Introduction
Colorectal cancer (CRC) is the second most commonly
diagnosed cancer in women and the third in men worldwide, with over
1.2 million new cancer cases and 608,700 deaths estimated to have
occurred in 2008 (1). Although it
is possible to cure colon cancer by surgery, the cure rate is
moderate to poor depending on the stage of the cancer (2). Patients with stage II and III
colorectal cancers remain at a high risk for tumor recurrence after
curative resection. Therefore, they may benefit from additional
treatment including adjuvant therapy. Chemotherapy and radiotherapy
have been mainly used as an initial treatment to shrink any cancer
and then commonly surgery is carried out to remove any tumors.
During chemotherapy, a significant obstacle to the successful
treatment of CRC patients is intrinsic or acquired drug resistance
in patients who initially respond to chemotherapy.
Many mechanisms of drug resistance such as
amplification or mutation of drug target genes, hypoxia,
heterogeneity of cell subpopulations and defective drug transport
or overexpression of p170 (protein of multidrug resistance), have
been identified and studied using principally tumor cell lines
(3,4). A major mechanism of drug resistance
in vitro is the overexpression of energy-dependent drug
efflux pumps known as the ATP-binding cassette (ABC) superfamily
including P-glycoprotein (MDR1), the multidrug resistance protein
(MRP) and ATP-binding cassette sub-family G member 2
(ABCG2) (5). They transport
various compounds such as lipids, bile acids, xenobiotics and
peptides for antigen presentation (6,7).
ABCG2, otherwise known as breast cancer
resistance protein (BCRP) and mitoxantrone resistant associated
gene (MXR), was identified in high
mitoxantrone-resistant-MCF-7/AdrVp and human colon cancer cell
line, S1-M1-80. ABCG2 contains a 655-amino acid polypeptide
transporter with six transmembrane domains and forms a homodimer.
Additionally, the ABCG2 protein was reported to be a 72-kDa
protein. As a half transporter, two nucleotide binding proteins are
required to perform as a drug efflux pump (8,9).
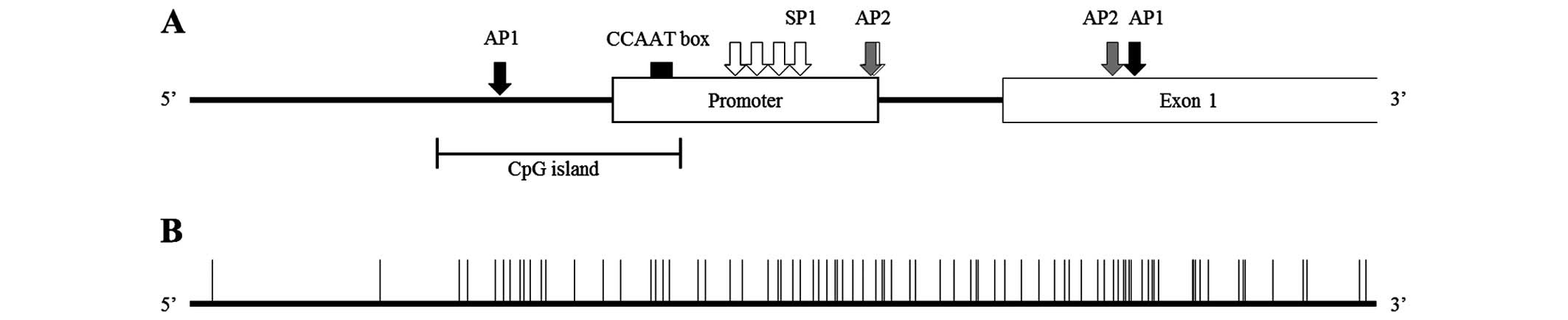
ABCG2 expression is regulated by a TATA-less promoter which
contains several SP1, AP1 and AP2 sites and putative CpG islands.
It has been noted that the potential CpG islands in the promoter
site may be regulated by methylation (7). Furthermore, the 5′ region upstream of
the basal promoter was revealed as both a positive and negative
regulatory domain (7,10).
ABCG2 expression has been shown to be
upregulated in some renal clear cell carcinomas and lung cancer,
breast cancer and multiple myeloma cell lines after treatment with
5-aza-2′-deoxycytidine (5-aza), a DNA demethylating agent (7,11–14).
Therefore, this observation suggested that the DNA methylation of
the ABCG2 promoter site, which consists of many CpG islands,
could play a role in the epigenetic regulation of gene expression
(10).
ABCG2 causes resistance to certain
chemotherapeutic drugs such as mitoxantrone, doxorubicin and
daunorubicin in breast cancers by releasing its substrates which
include topoisomerase I and II inhibitors (15). Furthermore, overexpression of
ABCG2 was found in drug-selected cell lines from breast,
colon, gastric, lung and ovary cancers (16). In a study using non-small cell lung
cancer (NSCLC), the chemotherapeutic response rate in patients was
found to be correlated with ABCG2 expression (17). In addition, 5-FU resistance was
increased in ABCG2-transfected MDCKII cells (18). Thus, drug resistance might be
induced by the regulation of ABCG2 expression in 5-FU,
irinotecan and oxaliplatin resistant cell lines. To investigate
whether the ABCG2 expression level and methylation status of
the promoter affect drug sensitivity in CRC cell lines, we
investigated the expression pattern of ABCG2 and the
methylation status of the ABCG2 promoter.
To show that ABCG2 expression is regulated by
promoter methylation in CRC cell lines, we analyzed the mRNA
expression of ABCG2 and methylation status of the
ABCG2 promoter in 32 CRC cell lines. Afterwards, we studied
whether ABCG2 expression and methylation status have an
influence on anti-cancer drug sensitivity using the cell
proliferation assay, WST-1 assay. Since drug sensitivity increased
in several demethylated CRC cell lines, the results of this study
suggest that DNA methylation of ABCG2 can be a drug
resistance marker for CRC patients who have resistance to
chemotherapeutic drugs.
Materials and methods
Cell culture
The 32 CRC cell lines were provided by the Korean
Cell Line Bank (KCLB, Seoul, Korea). All cell lines were cultured
in RPMI-1640 medium except for Caco-2 and WiDr. Caco-2 was
maintained in minimum essential medium and Dulbecco's modified
Eagle's medium was used for WiDr. Each medium was supplemented with
10% fetal bovine serum and 1.1% penicillin/streptomycin. Cells were
incubated in humidified incubators at 37°C with 5% CO2
and 95% air.
