumor
tissues. In the present study, ADAM12 and E2F1 were
highly expressed in the same SCLC tissue samples, indicating that
E2F1 may control the expression of ADAM12.
E2F1, as a classical transcriptional factor,
plays important roles in cell cycle regulation, cell proliferation
and apop-tosis (27). Numerous
studies suggest that E2F1 is involved in the invasion and
metastasis of tumor cells by regulating the expression of matrix
metalloproteinases (13,28), thrombos-pondin1 (29), platelet-derived growth factor
receptor (30) and vascular
endothelial growth factor receptor (31). The target genes regulated by
E2F1 in different cells were different when ChIP-on-ChIP or
ChIP-to-seq methods were used (32–35).
In our ChIP-to-seq database, ADAM12 was able to bind
E2F1, and ADAM12 expression was most significantly
decreased when there was depletion of E2F1. Therefore, we
explored the detailed mechanism of ADAM12 regulation.
Although there are three E2F1 binding sites in the
ADAM12 gene, their ability for E2F1 recruitment
differed. One reason may be a difference in the binding motif. The
present results showed that E2F1 regulated ADAM12
expression via the E2F1 binding site in the promoter region,
and this was shown to be a functional motif. The other two binding
sites were located in the upstream 20-kb and intron regions of
ADAM12. They were unable to promote the expression of
luciferase, but reduced the activity of the promoter. Although the
qPCR results showed that these three elements could recruit
E2F1, the strongest recruitment was by the promoter
sequence. This result possibly indicates that E2F1 has a
binding site preference due to the different binding motifs.
In conclusion, the present findings offer support
that E2F1 regulates the expression of ADAM12 by
binding the promoter and other cis-acting elements.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. 81302017) and the
Natural Science Foundation of Shandong (grant no. ZR2013HL004).
Abbreviations:
|
ADAM
|
A disintegrin and
metalloproteinase
|
|
SCLC
|
small cell lung cancer
|
References
|
1
|
Mazzocca A, Giannelli G and Antonaci S:
Involvement of ADAMs in tumorigenesis and progression of
hepatocellular carcinoma: Is it merely fortuitous or a real
pathogenic link? Biochim Biophys Acta. 1806:74–81. 2010.PubMed/NCBI
|
|
2
|
Jacobsen J, Visse R, Sørensen HP, Enghild
JJ, Brew K, Wewer UM and Nagase H: Catalytic properties of ADAM12
and its domain deletion mutants. Biochemistry. 47:537–547. 2008.
View Article : Google Scholar
|
|
3
|
Ieguchi K, Tomita T, Omori T, Komatsu A,
Deguchi A, Masuda J, Duffy SL, Coulthard MG, Boyd A and Maru Y:
ADAM12-cleaved ephrin-A1 contributes to lung metastasis. Oncogene.
33:2179–2190. 2014. View Article : Google Scholar
|
|
4
|
Kerna I, Kisand K, Suutre S, Murde M and
Tamm A, Kumm J and Tamm A: The ADAM12 is upregulated in synovitis
and postin-flammatory fibrosis of the synovial membrane in patients
with early radiographic osteoarthritis. Joint Bone Spine. 81:51–56.
2014. View Article : Google Scholar
|
|
5
|
Higashiyama S: Membrane-anchored
heparin-binding EGF-like growth factor processing by ADAM12 in
cardiac hypertrophy. Nihon Rinsho. 61:767–775. 2003.In Japanese.
PubMed/NCBI
|
|
6
|
Roy R, Rodig S, Bielenberg D, Zurakowski D
and Moses MA: ADAM12 transmembrane and secreted isoforms promote
breast tumor growth: A distinct role for ADAM12-S protein in tumor
metastasis. J Biol Chem. 286:20758–20768. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peduto L, Reuter VE, Sehara-Fujisawa A,
Shaffer DR, Scher HI and Blobel CP: ADAM12 is highly expressed in
carcinoma-associated stroma and is required for mouse prostate
tumor progression. Oncogene. 25:5462–5466. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shao S, Li Z, Gao W, Yu G, Liu D and Pan
F: ADAM-12 as a diagnostic marker for the proliferation, migration
and invasion in patients with small cell lung cancer. PLoS One.
9:e859362014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Le Pabic H, Bonnier D, Wewer UM, Coutand
A, Musso O, Baffet G, Clément B and Théret N: ADAM12 in human liver
cancers: TGF-beta-regulated expression in stellate cells is
associated with matrix remodeling. Hepatology. 37:1056–1066. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ray BK, Dhar S, Shakya A and Ray A:
Z-DNA-forming silencer in the first exon regulates human ADAM-12
gene expression. Proc Natl Acad Sci USA. 108:103–108. 2011.
View Article : Google Scholar :
|
|
11
|
Li H, Solomon E, Duhachek Muggy S, Sun D
and Zolkiewska A: Metalloprotease-disintegrin ADAM12 expression is
regulated by Notch signaling via microRNA-29. J Biol Chem.
