Introduction
Metastatic colorectal cancers (mCRCs) are known to
benefit from targeted therapy of epithelial growth factor receptor
(EGFR) inhibitor, including Cetuximab and Panitumumab. However,
Cetuximab is ineffective in patients harboring BRAF and KRAS
mutations, which accounted for 40 and 10% of CRC patients,
respectively (1–6). The phenomenon of mutually exclusivity
between mutated BRAF and KRAS in CRCs in a same tumor have been
particularly noted (7,8). The high correlation between BRAF and
KRAS mutation status and ERK1/2 activation has been proven in many
types of cancer included CRC. Nevertheless, once CRCs contain
mutation of either BRAF or KRAS genes, Cetuximab cannot suppress
the auto-signal transduction downstream of the signaling pathway,
and could even trigger uncontrolled abnormal cell growth,
proliferation and even metastases (4,9).
The miR-378 is a very short non-coding RNA, reported
to possibly play a crucial role in mediating gene expression in CRC
cells. It was considered to function as a tumor suppressor in
inhibiting tumor growth and invasion; aberrant expression of
miR-378 results in dysregulation of cell proliferation, increase
the tumor size, as well as the capability of tumor cell invasion
(10). Previous studies indicated
that miR-378 could act as a new biomarker in CRC, due to higher
expression level only observed in wtCRC (the CRC without
presenting BRAF or KRAS mutations) or normal of colonic tissues but
not in BRAF or KRAS mutated CRC cells (11,12).
Most recently, high expression of miR-378 can even suppress the
cell proliferation and induces apoptosis by targeting BRAF, as
observed by Wang et al (13). Although the correlation between
miR-378 and MAPK signaling pathway were revealed by substantial
evidence, for example, Nagalingam et al emphasized Ras
signaling could be one of the main targets of miR-378 in cardiac
hypertrophy (14), however, the
underlying mechanism of different subtypes of CRC, which correlated
with sensitivity to the targeting therapy or regulation of miR-378
still remain unknown.
The precursor of miR-378 is derived from the first
intron of peroxisome proliferator-activated receptor γ coactivator
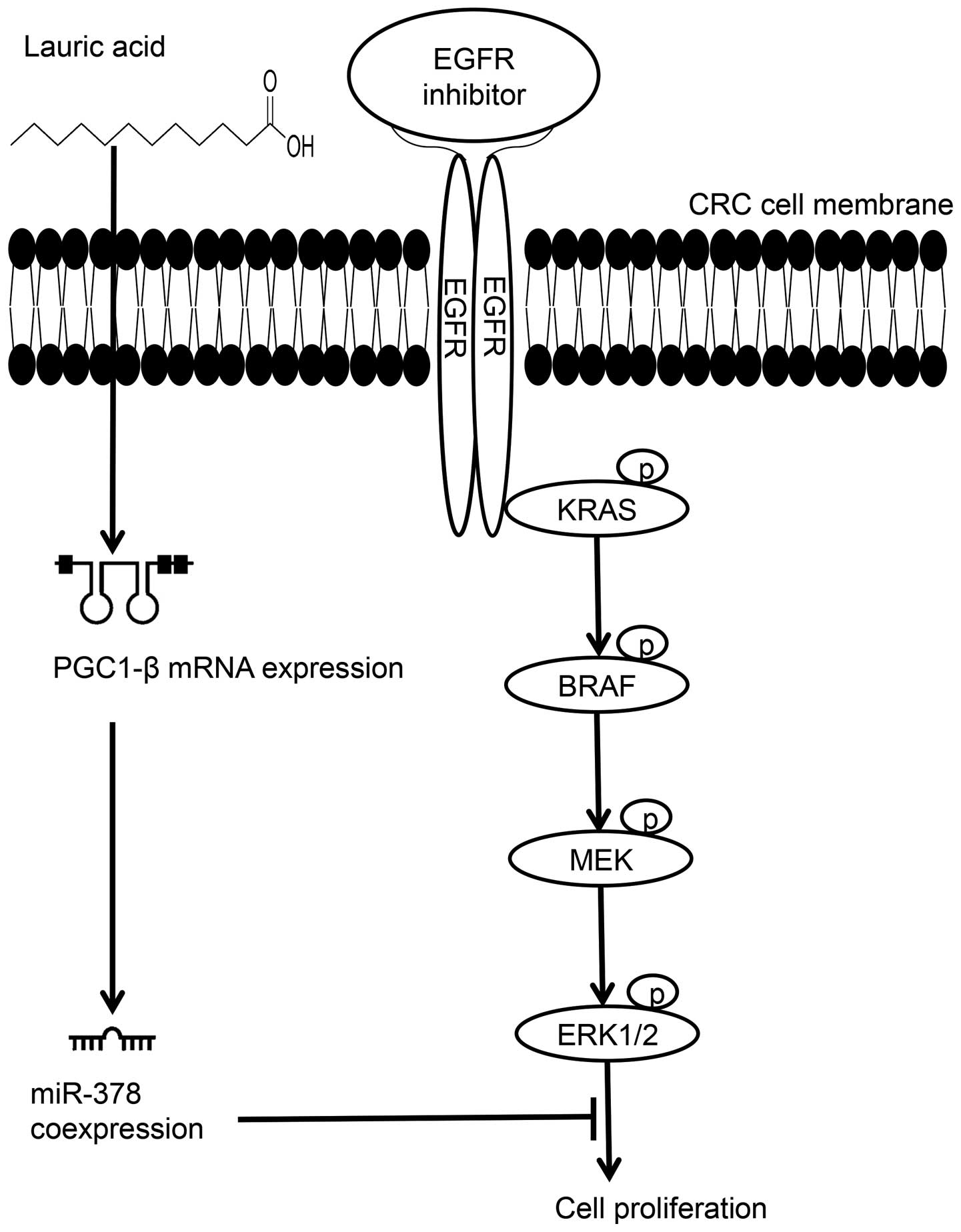
1β (PGC-1β) gene, a cellular energy-related gene. Expression of
PGC-1β is highly associated with saturated fatty acid intake
(Fig. 1) (15–19).
We therefore postulated that once CRC cells were induced by lauric
acid, the saturated fatty acid with a 12-carbon atom chain,
triggers the transcriptional activity of PGC-1β gene to accumulate
messenger RNA (mRNA) expression, consequently resulting in
increasing level of coexpressed products of miR-378 in cells
(Fig. 1); once elevated the
expression level of miR-378 in CRC mutants to the level of
wtCRC cells or normal cells, it may automatically
suppress cell growth or lead the mutated CRC cells restoring
sensitivity to Cetuximab. In the present study, the elevation of
miR-378 expression levels in BRAF or KRAS mutated CRC cells were
firstly restored by an in vitro transfection method; then
the cells were incubated in lauric acid growth medium, and their
response to Cetuximab was further measured via cellular viability.
In addition, we performed protein analysis to investigate the
potential role of miR-378 associated with MAPK signaling
pathway.