Genomic DNA extraction
Genomic DNA was extracted from the 32 CRC cell lines
using the G-DEX™ IIc genomic DNA extraction kit (Intron
Biotechnology, Gyeonggi, Korea) following the manufacturer's
instructions. Cells treated with trypsin were collected and then
suspended in cell lysis buffer. RNase A solution was added to the
cell lysates and they were incubated at 37°C. The protein
precipitation step was carried out by adding PPT buffer, vortexing
and then centrifuging the samples. The supernatant, which included
the DNA, was collected and inverted with 2-propanol and then, the
mixture was centrifuged at 13,000 rpm. The DNA pellet was dissolved
in DNA rehydration buffer after washing with 70% ethanol.
RNA isolation and cDNA synthesis
Cells were collected with trypsinization and
suspended in easy-BLUE™ (Intron Biotechnology). Total RNA was
isolated according to the manufacturer's instructions. For cDNA
synthesis, the QuantiTect Reverse Transcription kit (Qiagen, Venlo,
The Netherlands) was used. The mixture was composed of 1 µg
of total RNA, 2 µl gDNA wipe buffer and diethylpyrocarbonate
(DEPC) water to make a mixture with volume ≤14 µl. After
incubation at 42°C for 2 min, 4 µl of RT buffer, 1 µl
of the RT primer mix and 1 µl of RTase were mixed together
and incubated at 42°C for 45 min. The final reaction mixture was
maintained at 95°C for 2 min.
Bisulfite modification of genomic
DNA
For bisulfite modification, 2 µg of genomic
DNA from the 32 CRC cell lines were required. Bisulfite
modification was processed using the EZ DNA Methylation™ kit (Zymo
Research, Orange, CA, USA) following the manufacturer's
instructions.
Reverse transcriptase-PCR (RT-PCR)
To analyze the ABCG2 mRNA expression level, 1
µl of synthesized cDNA was amplified in a 14 µl PCR
mixture that contained 10X PCR buffer (with MgCl2),
dNTPs, forward and reverse primers (10 pmol/µl) (Table I), distilled water and i-Taq DNA
polymerase (Intron Biotechnology). The RT-PCR conditions consisted
of 5 min at 94°C for an initial denaturation, followed by 35 cycles
of 94°C for 30 sec, 65°C for 1 min, and 72°C for 30 sec and a final
elongation of 7 min at 72°C. The reaction was carried out using a
programmable thermal cycler (PCR System 9700, Applied Biosystems,
Foster City, CA, USA). The PCR products were fractionated on a 1.5%
agarose gel containing ethidium bromide.
 | Table IPrimer sequences for RT-PCR, qRT-PCR,
MS-PCR and bisulfite sequencing PCR. |
Table I
Primer sequences for RT-PCR, qRT-PCR,
MS-PCR and bisulfite sequencing PCR.
| Name | Sequences | Size (bp) | Refs. |
|---|
| MXR RT F |
5′-GTTTATCCGTGGTGTGTCTGG-3′ | 652 | |
| MXR RT R |
5′-CTGAGCTATAGAGGCCTGGG-3′ | | |
| ABCG2 qRT
F |
5′-CAGGTCTGTTGGTCAATCTCACA-3′ | 76 | (19) |
| ABCG2 qRT
R |
5′-TCCATATCGTGGAATGCTGAAG-3′ | | |
| ABCG2 M
F |
5′-TATTTATTTAATTTGTTTTGGGTGC-3′ | 141 | |
| ABCG2 M
R |
5′-TCATTAAACTAATCAATACCTCGTC-3′ | | |
| ABCG2 U
F |
5′-TTTATTTAATTTGTTTTGGGTGTGA-3′ | 139 | MethPrimer
software |
| ABCG2 U
R |
5′-TCATTAAACTAATCAATACCTCATC-3′ | | |
| ABCG2 BS
F |
5′-AAATTATTTATTTAATTTGTTTTGG-3′ | 282 | |
| ABCG2 BS
R |
5′-CCAACAAAACTAATACCACC-3′ | | |
Quantitative real-time PCR (qRT-PCR)
qRT-PCR was performed in a 386-well PCR plate
containing SYBR-Green Master Mix (Applied Biosystems), distilled
water, 10 ng of the cDNA templates and 900 nM of the ABCG2
forward and reverse primers (Table
I) (19). qRT-PCR analysis was
performed with the 7900HT Fast Real-Time PCR system (Life
Technologies Co, Carlsbad, CA, USA). The results were normalized to
the housekeeping gene, β-actin, and the cycle threshold (Ct) values
were determined. This experiment was repeated three times.
Methylation-specific PCR (MS-PCR)
The PCR reactions were performed at 94°C for 5 min,
and then 45 cycles of 94°C for 30 sec, 53°C for 1 min for the
methylated region and 54°C for 1 min for the unmethylated region,
and 72°C for 30 sec, and finally 72°C for 7 min for both PCR
reactions. To analyze the methylation of the ABCG2 promoter
region, 1 µl bisulfite modified DNA was amplified in a PCR
mixture that contained 10X PCR buffer, dNTPs, forward and reverse
primers for methylated or unmethylated DNA (10 pmol/µl)
(Table I), 5X Q-solution, distilled
water and Taq DNA polymerase (Qiagen).
Bisulfite sequencing analysis
The specific primers for the bisulfite sequencing
analysis were designed using MethPrimer software (http://www.urogene.org/methprimer/index1.html)
(Table I). The PCR reaction was
carried out at 94°C for 5 min, with 40 amplification cycles of 94°C
for 30 sec, 52°C for 1 min and 72°C for 30 sec with a final
extension step at 72°C for 7 min. The amplicons from the bisulfite
sequencing primers were inserted into the pGEM-T Easy vector
(Promega, Madison, WI, USA) for TA-cloning. Sequences from five
individual colonies for each CRC cell line were sequenced using
universal pUC/M13 primers and each sequence was analyzed using a
Taq dideoxy terminator cycle sequencing kit on an ABI 3730 DNA
sequencer (Applied Biosystems).
5-aza-2′-deoxycytidine treatment
For treatment with 5-aza, 2×105 cells/ml
were seeded in two 75 cm2 culture flasks. On the
following day, one of the flasks was treated with 3 µM of
5-aza (Sigma-Aldrich) and the other flask received the same volume
of DMSO as an untreated group for a 48-h incubation time.
Cell proliferation assay
Cells were seeded on a 96-well plate at
2×104 cells/well and incubated overnight at 37°C in 5%
CO2 and 95% air. On the following day, anticancer drugs
including 5-FU, irinotecan and oxaliplatin (all from Sigma-Aldrich)
were added separately into the well at 48 h after 5-aza treatment.