286:21500–21510. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Díaz B, Yuen A, Iizuka S, Higashiyama S
and Courtneidge SA: Notch increases the shedding of HB-EGF by
ADAM12 to poten-tiate invadopodia formation in hypoxia. J Cell
Biol. 201:279–292. 2013. View Article : Google Scholar
|
|
13
|
Li Z, Guo Y, Jiang H, Zhang T, Jin C,
Young CY and Yuan H: Differential regulation of MMPs by E2F1, Sp1
and NF-kappa B controls the small cell lung cancer invasive
phenotype. BMC Cancer. 14:2762014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ioannou M, Papamichali R, Kouvaras E,
Mylonis I, Vageli D, Kerenidou T, Barbanis S, Daponte A, Simos G,
Gourgoulianis K, et al: Hypoxia inducible factor-1 alpha and
vascular endothelial growth factor in biopsies of small cell lung
carcinoma. Lung. 187:321–329. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Harris HA, Murrills RJ and Komm BS:
Expression of meltrin-alpha mRNA is not restricted to fusagenic
cells. J Cell Biochem. 67:136–142. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kurisaki T, Masuda A, Osumi N, Nabeshima Y
and Fujisawa-Sehara A: Spatially- and temporally-restricted
expression of meltrin alpha (ADAM12) and beta (ADAM19) in mouse
embryo. Mech Dev. 73:211–215. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Borneman A, Kuschel R and Fujisawa-Sehara
A: Analysis for transcript expression of meltrin alpha in normal,
regenerating, and denervated rat muscle. J Muscle Res Cell Motil.
21:475–480. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Galliano MF, Huet C, Frygelius J, Polgren
A, Wewer UM and Engvall E: Binding of ADAM12, a marker of skeletal
muscle regeneration, to the muscle-specific actin-binding protein,
alpha -actinin-2, is required for myoblast fusion. J Biol Chem.
275:13933–13939. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao Y, Zhao Z, Gruszczynska-Biegala J and
Zolkiewska A: Role of metalloprotease disintegrin ADAM12 in
determination of quiescent reserve cells during myogenic
differentiation in vitro. Mol Cell Biol. 23:6725–6738. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abe E, Mocharla H, Yamate T, Taguchi Y and
Manolagas SC: Meltrin-alpha, a fusion protein involved in
multinucleated giant cell and osteoclast formation. Calcif Tissue
Int. 64:508–515. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Verrier S, Hogan A, McKie N and Horton M:
ADAM gene expression and regulation during human osteoclast
formation. Bone. 35:34–46. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu C, Li L, Zhao J, Fan Q, Tian WX and He
RQ: Effect of α2M on earthworm fibrinolytic enzyme III-1
from Lumbricus rubellus. Int J Biol Macromol. 31:71–77. 2002.
View Article : Google Scholar
|
|
23
|
Gilpin BJ, Loechel F, Mattei MG, Engvall
E, Albrechtsen R and Wewer UM: A novel, secreted form of human ADAM
12 (meltrin alpha) provokes myogenesis in vivo. J Biol Chem.
273:157–166. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tani N, Higashiyama S, Kawaguchi N,
Madarame J, Ota I, Ito Y, Ohoka Y, Shiosaka S, Takada Y and
Matsuura N: Expression level of integrin alpha 5 on tumour cells
affects the rate of metastasis to the kidney. Br J Cancer.
88:327–333. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kveiborg M, Albrechtsen R, Rudkjaer L, Wen
G, Damgaard-Pedersen K and Wewer UM: ADAM12-S stimulates bone
growth in transgenic mice by modulating chondrocyte proliferation
and maturation. J Bone Miner Res. 21:1288–1296. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bourd-Boittin K, Le Pabic H, Bonnier D,
L'Helgoualc'h A and Théret N: RACK1, a new ADAM12 interacting
protein. Contribution to liver fibrogenesis. J Biol Chem.
283:26000–26009. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wyllie AH: E2F1 selects tumour cells for
both life and death. J Pathol. 198:139–141. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Johnson JL, Pillai S, Pernazza D, Sebti
SM, Lawrence NJ and Chellappan SP: Regulation of matrix
metalloproteinase genes by E2F transcription factors: Rb-Raf-1
interaction as a novel target for metastatic disease. Cancer Res.
72:516–526. 2012. View Article : Google Scholar :
|
|
29
|
Ji W, Zhang W and Xiao W: E2F-1 directly
regulates thrombos-pondin 1 expression. PLoS One. 5:e134422010.
View Article : Google Scholar
|
|
30
|
Minato Y, Tashiro E, Kanai M, Nihei Y,
Kodama Y and Imoto M: Transcriptional regulation of a new variant
of human platelet-derived growth factor receptor alpha transcript
by E2F-1. Gene. 403:89–97. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pillai S, Kovacs M and Chellappan S:
Regulation of vascular endothelial growth factor receptors by Rb
and E2F1: Role of acetylation. Cancer Res. 70:4931–4940. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lavrrar JL and Farnham PJ: The use of
transient chromatin immunoprecipitation assays to test models for
E2F1-specific transcriptional activation. J Biol Chem.
279:46343–46349. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jin VX, Rabinovich A, Squazzo SL, Green R
and Farnham PJ: A computational genomics approach to identify
cis-regulatory modules from chromatin immunoprecipitation
microarray data - a case study using E2F1. Genome Res.
16:1585–1595. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Weinmann AS, Bartley SM, Zhang T, Zhang MQ
and Farnham PJ: Use of chromatin immunoprecipitation to clone novel
E2F target promoters. Mol Cell Biol. 21:6820–6832. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wells J, Graveel CR, Bartley SM, Madore SJ
and Farnham PJ: The identification of E2F1-specific target genes.
Proc Natl Acad Sci USA. 99:3890–3895. 2002. View Article : Google Scholar : PubMed/NCBI
|