Materials and methods
Confirmation of BRAF and KRAS mutational
status of the cell lines
Seven colon cancer cell lines were used, these were
HCT-116, HCT-15, SW-480, SW-620, WiDr, HT-29 and Caco-2. The
mutation statuses of both BRAF and KRAS of each cell line was
confirmed by sequencing analysis, and further searched according to
the COSMIC database (http://www.sanger.ac.uk/genetics/CGP/cosmic/) before
the experiments. KRAS mutation with G13D substitution in exon 2 was
confirmed in HCT-116 and HCT-15, SW480 and SW620 are KRAS mutated
with G12V in exon 2. HT-29 and WiDr are BRAF mutated with V600E in
exon 15. The mutation of BRAF and KRAS did not coexist in any of
the cell line used. Caco-2 was confirmed as non-BRAF or non-KRAS
mutation, labeled as wtCRC, which is reported to be
sensitive to Cetuximab according to clinical experience (20–23),
was used as internal control when needed. The cells were cultured
in RPMI or Dulbecco's modified Eagle's medium (DMEM) growth medium
with 10–20% fetal bovine serum (FBS). Indeed, for the time of
lauric acid treatment in vitro, we could only select the
more aggressive cell growth, such as HCT116, SW480 and HT29, in
order to further obtained proteins for detection and observed
differences in cell viability.
Elevated level of miR-378 in cells by
direct in vitro miR-378 transfection
Directly transfected miR-378 to all CRC cell lines
were performed by seeding into a 6-well plate with 1×105
cells/well and cultured overnight at 37°C with 5% CO2.
miR-378 transfection complexes were then prepared by mixing with
miR-378 mimic and HiPerFect Transfection Reagent (cat. no. 301704)
(both from Qiagen, Valencia, CA, USA) under instructions of the
manufacturer's protocol. To confirm successful miR-378
transfection, the commercially available positive control AllStars
Hs Cell Death control siRNA (cat. no. 1027298), and Allstars
negative control siRNA validated non-silencing siRNA (cat. no.
1027280) (both from Qiagen) were included in every batch of the
experiments. Before and after transfection of miR-378, the
expression level was detected by qRT-PCR, and the commercial
available normal colon RNA extraction (BioChain, Newark, CA, USA)
was used as baseline comparison.
Elevated level of miR-378 in cells by
indirectly incubating cells in lauric acid medium
In order to indirectly stimulate cells to coexpress
miR-378 from its host gene PGC-1β by feeding lauric acid (15–19),
the three cell lines (HT29, HCT116 and SW480) with aggressive
growth patterns were selected and cultured in a gradient lauric
acid concentration (0.15, 0.3, 0.45 and 0.6 mM) of growth medium
for 96 h. The metabolic rates of lauric acid in cells were then
further measured before the RNA expression level of PGC-1β, and
coexpressed miR-378 levels were investigated.
RNA extraction and quantification of
miR-378
The expression level of miR-378 in cells was
detected at original status cells, before and after miR-378
transfection in vitro or indirectly induced miR-378 by
lauric acid feeding. Extraction of total RNA was performed using
TRIzol (Invitrogen Inc., Carlsbad, CA, USA), cDNA of miR-378 was
synthesized according to the TaqMan microRNA assay protocol
(Applied Biosystems, Foster City, CA, USA). Quantitative real-time
polymerase chain reaction (qRT-PCR) was performed using TaqMan
Universal Master Mix, and 20 times volume of miR-378 primer (TaqMan
microRNA assay). The expression level of miR-378 was quantified by
comparative CT method with an iCycler iQ Real-Time Detection System
(Bio-Rad, Hercules, CA, USA) according to the manufacturer's
instruction, the expression level was quantified as the relative
quantitative (RQ) level, in which RNU-44 was used as an endogenous
control. Normal colon total RNA extract (BioChain) was utilized as
normal control in the experiments.
Optimal concentration of
Cetuximab-resistant test
The optimal concentration of Cetuximab (Merck
Serono) resistant test was determined by flow cytometry with
Annexin V-FITC apoptosis detection test (Annexin V-FITC apoptosis
detection kit II; BD Pharmingen™) after incubation of cells with
Cetuximab for 24 h through a gradient concentration of 0, 20, 50,
100, 120 and 200 µg/ml. The final drug concentration of 100
µg/ml was determined to be the minimal concentration that
provided an effective steady apoptotic effect on all cells.
Cell viability analysis
To determine the number of viable cells in culture,
CellTiter 96® AQueous One Solution Cell Proliferation
Assay (Promega) was performed before and after miR-378
transfection, lauric acid and Cetuximab treatment. In brief, a
total of 5×103 cells were cultured in a 96-well
plate/well; and then cultured with different concentration of
lauric acid (0.15, 0.3, 0.45 and 0.6 mM) for 96 h. For the drug
testing, 100 µg/ml Cetuximab was added and for another 72 h.
Before detection of cell viability, 20 µl of CellTiter
96® AQueous One Solution Reagent was added and cultured
at 37°C in an incubator for 1 h, and the the absorbance at 490 nm
was recorded with a 96-well plate reader.
Activation of MAPK pathway analysis for
the transfected miR-378 cells
The bioinformatic prediction suggested that factors
of the mitogen-activated protein kinase (MAPK) pathway are enriched
among miR-378 targets. Therefore, western blotting was performed to
analyze the MAPK pathway related proteins before and after
transfection of miR-378 cells according to the standard protocol.
In brief, cells were washed with phosphate-buffered saline (PBS)
and then trypsinized (0.05% trypsin w/v with 0.02% EDTA). The
pellets were lysed in buffer [50 mM Tris-HCl, 10 mM EDTA, 1% v/v
Triton X-100, 1% phenylmethylsulfonyl fluoride (PMSF), 0.05 mM
pepstatin A and 0.2 mM leupeptin], and after mixing with sample
loading buffer (50 mM Tris-HCl, pH 6.8, 10% w/v sodium dodecyl
sulfate, 10% v/v glycerol, 10% v/v 2-mercaptoethanol and 0.04%
bromophenol blue) at a ratio of 4:1 were denatured at 95°C for 5
min. Protein (30 µg) was then loaded into 10%
SDS-polyacrylamide (SDS-PAGE) gels and subjected to electrophoresis
(120 V, 75 min). The separated proteins were transferred to
nitrocellulose membranes (Bio-Rad; 40 min, 350 mA/gel). The blots
were incubated and slightly shacken in 5% non-fat milk/TBS-Tween-20
blocking buffer for 1 h, followed by overnight incubation at 4°C
with different ratio dilution of the primary antibodies: 1:500
dilution of anti-KRAS (mouse monoclonal, ab55391), 1:2,500 dilution
of anti-BRAF (rabbit monoclonal, ab33899), 1:2,000 dilution of
anti-MEK (mouse monoclonal, ab69502), 1:2,500 dilution of anti-ERK
(mouse monoclonal, ab36991) (Abcam, Cambridge, MA, USA). After
washing with TBS, the blots were incubated at room temperature for
1 h with the secondary antibody. Protein detection and
quantification by densitometric analysis were performed after
normalization with β-actin (mouse monoclonal, NB600-501; Novus
Biologicals, Littleton, CO, USA).
Enzyme-linked immunosorbent assay using
ERK 1/2 protein in the lauric acid fed cells
Amounts of proteins derived from 0.45 mM lauric acid
fed CRC cells (HT29, HCT116 and SW480), with or without 0.6
µM Cetuximab, were analyzed using ERK 1/2 (Total)
InstantOne™ enzyme-linked immunosorbent assay (ELISA) according to
the instructions (Affymetrix, eBioscience®).