Cell proliferation reagent EZ-Cytox (DoGen, Seoul, Korea) was added
to each well after a 72 h incubation time from the addition of the
anticancer drugs. Then, the plates were incubated at 37°C for 4 h,
and the absorbance was measured with a Multiscan FC microplate
photometer (Thermo Scientific Inc., Bremen, Germany) at 450 nm.
This assay was performed in triplicate wells.
Statistical analysis
Numerical data for all graphs are expressed as the
mean ± standard deviation (SD). P<0.05 was considered to
indicate a statistically significant difference, and statistical
analysis was carried out with SPSS software version 20.0.
Results
Expression of ABCG2 in the CRC cell
lines
CRC cell lines were examined by RT-PCR and qRT-PCR
to identify the mRNA expression level of ABCG2. After gel
electrophoresis, obtained RT-PCR bands were processed by ImageJ
(http://rsbweb.nih.gov/ij/) as the rate
of ABCG2 expression using the formula: ABCG2
expression = (amplified ABCG2/amplified β-actin) × 100. The
ABCG2 mRNA band was detected in 23 cell lines (range of
expression rate from 2.4 to 116.7, data not shown) but not in 9
cell lines (Fig. 1A and Table II). Additionally, we classified the
groups into high (>1 of the relative expression level) and low
(<1 of relative expression level) groups according to the
relative expression level shown by qRT-PCR (Fig. 1B) [Relative expression level =
(ABCG2 expression level/β-actin expression level) × 100;
Table II]. SNU-61, SNU-175,
SNU-503, SNU-769A, SNU-1033, SNU-C1, SNU-C2A, SNU-C5, Caco-2, DLD1,
Colo320, HCT 8, HCT 116, HT 29, LoVo and WiDr showed a relatively
higher mRNA expression level of ABCG2 and SNU-81, SNU-283,
SNU-407, SNU-769B, SNU-1047, SNU-1197, SNU-C4, Colo201, Colo205,
HCT 15, LS174T, NCI-H716, SW403, SW480 and SW1116 cell lines
belonged to the low group. The relative expression level was not
detected in 4 cell lines: SNU-C4, Colo201, LS174T and SW480. Taken
together, there were 8 cell lines that had low or no mRNA
expression for ABCG2 in the RT-PCR and qRT-PCR analyses:
SNU-283, SNU-769B, SNU-C4, Colo201, LS174T, NCI H716, SW403 and
SW480.
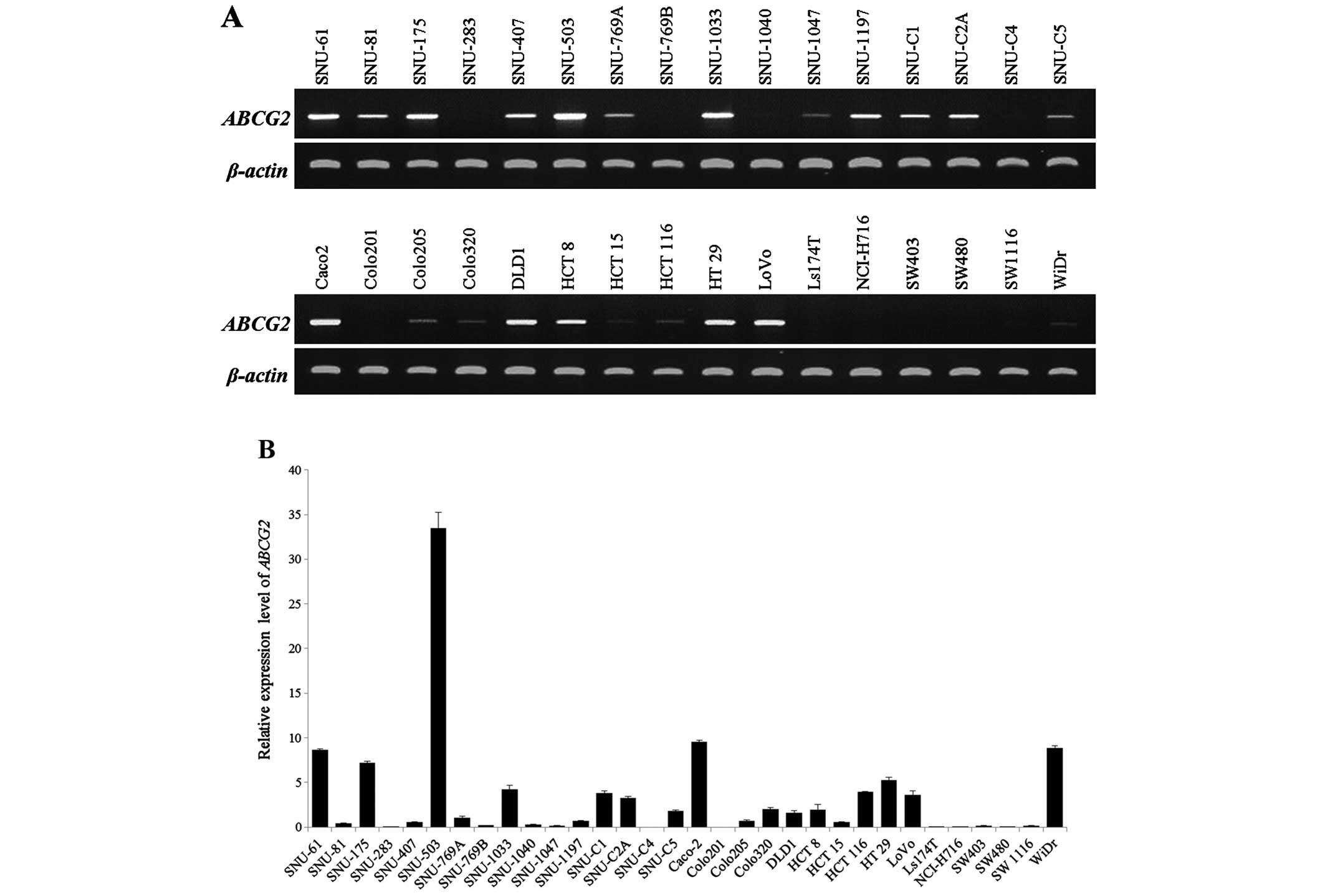 | Figure 1Expression analysis of the
ABCG2 gene was performed in 32 colorectal cancer cell lines
by RT-PCR and qRT-PCR. (A) RT-PCR analysis for screening the
ABCG2 mRNA expression level in 32 colorectal cancer cell
lines. ABCG2 expression was shown in 23 cell lines (SNU-61,
SNU-81, SNU-175, SNU-407, SNU-503, SNU-769A, SNU-1033, SNU-1047,
SNU-1197, SNU-C1, SNU-C2A, SNU-C5, Caco-2, Colo205, Colo320, DLD1,
HCT 8, HCT 15, HCT 116, HT 29, LoVo, SW1116 and WiDr) but not in 9
cell lines (SNU-283, SNU-769B, SNU-1040, SNU-C4, Colo201, LS174T,
NCI-H716, SW403 and SW480). (B) Quantitative differences in
ABCG2 mRNA expression as determined by qRT-PCR analysis. |
 | Table IICorrelation between the promoter
methylation status and ABCG2 expression. |
Table II
Correlation between the promoter
methylation status and ABCG2 expression.