Statistical analysis
In order to eliminate any background variation among
the experiments, each original CRC cell line was used as a
reference group to normalize the following five groups: cells
treated with Cetuximab only, cells transfected with miR-378 only,
miR-378 transfected cells combined with Cetuximab treatment, lauric
acid incubated only, and lauric acid incubated combined with
Cetuximab treatment. Analysis were performed with SPSS 15.0
statistics software (SPSS, Inc., Chicago, IL, USA). The difference
between miR-378 expression in colon cancer cell lines and normal
colon extract was analyzed by single variant post hoc test. Paired
comparisons of percentage cell viability analysis of original
cells, miR-378 transfected cells with, and without Cetuximab
treatment were analyzed with Student's t-test. A p-value of ≤0.05
was considered to indicate a statistically significant result.
Results
Significantly elevated expression level
of miR-378 in cells by directly transfected miR-378 or indirectly
induced by lauric acid
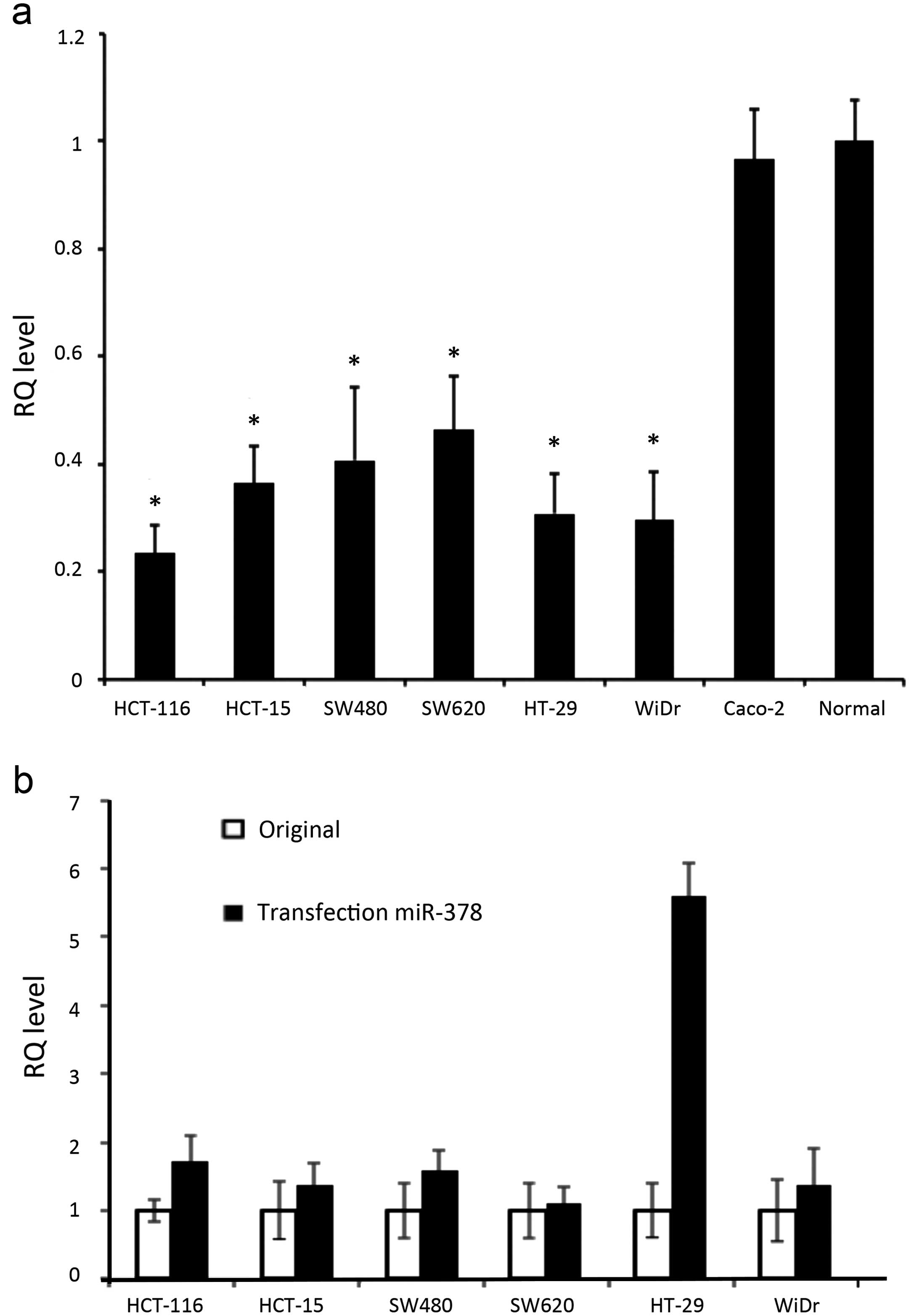
The expression levels of miR-378 in all original
mutants were significantly lower than the normal control and Caco-2
(p<0.0001) (Fig. 2a). We
succeeded in transfecting miR-378 to all CRC cell lines, confirmed
by the parallel experiments of AllStars Hs Cell Death Control
siRNA. All CRC cells significantly increased their expression
levels of miR-378 after in vitro transfection, in which
HT-29 showed the highest efficiency with 5.6-fold and 1.1-fold
elevation for SW620 (Fig. 2b).
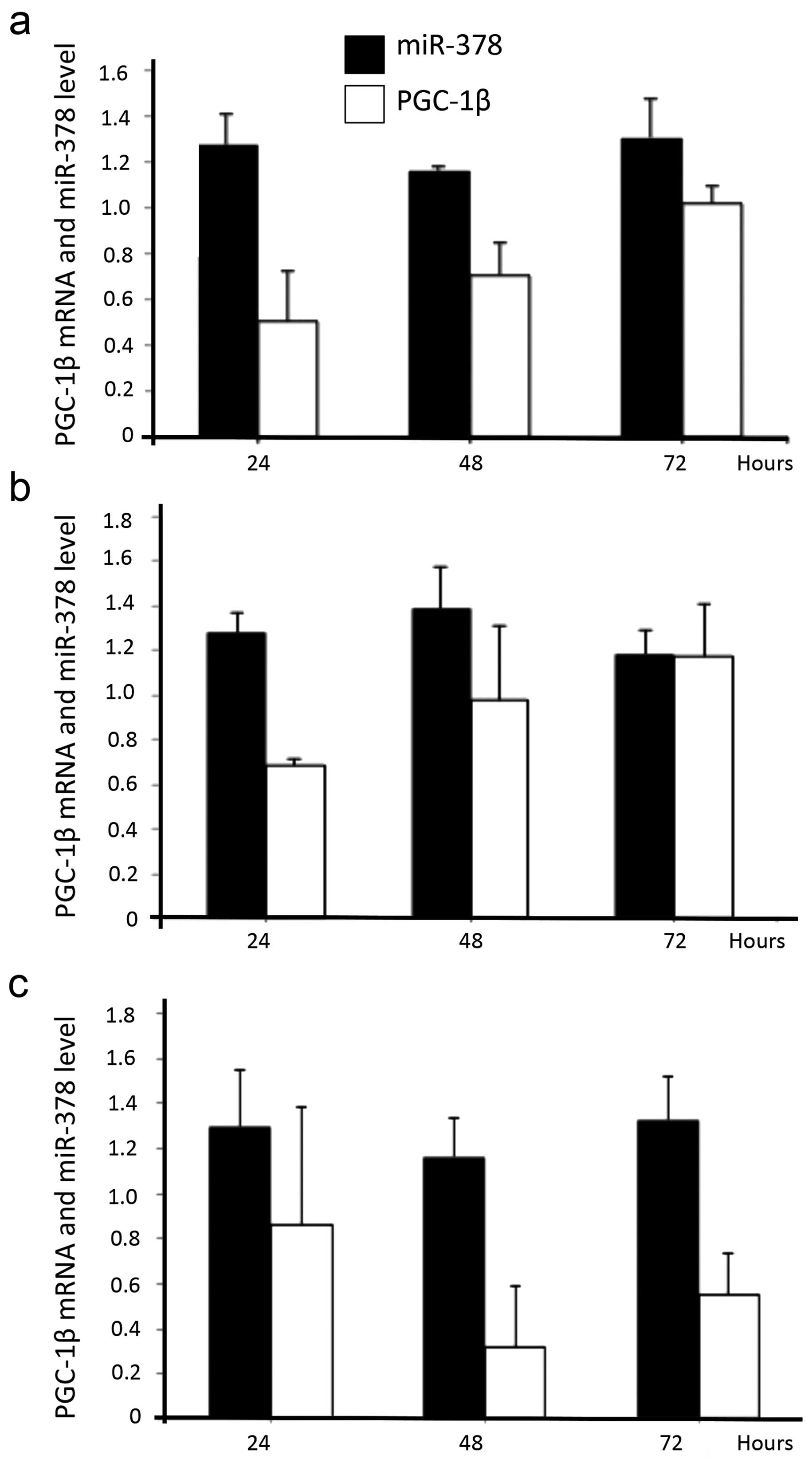
Similarly, three selected aggressive growth pattern cells (HT29,
HCT116 and SW480) after incubated in lauric acid culture medium for
24–72 h, also presented increased mRNA level of PGC-1β and also
coexpressed miR-378 in cells (Fig.
3a–c).
All cells restored sensitivity to
Cetuximab after elevation of the level miR-378 in cells
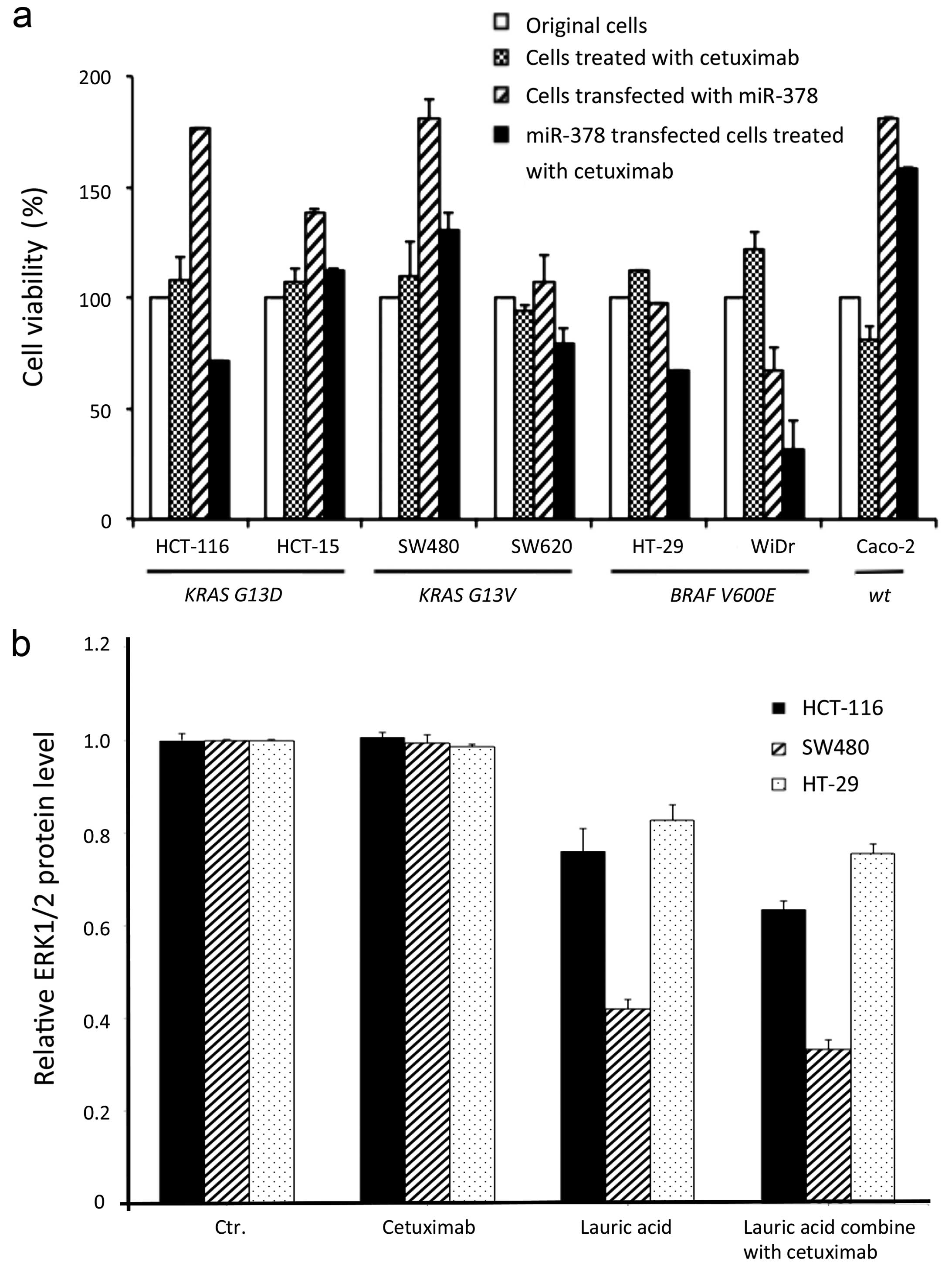
The cell viability was significantly decreased after
treatment with Cetuximab both in miR-378 transfected mutants and
lauric acid incubated cells (Figs.
4a and 5; Table I). Notably, the cell viability of
miR-378 transfected KRAS mutants HCT116 and SW620, were
significantly decreased after treatment with Cetuximab when
compared to the cells treated with Cetuximab only (p=0.007 and
0.39, respectively); a similar response also occurred in the BRAF
mutants, HT29 (p<0.0001) and WiDr (p=0.0003). Although there was
no statistical significance in the HCT15 and SW480 cells,
increasing cell viability should be not ignored when miR-378
transfected before treatment with Cetuximab (Fig. 4a; Table
I).
 | Table IStatistical significance of changes
in cell viability was analyzed by comparison between the status of
two different cells, included untreated original cells, cells
treated with Cetuximab, cells transfected with miR-378, miR-378
transfected cells treated with Cetuximab. |
Table I
Statistical significance of changes
in cell viability was analyzed by comparison between the status of
two different cells, included untreated original cells, cells
treated with Cetuximab, cells transfected with miR-378, miR-378
transfected cells treated with Cetuximab.