| Cell lines | Methylation | Unmethylation |
Expression
(%) |
Methylation
(%) |
|---|
| SNU-61 | + | − | 8.7 | 1.9 |
| SNU-81 | + | + | 0.4 | 0.0 |
| SNU-175 | + | + | 7.2 | 3.8 |
| SNU-283 | − | − | 0.1 | 14.3 |
| SNU-407 | + | + | 0.5 | 2.9 |
| SNU-503 | + | + | 33.5 | 2.9 |
| SNU-769A | + | + | 1.0 | 57.1 |
| SNU-769B | + | + | 0.2 | 0.0 |
| SNU-1033 | + | − | 4.3 | 0.0 |
| SNU-1040 | − | − | 0.3 | 0.0 |
| SNU-1047 | + | + | 0.2 | 0.0 |
| SNU-1197 | + | + | 0.7 | 0.0 |
| SNU-C1 | + | − | 3.8 | 7.6 |
| SNU-C2A | + | + | 3.3 | 5.7 |
| SNU-C4 | + | + | 0.0 | 28.6 |
| SNU-C5 | + | − | 1.8 | 1.9 |
| Caco-2 | + | − | 9.6 | 0.0 |
| Colo201 | + | − | 0.0 | 2.9 |
| Colo205 | + | − | 0.7 | 19.0 |
| Colo320 | + | − | 2.0 | 17.1 |
| DLD1 | + | − | 1.6 | 0.0 |
| HCT 8 | + | + | 2.0 | 0.0 |
| HCT 15 | + | + | 0.6 | 1.0 |
| HCT 116 | + | + | 3.9 | 1.0 |
| HT 29 | + | − | 5.3 | 0.0 |
| LoVo | + | − | 3.6 | 30.5 |
| LS174T | + | + | 0.0 | 24.8 |
| NCI-H716 | + | + | 0.1 | 45.7 |
| SW403 | + | − | 0.2 | 1.0 |
| SW480 | + | + | 0.0 | 4.8 |
| SW1116 | + | − | 0.1 | 4.8 |
| WiDr | + | − | 8.9 | 0.0 |
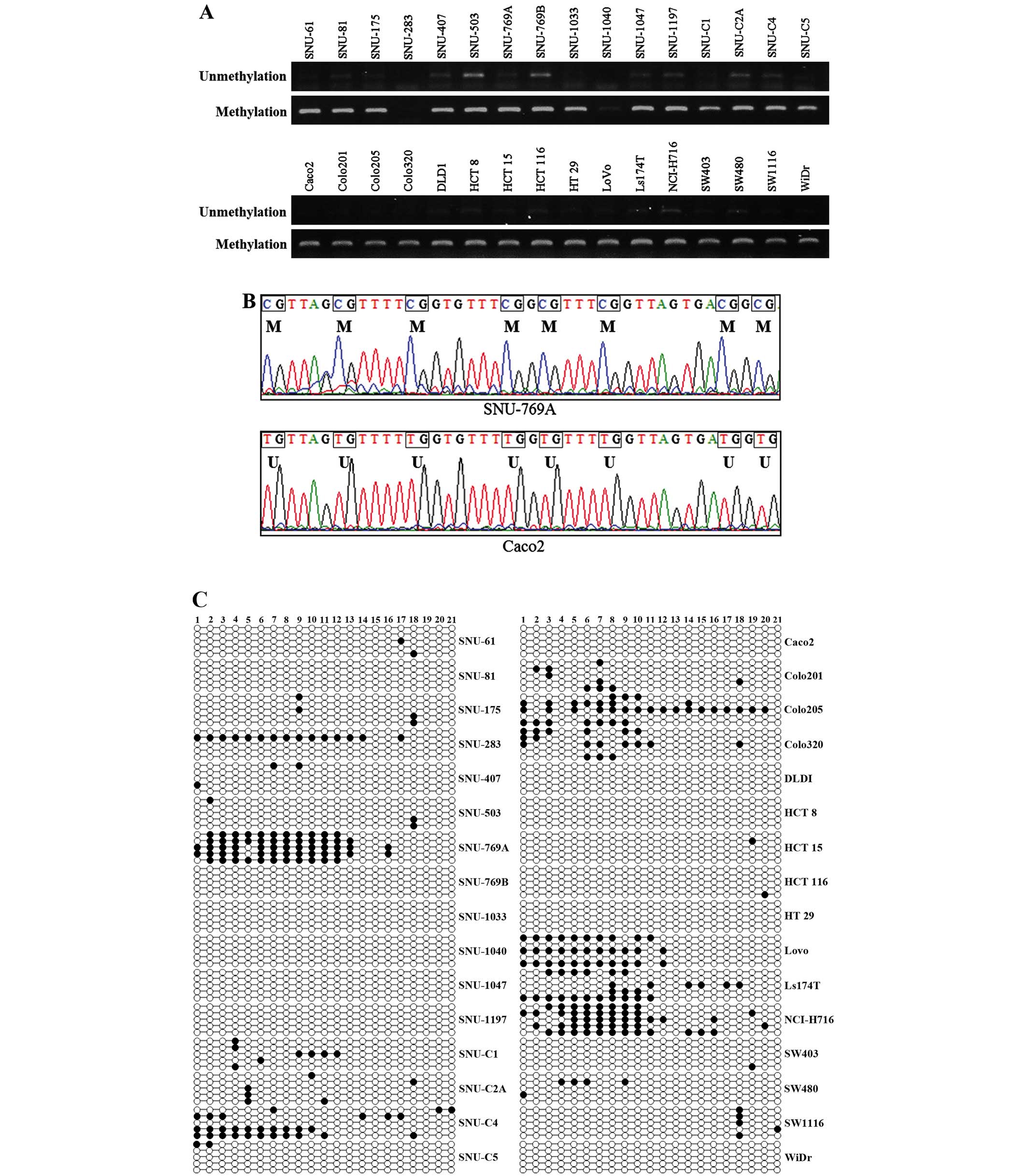
Evaluation of the promoter methylation
status of the ABCG2 gene by MS-PCR and bisulfite sequencing
analysis
To determine whether ABCG2 expression is
related to epigenetic changes such as CpG methylation of the
promoter site, we investigated the methylation status of the
ABCG2 promoter site in 32 CRC cell lines with MS-PCR and
bisulfite sequencing analysis. Genomic DNA, which was modified with
sodium bisulfite, had all unmethylated cytosines converted to
uracils but methylated cytosines remained unchanged. The specifi
cally designed primers (Table I)
for MS-PCR amplified the unmethylated and methylated sequences
located from −273 to −414 which contained 21 CpG islands (Fig. 2 and Table III). Methylated DNAs were detected
in all cell lines except for SNU-283, and there was a weak
methylated band in SNU-1040 (Fig.