| Mutation type/cell
line | Treated with
Cetuximab | Transfected with
miR-378 | miR-378 transfected
cell treated with Cetuximab |
|---|
| KRAS mutation | | | |
|
HCT-116 | | | |
| Untreated
cell | 0.294 | 0.0003 | 0.0002 |
| Transfected with
miR-378 | | | 0.0001 |
| Treated with
Cetuximab | | | 0.007 |
| HCT-15 | | | |
| Untreated
cell | 0.084 |
<0.0001 |
<0.0001 |
| Transfected with
miR-378 | | |
<0.0001 |
| Treated with
Cetuximab | | | 0.081 |
| SW480 | | | |
| Untreated
cell | 0.508 | 0.008 | 0.042 |
| Transfected with
miR-378 | | | 0.008 |
| Treated with
Cetuximab | | | 0.160 |
| SW620 | | | |
| Untreated
cell | 0.01 | 0.334 | 0.005 |
| Transfected with
miR-378 | | | 0.001 |
| Treated with
Cetuximab | | | 0.039 |
| BRAF mutation | | | |
| HT-29 | | | |
| Untreated
cell | 0.01 | 0.03 | 0.001 |
| Transfected with
miR-378 | | |
<0.0001 |
| Treated with
Cetuximab | | |
<0.0001 |
| WIDR | | | |
| Untreated
cell | 0.074 | 0.059 | 0.025 |
| Transfected with
miR-378 | | | 0.028 |
| Treated with
Cetuximab | | | 0.0003 |
| Wild-type | | | |
| Caco-2 | | | |
| Untreated
cell | 0.015 | 0.0004 | 0.0004 |
| Transfected with
miR-378 | | | 0.012 |
| Treated with
Cetuximab | | |
<0.0001 |
MEK and ERK1/2 protein detection after
increasing miR-378 expression level in cells
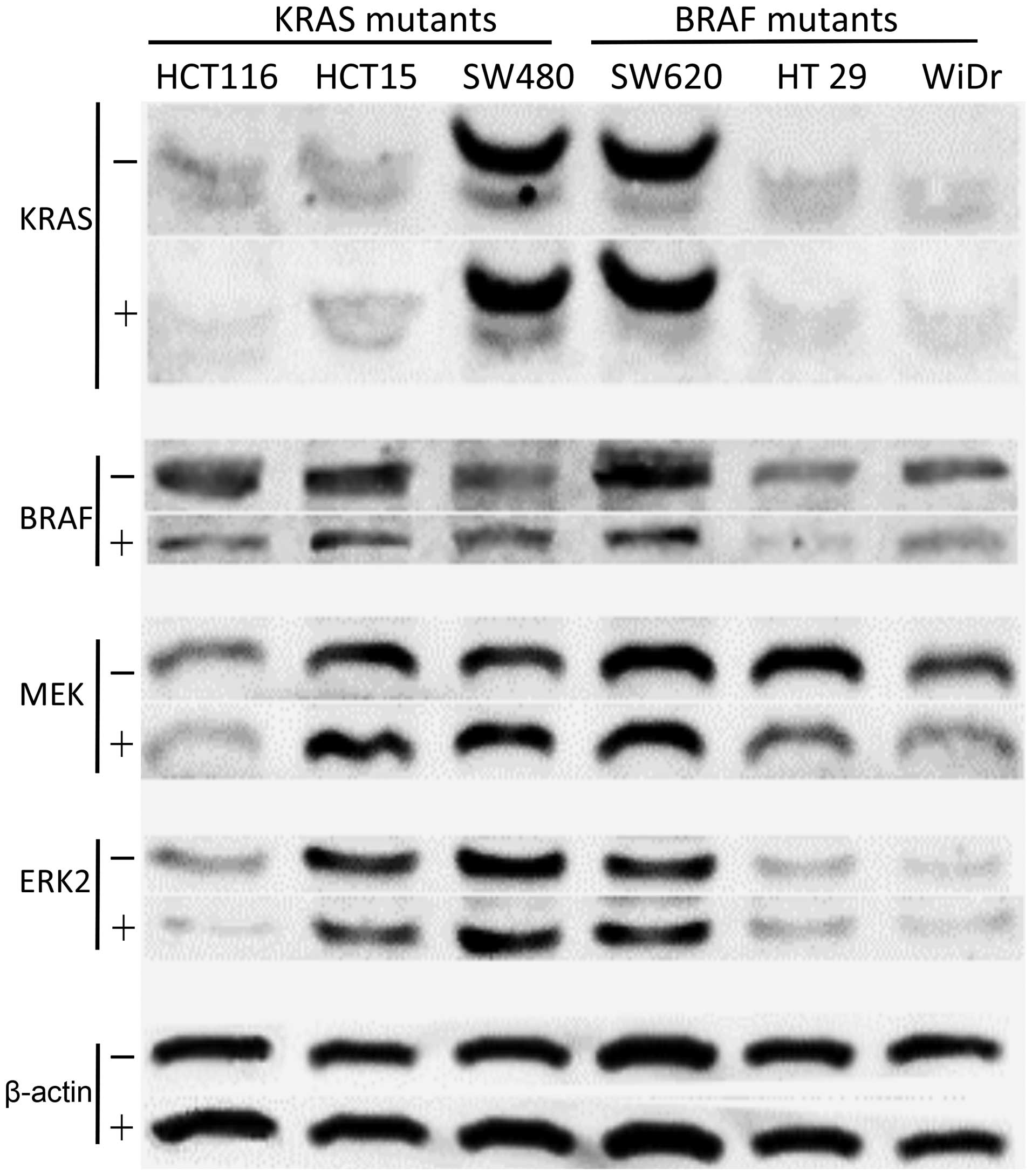
To investigate the MAPK pathway proteins, western
blot analysis was performed. Despite KRAS protein, lower protein
products of BRAF were observed in either KRAS or BRAF mutants.
Similar results were found in EKR2 protein. However, lower MEK
protein was observed in all mutants, except in two KRAS mutants
(Fig. 6). Parallel experiments of
ERK1/2 proteins were performed by ELISA for the lauric acid induced
coexpressed miR-378 in three selected aggressive growth CRC cells
(HT29, HCT116 and SW480). Lower protein expression of ERK1/2 were
observed after CRC cells were incubated in lauric acid medium and
compared to the original cells or Cetuximab-treated only cells
(Fig. 4b). Apparently, decreased
percentage of cell survival followed by increasing concentration of
lauric acid that was absorbed by cells, and even regardless of the
mutation types of the cells, all the cells incubated in 0.6 mM
lauric acid growth medium presented significant sensitivity at all
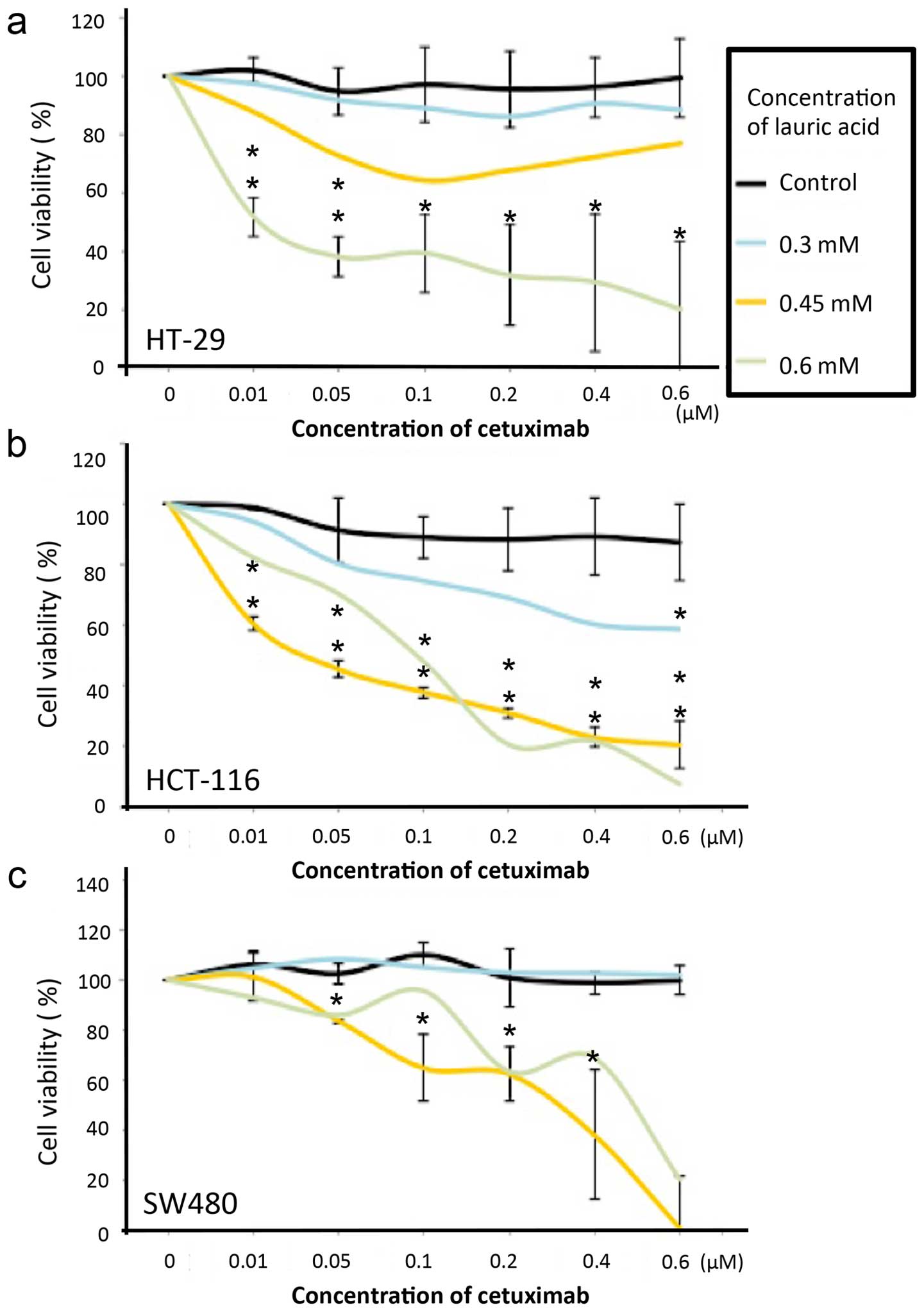
dosages of Cetuximab tested (p<0.001−0.005) (Fig. 5a–c).
Discussion
To improve the benefit to CRC patients with BRAF or
KRAS mutations in target drug-Cetuximab therapy, we restored the
expression level of miR-378 in BRAF or KRAS mutated CRC cells in
two ways: i) directly in vitro transfected miR-378 into
cells (Fig. 2b); ii) induced the
transcriptional activity of PGC1-β gene to produce mRNA by
incubating cells in lauric acid growth medium, and consequently
indirectly stimulated coexpression of miR-378 in cells (Fig. 3). We demonstrated lower cell
viabilities were strongly linked to higher expression level of
miR-378 present in CRC mutated cells, and it further significantly
improved the sensitivity of the mutants to Cetuximab (Fig. 4a and Table I). To uncover the possible
underlying mechanism between miR-378 and Cetuximab, the association
of the proteins with MAPK pathway was investigated. Generally, our
findings demonstrated coincident results from parallel experiments
both directly in vitro transfected miR-378 into CRC cells
and indirectly induced miR-378 by lauric acid (Figs. 4b and 6). Hence, lower level products of MEK, ERK
proteins and decreasing cell viability provided evidence to
indicate miR-378 inhibited cell proliferations and triggered cell
apoptosis (Weng et al, unpublished data), particularly in
BRAF mutants (Fig. 4); the
phenomena led us to evaluate and address the characteristics of
miR-378 in modulating certain molecules in MAPK signaling pathway
in CRCs.
The linkage of miR-378 bound MAPK signaling pathway
associate with the targeting drug-Cetuximab and CRC the mutants
interaction was hypothesized in the present study. Thus, we firstly
confirmed the status of BRAF and KRAS mutation of all CRC cells,
then tested insensitive to Cetuximab. The results showed the
mutants did not respond to the drug, except wtCRC
(Caco-2) (Fig. 4a). We then further
observed the original basal miR-378 expression levels of all BRAF
and KRAS mutants, both were significantly lower than normal control
cells and wtCRC cells (Fig.
2a and Table I). A similar
finding of lower expression of miR-378 in the BRAF mutated CRC
cells were recently demonstrated by Wang et al (13). Our results supported by previous
reports that pinpointed the expression of miR-378 in mutated CRC
tumors differ from normal tissues or wtCRCs (11). Obviously, after the cells were
transfected with miR-378 in vitro to mutants, all BRAF and
KRAS mutated cells reversed the drug sensitivity to Cetuximab;
simultaneously presented decreasing cell viabilities
(p<0.028−0.0001) (Fig. 2a and
Table I). Though all the cells
presented significant response to Cetuximab after transfection of
miR-378 into cells; only half of the KRAS mutants (HCT116 and
SW620) showed 'real' significant re-sensitization to Cetuximab when
compared the the cell viability of Cetuximab treated-only cells
(Fig. 4a and Table I). We assumed that possible subtypes
of KRAS mutant CRCs may exist with unknown molecular
characteristics. This phenomenon strongly suggested miR-378 plays a
crucial role in modulating mutated CRC cells to respond to
Cetuximab. Apparently, stable high expression level of miR-378 is
required for CRC cells response to Cetuximab, irrespective of their
BRAF or KRAS status (Fig. 4a,
Table I) (24).
We noted a contradictory effect on the cell survival
between BRAF and KRAS mutated CRC cells after performing miR-378
transfection in vitro. The cell growth inhibition was
observed in the BRAF mutants; contrarily, increasing cell growth
occurred in all KRAS mutants as well as in Caco-2 (Fig. 4a). Similar findings have been
discussed in the study of Mosakhani et al, indicating the
roles of miR-378 may act as an effector to stimulate cell
proliferation, although the reason remain still unknown (11). It is also known that miR-378 acts as
a Myc target, and modulates the c-Myc/TOB2/cyclin D1 transformation
signaling pathway. Later on miR-378 is involved in activating Ras
and EGFR signaling transduction, and acts as a downstream effector
of the oncogenic EGFR-Ras-ERK pathway (25). Once a mutation occur in one of the
genes it could lead to CRC, although they are considered as
independent prognostic factors (26). The contrary evidence was provided by
recent finding that emphasized miR-378-5p
(5-cuccugacuccagguccugugu-26, the sequence same as the present
study) suppresses cell proliferation and induces apoptosis in CRCs
by targeting BRAF (13), similar
data from the present study also show obviously lower BRAF protein
expression in the BRAF-mutated CRCs after transfection of miR-378
in vitro (Fig. 6).