3A). Unmethylated DNAs were amplified weakly in most of the
cell lines except for SNU-283 and SNU-1040 which did not show any
methylated DNA bands. There were 10 cell lines (SNU-769B, SNU-1047,
SNU-C4, Colo201, HCT 15, LS174T, NCI-H716, SW403, SW480 and SW1116)
that had low or no expression of ABCG2 mRNA and methylated
DNAs. The expression levels of both ABCG2 mRNA and amplified
methylated DNAs were observed in the other 20 cell lines. In
SNU-C4, Colo201, LS174T and SW480, methylated bands were present
but ABCG2 gene expression was not detected in RT-PCR and
qRT-PCR (Fig. 1) at the same time.
The CpG island region (−136 to −417) that contains 21 CpG
dinucleotide sites (Fig. 2 and
Table III) and part of the
promoter for the ABCG2 gene was amplified with a bisulfite
sequencing specific primer set (Table
I). Part of the CpG island sequence was determined in Fig. 3B. SNU-769A represented the
methylated CpG dinucleotide sequence and Caco-2 represented the
unmethylated sequence around the seven CpG islands. The methylation
status of the CpG island in the ABCG2 promoter is shown in
Fig. 3C. To compare the
ABCG2 mRNA and methylation status of the promoter, the
percentage of methylation was analyzed (Table II). The percentage of promoter
methylation of ABCG2 in 8 cell lines was >10% and
SNU-769A (57.1%), NCI-H716 (45.7%) and SNU-C4 (28.6%) were verified
as having a hypermethylated ABCG2 promoter. SNU-C4, LS174T
and NCI-H716 had >20% methylation in the promoter and
simultaneously low or no ABCG2 gene expression was detected
(Fig. 1).
 | Table IIIList of transcriptional regulation
sites and genomic regions in ABCG2. |
Table III
List of transcriptional regulation
sites and genomic regions in ABCG2.
| Potential site | Genomic
position |
|---|
| Promoter site | −36 to −266 |
| XBBF | −363 to −378 |
| CpG island | −249 to −402 |
| SP1 site | −210 to −222 |
| −178 to −187 |
| −151 to −160 |
| −116 to −127 |
| −37 to −49 |
| AP1 site | −349 to −360 |
| +124 to +136 |
| CCAAT box | −275 to −280 |
| AP2 site | −38 to −50 |
| +107 to +118 |
| Exon 1 | +1 to +532 |
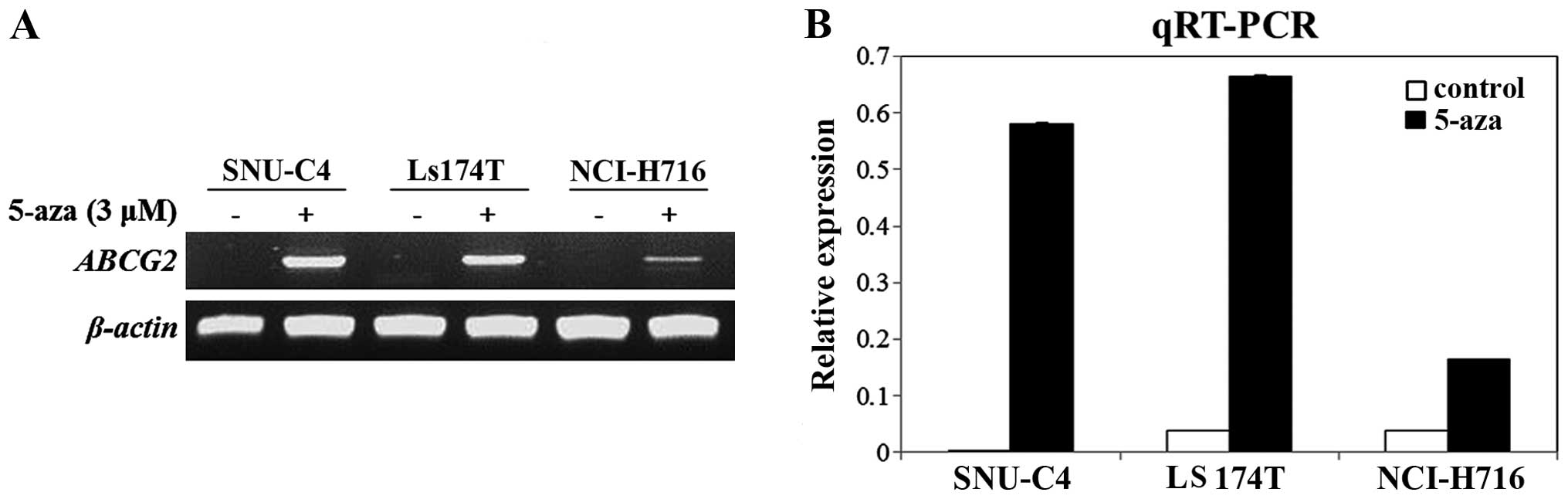
Recovery of ABCG2 mRNA expression after
treatment with 5-aza
To determine whether DNA methylation affects
ABCG2 expression, we chosen three CRC cell lines (SNU-C4,
LS174T and NCI-H716) that showed methylated DNAs in the MS-PCR,
>20% methylated CpG dinucleotides in the bisulfite sequencing
analysis and weak or no ABCG2 mRNA expression. In all three
cell lines, ABCG2 mRNA expression was recovered when the
cell lines were cultured with 3 µM of 5-aza for 48 h
(Fig. 4). Furthermore, there was no
significant re expression of ABCG2 when the LS174T and
NCI-H716 cell lines were treated with trichostatin A (TSA, histone
deacetylase inhibitor) (data not shown). Therefore, re-expression
of ABCG2 mRNA resulted from demethylation mediated by 5-aza,
not acetylation.