Therefore, it may be the reason why transfection of miR-378 into
cells trigger cell proliferation in KRAS mutants, but not in BRAF
mutants, i.e. due to the molecular alterations and miR-378
modulating in MAPK pathway (Fig. 4a
and Table I). Indeed, increasing
evidence supports that miR-378 is widely accepted as having diverse
roles in carcinogenesis, and involved in the regulation of tumor
either in proliferation or apoptosis; and may act as an oncogene or
tumor suppressor depending on the targeting of mRNAs or tumors
(27–32). Accordingly, we speculated that
miR-378 may behave as an effector to manipulate several proteins
that involve in the MAPK signaling pathway, and/or block the
signaling transduction to preclude the possibility of promotional
tumorigenesis. This phenomenon provides information suggesting
clinicians may need to make the treatment distinct between BRAF-and
KRAS-mutation patients due to their underlying molecular
differences. Despite increased cell growth of the KRAS mutants
induced by increasing miR-378 amount in cells, it was still
overcome by an even higher efficiency in restoring the sensitivity
to Cetuximab of miR-378 transfected KRAS mutants (Fig. 4a and Table I).
Based on above findings, we tried to find possible
natural resources for clinically practical usage. The precursor of
miR-378 coexpressed with PGC-1β mRNA, and was easily induced by
saturated oil as previously documented (23,29).
The miR-378 is located in the first intron of the PGC-1β gene, and
coordinately expressed with PGC1 (24). The expression of PGC-1β is highly
inducible in response to the dietary intake of saturated fats in
vivo (Fig. 1) (19). Herewith, we elevated the expression
level of miR-378 in CRC cells using saturated lauric fatty acid,
which contains a 12-carbon atom medium chain and was selected to
apply in a series of experiments. Lauric acid is a common fatty
acid easily be found in milk, and much more in many vegetable fats,
particularly in coconut and palm kernel oils (22). Although it is a saturated fatty
acid, it was characterized as having a more favorable effect on
total high-density lipoprotein (HDL) cholesterol than any other
fatty acid, either saturated or unsaturated (33). Therefore, it has been generally
used, and believed it could be applied in medical treatment, such
as viral infections, yeast infections and anti-bacteria (34). More evidence associated to CRC was
mentioned by Fauser et al, who emphasized induction of
apoptosis by the lauric acid in the CRC cells were potentially
resulted from oxidative stress (35). In the present study, in spite of
in vitro transfected miR-378, we used lauric acid to
stimulate the transcriptional activity of the PGC1-β gene and due
to the strict experimental time schedule for lauric acid incubation
and harvest, we therefore only selected three CRC cell lines with
aggressive growth curve (HT29, HCT116 and SW480) for testing. After
grading scale of lauric acid dosage supplied to cells, the
elevation of the miR-378 levels in cells were followed by increase
in expression of mRNA of PGC-1β; the inhibition of protein ERK1/2
was present in all cells, consequently leading to decrease of cell
proliferation (Figs. 3 and 4b). Similar data supported by Fauser et
al also proved successfully induced apoptosis of CRC cells by
lauric acid treated in vitro (35). Our findings imply a possible
strategy that should be kept in mind, the mutated CRC cells may
simply apply with elevated amount of miR-378 in cells without
combining with targeting drug treatment and still efficiently
limited the tumor growth (Fig. 4b).
Nevertheless, after the lauric acid induced either BRAF or KRAS,
mutants tend to present similar results, where decreasing cell
viability was followed by increasing the concentration of Cetuximab
treatment (Fig. 5). Moreover, in
all mutants lower ERK1/2 protein expression was observed either in
lauric acid induced growth medium-only or lauric acid combined with
Cetuximab treatment (Fig. 4b).
These findings provide clinicians a straight, practical and not
harmful usage to remedy current difficulties in applying targeted
therapy in CRC patients with BRAF or KRAS mutations.
To uncover the mechanism of miR-378 activity
correlated with the protein MAPK pathway. KRAS, BRAF, MEK and
ERK1/2 proteins were included, and the protein quantities were
measured before and after miR-378 transfection in vitro or
lauric acid was induced, and then compared the cell viability
between with or without Cetuximab treatment (Figs. 4b and 6). Variant levels of protein inhibition
were observed in both MEK and ERK proteins when directly
transfected with miR-378 in CRC cells, particularly the BRAF
mutants (Fig. 6). Moreover, the
consistent results of lower ERK1/2 protein expression were also
demonstrated in lauric acid-induced cells either in BRAF- or
KRAS-mutants (Fig. 4b).
In conclusion, the present study revealed a novel
molecular mechanism that tightens the bonds between miR-378 and
MAPK pathway, and the association with the efficiency of Cetuximab
treatment. The fact that lauric acid induced miR-378 expression in
cells affects the sensitivity of the mutated CRC cells to
Cetuximab. Hence, high expression level of miR-378 may serve as a
treatment modality for CRCs, particularly in BRAF mutation CRC;
combination with Cetuximab may even impact the CRC patient
treatments. Hopefully, our findings could provide a useful strategy
to promote the use of Cetuximab on BRAF or KRAS mutated CRCs.
Acknowledgments
The authors would like to thank the grant
NTUT-MMH-104-7 support, Miss Jing-Jung Chen, Mr. Yu-Wen Wu and Mr.
Cheng-Chi Wang (National Taipei University of Technology, Taiwan)
for their instructions of statistical analysis and excellent
assistance with laboratory work.
References
|
1
|
Lièvre A, Bachet JB, Le Corre D, Boige V,
Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, et al:
KRAS mutation status is predictive of response to cetuximab therapy
in colorectal cancer. Cancer Res. 66:3992–3995. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Di Fiore F, Blanchard F, Charbonnier F, Le
Pessot F, Lamy A, Galais MP, Bastit L, Killian A, Sesboüé R, Tuech
JJ, et al: Clinical relevance of KRAS mutation detection in
metastatic colorectal cancer treated by Cetuximab plus
chemotherapy. Br J Cancer. 96:1166–1169. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khambata-Ford S, Garrett CR, Meropol NJ,
Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin
AK, et al: Expression of epiregulin and amphiregulin and K-ras
mutation status predict disease control in metastatic colorectal
cancer patients treated with cetuximab. J Clin Oncol. 25:3230–3237.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Amado RG, Wolf M, Peeters M, Van Cutsem E,
Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et
al: Wild-type KRAS is required for panitumumab efficacy in patients
with metastatic colorectal cancer. J Clin Oncol. 26:1626–1634.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Benvenuti S, Sartore-Bianchi A, Di
Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S and Bardelli
A: Oncogenic activation of the RAS/RAF signaling pathway impairs
the response of metastatic colorectal cancers to anti-epidermal
growth factor receptor antibody therapies. Cancer Res.