Drug sensitivity is reversed by 5-aza
treatment in several CRC cell lines
To determine whether mRNA re-expression by
demethylation affects anticancer drug sensitivity, we performed the
WST-1 assay using 5-aza-treated CRC cell lines which expressed a
low mRNA level under relative expression level 1 and had >20%
methylation of the promoter (Table
II). Selected cell lines, SNU-C4, LS174T and NCI-H716, were
treated with chemotherapeutic drugs in a dose-dependent manner used
to treat CRC patients known as ABCG2 substrates: 5-FU,
irinotecan and oxaliplatin. Drug sensitivity was measured inversely
by cell viability depending on the absorbance at 450 nm. In the
SNU-C4, LS174T and NCI-H716 cell lines treated with 5-aza, the cell
viability was significantly increased in the presence of 5-FU,
irinotecan and oxaliplatin at all drug concentrations (Fig. 5).
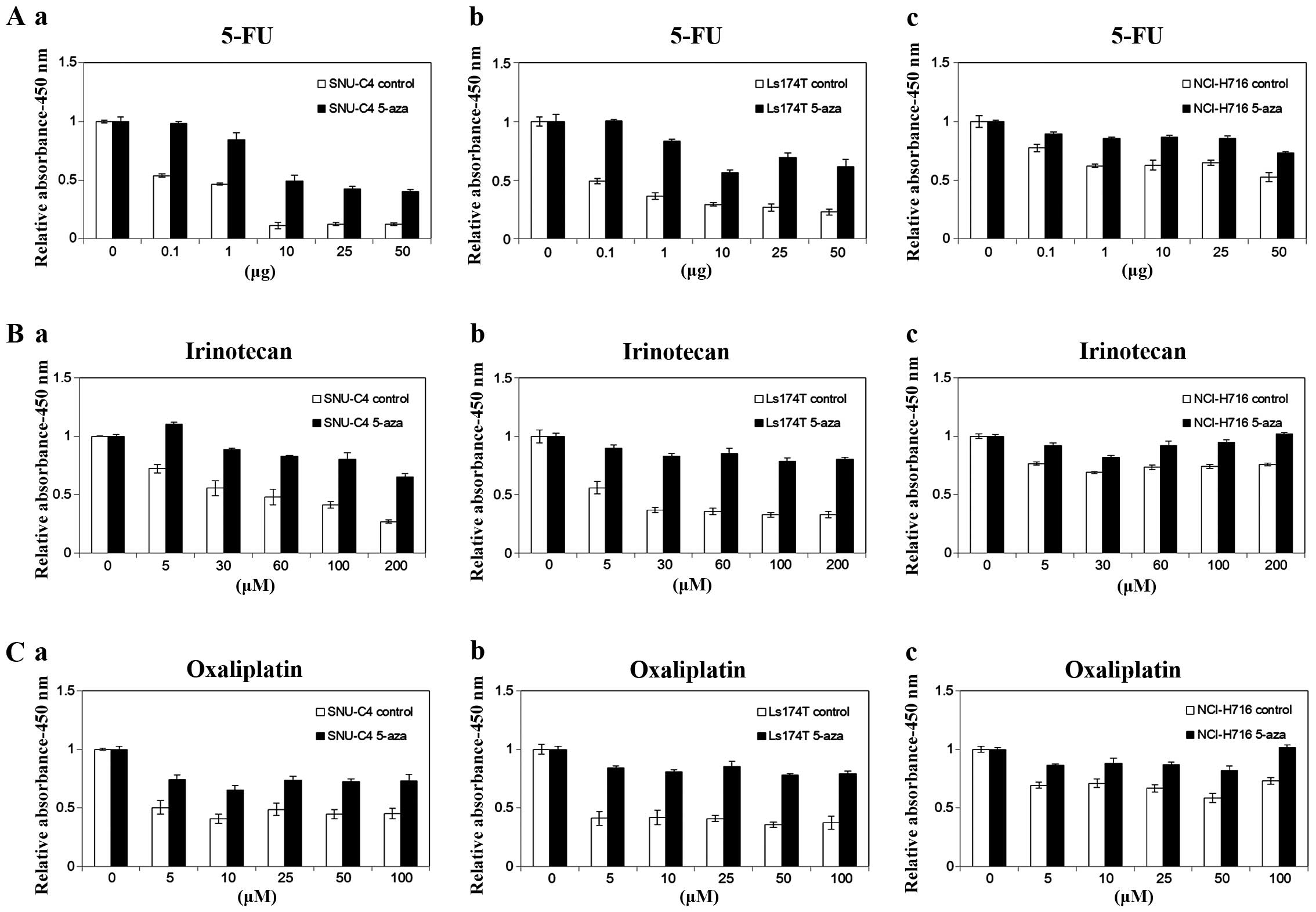 | Figure 5A comparison of cell viability for
anticancer drugs in the colorectal cancer cell lines with or
without 5-aza treatment. (a) SNU-C4, (b) LS174T and (c) NCI-H716
were treated with (A) 5-FU (0, 0.1, 1, 10, 25 and 50 µg),
(B) irinotecan (0, 5, 30, 50, 100 and 200 µM) and (C)
oxaliplatin (0, 5, 10, 25, 50 and 100 µM) for 72 h after
demethylation by 5-aza, and cell viability was determined using
WST-1 assay. |
5-aza potentiated the cell viability together with
5-FU (1.83-fold to 4.33-fold increase with 10 µg),
irinotecan (1.52-fold to 2.43-fold increase with 200 µM) and
oxaliplatin (1.48-fold to 1.62-fold increase with 50 and 100
µM) in the SNU-C4 cell line. In LS174T with 5-aza, cell
viability was maximally increased at 50 µg of 5-FU
(2.67-fold), 200 µM of irinotecan (2.45-fold) and 25
µM of oxaliplatin (2.18-fold). The cell viability of
5-aza-treated NCI-H716 cells reached the greatest level at 50
µg of 5-FU (1.40-fold), 200 µM of irinotecan
(1.35-fold) and 50 µM of oxaliplatin (1.40-fold). SNU-C4
(2.92-fold increase with 5-FU) and LS174T (2.22-fold increase with
irinotecan and 2.08-fold increase with oxaliplatin) showed a
maximum increase in cell viability for each anticancer drug, and a
minimal increase was detected in the NCI-H716 cells (1.33-fold
increase with 5-FU, 1.25-fold increase with irinotecan and
1.32-fold increase with oxaliplatin) according to the average cell
viability. Additionally, increments in cell viability were observed
at the greatest level when the cell lines were treated with 5-FU
(2.18-fold) and oxaliplatin (1.65-fold) had the lowest level for
the average enhanced cell viability. Taken together, 5-aza
treatment which induces the demethylation of ABCG2 in
several colorectal cell lines has an effect on the decrease in drug
sensitivity.
Discussion
Studies have reported that overexpression of
ABCG2 is associated with anticancer drug resistance by
mediating drug efflux. MCF-7/AdrVp cells are a multidrug-resistant
human breast cancer subline which does not express P-gp or MRP1,
known as multidrug resistance transporters, but does express
ABCG2. In this cell line, the multidrug resistance phenotype
is acquired by ABCG2 overexpression (15). The expression of ABCG2 is
regulated by DNA methylation, which has been known to be
responsible for inhibiting gene expression. Methylation of the
transcriptional regulatory region including the transcriptional
binding sites induces the transcriptional repression of several
genes (20,21). In a prior study on lung cancer
cells, it was discovered that methylation of the ABCG2
promoter was inversely correlated with its expression (13). Following treatment with
5′-aza-2′-deoxycytidine, the DNA demethylation agent, ABCG2
expression was re-activated. This indicated that DNA methylation of
the promoter site, which consists of many CpG islands, could play a
central role in the epigenetic regulation of ABCG2 gene
expression (11).
To study the correlation between the methylation
patterns of the ABCG2 promoter region and gene expression in
CRC cell lines, we performed MS-PCR and bisulfite sequencing
analysis. The ABCG2 mRNA levels were examined by RT-PCR and
quantitative real-time PCR. First, we classified the CRC cell lines
into high or low ABCG2 expression groups according to the
relative expression level shown by qRT-PCR (Fig. 1B). The mean relative ABCG2
expression value of the high group was >2. Then, we selected
cell lines which had a hypermethylated promoter site identified by
MS-PCR (Fig. 3A) and bisulfite
sequencing analysis (Fig. 3C). As a
result, SNU-C4, LS174T and NCI-H716 cells were selected as they
exhibited low expression of the ABCG2 gene less than the
relative expression level 1 and had >20% methylated CpG
dinucleotides in the promoter site (Table II). The three cell lines were
treated with demethylating agent 5-aza to determine whether DNA
demethylation increases ABCG2 mRNA expression. After
treatment of the cell lines SNU-C4, LS174T and NCI-H716 with 3
µM 5-aza for 48 h, ABCG2 mRNA was re-expressed in all
three cell lines (Fig. 4).
Consequently, demethylation of the CpG dinucleotides in the
ABCG2 promoter upregulated ABCG2 gene expression. In
other words, the promoter was negatively regulated by DNA
methylation in several CRC cell lines. However, we demonstrated
that SNU-769A moderately expressed the ABCG2 gene and had
hypermethylation of promoter CpG islands (Table II). As referred to earlier in the
study, 1 allele of the chromosome was methylated but another allele
was not methylated in the moderate ABCG2-expressing cells
(NCI-H460, NCI-H441 and NCI-H358 cell lines) (13). Therefore, there is a possibility
that 1 allele might be methylated in SNU-769A. However, to make
sure of this speculation, additional DNA sequencing is required to
analyze both alleles of SNU 769A. Additionally, there were somewhat
different cases. For instance, in SNU 283 and SW480 cells, the
ABCG2 mRNA was merely expressed and was not observed to be
hypermethylation. In this case, we speculate that there are other
pathways which regulate the expression of ABCG2, such as
histone acetylation or methylation. Further study is warranted to
verify this speculation.
Various epigenetic modification types affect the
regulation of genes such as acetylation at Lys and methylation at
Arg and Lys. When the LS174T and NCI-H716 cell lines were treated
with TSA, there was no significant re-expression of the
ABCG2 gene (data not shown). Taken together, these results
suggested that methylation of the ABCG2 promoter region
might have an influence on ABCG2 expression but acetylation
might not be related to the regulation of the gene in various CRC
cell lines. However, it is necessary to perform additional
experiments such as the ChIP assay to determine whether other
mechanisms or proteins are involved in the regulatory steps of
ABCG2 expression since methylation is not the only mechanism
of epigenetic regulation.
In a previous study, it was shown that the
development of drug resistance was not dependent on P-gp or MRP but
was related to the upregulated protein expression of ABCG2
in a mitoxantrone-resistant HT 29 colon carcinoma cell line
(22). Likewise, ABCG2 was
overexpressed in irinotecan and oxaliplatin resistant cell lines,
and 5-FU resistance was increased in ABCG2-transfected
MDCKII cells (18,23). 5-FU, irinotecan and oxaliplatin are
substrates for ABCG2 (24).
In summary, these studies suggest that overexpression of
ABCG2 contributes to drug resistance in cancer cells.
After we confirmed that demethylation can enhance
ABCG2 gene expression in the SNU-C4, LS174T and NCI-H716
cell lines, we investigated whether drug sensitivity can be
affected by the increased ABCG2 gene expression following
5-aza-induced demethylation. Cell viability was measured by WST-1
assay and inversely indicates drug sensitivity. SNU-C4, LS174T and
NCI-H716 cells were treated with 5-FU, irinotecan and oxaliplatin
for 72 h after a 48-h treatment with 5-aza. The reversible effects
of drug sensitivity appeared significantly in all cell lines
treated with 5-aza (Fig. 5). 5-aza
maximally potentiated the cell viability of 5-FU (4.33-fold at 10
µg) in SNU-C4 cells, irinotecan (2.45-fold at 200 µM)
in LS174T cells and oxaliplatin (2.18-fold at 50 µM) in
LS174T cells. In NCI-H716, a minimal increase was measured
according to the average increased cell viability (1.33-fold for
5-FU, 1.25-fold for irinotecan and 1.32-fold for oxaliplatin).
Since inverse cell viability is considered equivalent to drug
sensitivity, we concluded that drug sensitivity was decreased in
the 5-aza-treated CRC cell lines despite the differences in the
increased levels of cell viability. The reason why there were
differences in the increased levels of cell viability is thought to
be due to distinctions in the expression level of the ABCG2
mRNA in each cell line. Actually, the increments for the ratio of
ABCG2 expression in the 5-aza-treated cell lines were
1.66-fold in SNU-C4, 25.16-fold in LS174T and 6.89-fold in NCI-H716
cells. Taken together, we found that the 5-aza-induced
demethylation of the promoter site in some colorectal cell lines
might have an effect on the decrease in drug sensitivity through
the positive regulation of ABCG2 mRNA expression based on
various tests. According to the results, overexpression of the
ABCG2 gene as well as the ABCG2 methylation status
may be useful as a marker of drug resistance in CRC patients
regarding those regimens, and it is possible to understand
individual specific drug sensitivity for each CRC patient. Thus,
the present study is meaningful in terms of anticancer treatment as
appropriate therapy could be provided to CRC patients.