67:2643–2648. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Di Nicolantonio F, Martini M, Molinari F,
Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L,
Frattini M, Siena S, et al: Wild-type BRAF is required for response
to panitumumab or cetuximab in metastatic colorectal cancer. J Clin
Oncol. 26:5705–5712. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gibbs JBSI, Sigal IS, Poe M and Scolnick
EM: Intrinsic GTPase activity distinguishes normal and oncogenic
ras p21 molecules. Proc Natl Acad Sci USA. 81:5704–5708. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garnett MJ and Marais R: Guilty as
charged: B-RAF is a human oncogene. Cancer Cell. 6:313–319. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karapetis CS, Khambata-Ford S, Jonker DJ,
O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD,
Robitaille S, et al: K-ras mutations and benefit from cetuximab in
advanced colorectal cancer. N Engl J Med. 359:1757–1765. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang GJ, Zhou H, Xiao HX, Li Y and Zhou
T: MiR-378 is an independent prognostic factor and inhibits cell
growth and invasion in colorectal cancer. BMC Cancer. 14:109–118.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mosakhani N, Sarhadi VK, Borze I,
Karjalainen-Lindsberg ML, Sundström J, Ristamäki R, Osterlund P and
Knuutila S: MicroRNA profiling differentiates colorectal cancer
according to KRAS status. Genes Chromosomes Cancer. 51:1–9. 2012.
View Article : Google Scholar
|
|
12
|
Faltejskova P, Svoboda M, Srutova K,
Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K,
Kiss I, et al: Identification and functional screening of microRNAs
highly deregulated in colorectal cancer. J Cell Mol Med.
16:2655–2666. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Z, Ma B, Ji X, Deng Y, Zhang T, Zhang
X, Gao H, Sun H, Wu H, Chen X, et al: MicroRNA-378-5p suppresses
cell proliferation and induces apoptosis in colorectal cancer cells
by targeting BRAF. Cancer Cell Int. 15:402015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nagalingam RS, Sundaresan NR, Gupta MP,
Geenen DL, Solaro RJ and Gupta M: A cardiac-enriched microRNA,
miR-378, blocks cardiac hypertrophy by targeting Ras signaling. J
Biol Chem. 288:11216–11232. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Walczak R and Tontonoz P: Setting fat on
fire. Nat Med. 9:1348–1349. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Crunkhorn S, Dearie F, Mantzoros C, Gami
H, da Silva WS, Espinoza D, Faucette R, Barry K, Bianco AC and
Patti ME: Peroxisome proliferator activator receptor gamma
coactivator-1 expression is reduced in obesity: Potential
pathogenic role of saturated fatty acids and p38 mitogen-activated
protein kinase activation. J Biol Chem. 282:15439–15450. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gerin I, Bommer GT, McCoin CS, Sousa KM,
Krishnan V and MacDougald OA: Roles for miRNA-378/378* in adipocyte
gene expression and lipogenesis. Am J Physiol Endocrinol Metab.
299:198–206. 2010.
|
|
18
|
Carrer M, Liu N, Grueter CE, Williams AH,
Frisard MI, Hulver MW, Bassel-Duby R and Olson EN: Control of
mitochondrial metabolism and systemic energy homeostasis by
microRNAs 378 and 378. Proc Natl Acad Sci USA. 109:15330–15335.
2012. View Article : Google Scholar
|
|
19
|
Lin J, Yang R, Tarr PT, Wu PH, Handschin
C, Li S, Yang W, Pei L, Uldry M, Tontonoz P, et al: Hyperlipidemic
effects of dietary saturated fats mediated through PGC-1beta
coactivation of SREBP. Cell. 120:261–273. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wilson PM and Lenz HJ: Integrating
biomarkers into clinical decision making for colorectal cancer.
Clin Colorectal Cancer. 9(Suppl 1): S16–S27. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cushman-Vokoun AM, Stover DG, Zhao Z,
Koehler EA, Berlin JD and Vnencak-Jones CL: Clinical utility of
KRAS and BRAF mutations in a cohort of patients with colorectal
neoplasms submitted for microsatellite instability testing. Clin
Colorectal Cancer. 12:168–178. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dollah S, Abdulkarim SM, Ahmad SH,
Khoramnia A and Ghazali HM: Physicochemical properties and
potential food applications of Moringa oleifera seed oil blended
with other vegetable oils. J Oleo Sci. 63:811–822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nelson VM and Benson AB III: Status of
targeted therapies in the adjuvant treatment of colon cancer. J
Gastrointest Oncol. 4:245–252. 2013.PubMed/NCBI
|
|
24
|
Yu J, Kong X, Liu J, Lv Y, Sheng Y, Lv S,
Di W, Wang C, Zhang F and Ding G: Expression profiling of
PPARγ-regulated microRNAs in human subcutaneous and visceral
adipogenesis in both genders. Endocrinology. 155:2155–2165. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Feng M, Li Z, Aau M, Wong CH, Yang X and
Yu Q: Myc/miR-378/TOB2/cyclin D1 functional module regulates
oncogenic transformation. Oncogene. 30:2242–2251. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kruszewski W, Kowara R, Rzepko R, Warezak
C, Zieliński J, Gryglewski G, Kopacz A, Jastrzebski T and Pawełczyk
T: K-RAS point mutation, and amplification of C-MYC and C-ERBB2 in
colon adenocarcinoma. Folia Histochem Cytobiol. 42:173–179.
2004.PubMed/NCBI
|
|
27
|
Chen LT, Xu SD, Xu H, Zhang JF, Ning JF
and Wang SF: MicroRNA-378 is associated with non-small cell lung
cancer brain metastasis by promoting cell migration, invasion and
tumor angiogenesis. Med Oncol. 29:1673–1680. 2012. View Article : Google Scholar
|
|
28
|
Deng H, Guo Y, Song H, Xiao B, Sun W, Liu
Z, Yu X, Xia T, Cui L and Guo J: MicroRNA-195 and microRNA-378
mediate tumor growth suppression by epigenetical regulation in
gastric cancer. Gene. 518:351–359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eichner LJ, Perry MC, Dufour CR, Bertos N,
Park M, St-Pierre J and Giguère V: miR-378* mediates
metabolic shift in breast cancer cells via the PGC-1β/ERRγ
transcriptional pathway. Cell Metab. 12:352–361. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee DY, Deng Z, Wang CH and Yang BB:
MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis
by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA.
104:20350–20355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li LH, Gao Q, Wang XY and Guo ZJ: miR-378
suppresses HBV-related hepatocellular carcinoma tumor growth by
directly targeting the insulin-like growth factor 1 receptor.
Zhonghua Gan Zang Bing Za Zhi. 21:609–613. 2013.In Chinese.
PubMed/NCBI
|
|
32
|
Scapoli L, Palmieri A, Lo Muzio L,
Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M and
Carinci F: MicroRNA expression profiling of oral carcinoma
identifies new markers of tumor progression. Int J Immunopathol
Pharmacol. 23:1229–1234. 2010.
|
|
33
|
Mensink RP, Zock PL, Kester AD and Katan
MB: Effects of dietary fatty acids and carbohydrates on the ratio
of serum total to HDL cholesterol and on serum lipids and
apolipoproteins: A meta-analysis of 60 controlled trials. Am J Clin
Nutr. 77:1146–1155. 2003.PubMed/NCBI
|
|
34
|
Zhao L, Hu Y, Xu D and Cai K: Surface
functionalization of titanium substrates with chitosan-lauric acid
conjugate to enhance osteoblasts functions and inhibit bacteria
adhesion. Colloids Surf B Biointerfaces. 119:115–125. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fauser JK, Matthews GM, Cummins AG and
Howarth GS: Induction of apoptosis by the medium-chain length fatty
acid lauric acid in colon cancer cells due to induction of
oxidative stress. Chemotherapy. 59:214–224. 2013. View Article : Google Scholar : PubMed/NCBI
|