In conclusion, we identified how the promoter
methylation status of ABCG2 regulates pharmaceutical
resistance in CRC cell lines. ABCG2 plays a role in drug
efflux in many types of cancers. We found that demethylation
upregulated ABCG2 gene expression and the enhanced
expression was negatively correlated to anticancer drug sensitivity
in various CRC cell lines. However, these findings should be
verified through additional study concerning other epigenetic
mechanisms or clinical trials.
Acknowledgments
This study was supported by the Priority Research
Center Program (2009-0093820) through a National Research
Foundation of Korea grant funded by the MSIP.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
André T, Boni C, Mounedji-Boudiaf L,
Navarro M, Tabernero J, Hickish T, Topham C, Zaninelli M, Clingan
P, Bridgewater J, et al Multicenter International Study of
Oxaliplatin/5-Fluorouracil/Leucovorin in the Adjuvant Treatment of
Colon Cancer (MOSAIC) Investigators: Oxaliplatin, fluorouracil and
leucovorin as adjuvant treatment for colon cancer. N Engl J Med.
350:2343–2351. 2004. View Article : Google Scholar
|
|
3
|
Violette S, Poulain L, Dussaulx E, Pepin
D, Faussat AM, Chambaz J, Lacorte JM, Staedel C and Lesuffleur T:
Resistance of colon cancer cells to long-term 5-fluorouracil
exposure is correlated to the relative level of Bcl-2 and Bcl-X(L)
in addition to Bax and p53 status. Int J Cancer. 98:498–504. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cazin JL, Gosselin P, Cappelaere P, Robert
J and Demaille A: Drug resistance in oncology: From concepts to
applications. J Cancer Res Clin Oncol. 119:76–86. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eijdems EW, De Haas M, Coco-Martin JM,
Ottenheim CP, Zaman GJ, Dauwerse HG, Breuning MH, Twentyman PR,
Borst P and Baas F: Mechanisms of MRP over-expression in four human
lung-cancer cell lines and analysis of the MRP amplicon. Int J
Cancer. 60:676–684. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bailey-Dell KJ, Hassel B, Doyle LA and
Ross DD: Promoter characterization and genomic organization of the
human breast cancer resistance protein (ATP-binding cassette
transporter G2) gene. Biochim Biophys Acta. 1520:234–241. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Litman T, Brangi M, Hudson E, Fetsch P,
Abati A, Ross DD, Miyake K, Resau JH and Bates SE: The
multidrug-resistant phenotype associated with overexpression of the
new ABC half transporter, MXR (ABCG2). J Cell Sci. 113:2011–2021.
2000.
|
|
9
|
Xu J, Liu Y, Yang Y, Bates S and Zhang JT:
Characterization of oligomeric human half-ABC transporter
ATP-binding cassette G2. J Biol Chem. 279:19781–19789. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Turner JG, Gump JL, Zhang C, Cook JM,
Marchion D, Hazlehurst L, Munster P, Schell MJ, Dalton WS and
Sullivan DM: ABCG2 expression, function, and promoter methylation
in human multiple myeloma. Blood. 108:3881–3889. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
To KK, Zhan Z and Bates SE: Aberrant
promoter methylation of the ABCG2 gene in renal carcinoma. Mol Cell
Biol. 26:8572–8585. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
To KK, Zhan Z, Litman T and Bates SE:
Regulation of ABCG2 expression at the 3′ untranslated region of its
mRNA through modulation of transcript stability and protein
translation by a putative microRNA in the S1 colon cancer cell
line. Mol Cell Biol. 28:5147–5161. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nakano H, Nakamura Y, Soda H, Kamikatahira
M, Uchida K, Takasu M, Kitazaki T, Yamaguchi H, Nakatomi K,
Yanagihara K, et al: Methylation status of breast cancer resistance
protein detected by methylation-specific polymerase chain reaction
analysis is correlated inversely with its expression in
drug-resistant lung cancer cells. Cancer. 112:1122–1130. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mirza S, Sharma G, Pandya P and Ralhan R:
Demethylating agent 5-aza-2-deoxycytidine enhances susceptibility
of breast cancer cells to anticancer agents. Mol Cell Biochem.
342:101–109. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Doyle LA, Yang W, Abruzzo LV, Krogmann T,
Gao Y, Rishi AK and Ross DD: A multidrug resistance transporter
from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA.
95:15665–15670. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Allen JD and Schinkel AH: Multidrug
resistance and pharmacological protection mediated by the breast
cancer resistance protein (BCRP/ABCG2). Mol Cancer Ther. 1:427–434.
2002.PubMed/NCBI
|
|
17
|
Yoh K, Ishii G, Yokose T, Minegishi Y,
Tsuta K, Goto K, Nishiwaki Y, Kodama T, Suga M and Ochiai A: Breast
cancer resistance protein impacts clinical outcome in
platinum-based chemotherapy for advanced non-small cell lung
cancer. Clin Cancer Res. 10:1691–1697. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
König SK, Herzog M, Theile D, Zembruski N,
Haefeli WE and Weiss J: Impact of drug transporters on cellular
resistance towards saquinavir and darunavir. J Antimicrob
Chemother. 65:2319–2328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Langmann T, Mauerer R, Zahn A, Moehle C,
Probst M, Stremmel W and Schmitz G: Real-time reverse
transcription-PCR expression profiling of the complete human
ATP-binding cassette transporter superfamily in various tissues.
Clin Chem. 49:230–238. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hendrich B and Bird A: Identification and
characterization of a family of mammalian methyl-CpG binding
proteins. Mol Cell Biol. 18:6538–6547. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Watt F and Molloy PL: Cytosine methylation
prevents binding to DNA of a HeLa cell transcription factor
required for optimal expression of the adenovirus major late
promoter. Genes Dev. 2:1136–1143. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Perego P, De Cesare M, De Isabella P,
Carenini N, Beggiolin G, Pezzoni G, Palumbo M, Tartaglia L, Pratesi
G, Pisano C, et al: A novel 7-modified camptothecin analog
overcomes breast cancer resistance protein-associated resistance in
a mitoxantrone-selected colon carcinoma cell line. Cancer Res.
61:6034–6037. 2001.PubMed/NCBI
|
|
23
|
Boyer J, McLean EG, Aroori S, Wilson P,
McCulla A, Carey PD, Longley DB and Johnston PG: Characterization
of p53 wild-type and null isogenic colorectal cancer cell lines
resistant to 5-fluorouracil, oxaliplatin and irinotecan. Clin
Cancer Res. 10:2158–2167. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan J, Lv H, Peng B, Wang C, Yu Y and He
Z: Role of BCRP as a biomarker for predicting resistance to
5-fluorouracil in breast cancer. Cancer Chemother Pharmacol.
63:1103–1110. 2009. View Article : Google Scholar
|