Introduction
Colorectal cancer (CRC) has high morbidity and
mortality as one of the most common malignant tumors of human
(1,2). It is the second ranking newly
diagnosed cancer and the second leading cause of overall cancer
deaths (3). Radiotherapy occupies
an important position in the treatment of CRC. Radiotherapy
decreases the rate of local relapse and improves survival for stage
II and III CRC (4). However,
radioresistance is still an obstacle of radiotherapy in CRC.
Therefore, it is necessary to improve our understanding of the
occurrence and development of the disease and accurately predict
tumor radioresistance.
Molecular techniques reveal that several genes are
differently activated in CRC. Most attention has focused on the
biological characteristics of protein-coding genes while ignoring
non-coding RNA (ncRNA), the majority of human genome (5,6).
NcRNAs are divided into long ncRNAs (lncRNAs) and small ncRNAs
based on size (7). LncRNAs are
frequently >200 nt in length (8). Increasing research suggests that
aberrant lncRNA expression may be a major contributor to
tumorigenesis in numerous cancer types (9–12).
Recent finding suggested that HOTAIR functioned as a
molecular scaffold to target PRC2 and LSD1 and ultimately caused
epigenetic gene silencing promoting cancer metastasis (13). Aberrant HOTAIR expression
significantly affects survival and prognosis of various types of
cancers such as hepatocellular, breast, gastric, and pancreatic
cancer (14). Upregulation of
HOTAIR promotes metastasis and poor prognosis of esophageal
squamous cell carcinoma (15). A
recent study showed that suppressed expression of HOTAIR inhibited
proliferation and tumorigenicity in renal carcinoma cells (16). However, the potential involvement of
HOTAIR in radiosensitivity of CRC is so far unknown. In the present
study, therefore, we explored the influence of HOTAIR knockdown on
cell proliferation, migration, invasion, apoptosis and
radiosensitivity.
Materials and methods
Clinical specimens
Fifty-three cases of fresh colorectal cancer (CRC)
tissues and matched adjacent normal colorectal tissues were
collected immediately after surgical resection and stored at −80° C
for further analysis. This study was conducted in accordance with
the Declaration of Helsinki and with approval from the Ethics
Committee of Soochow University. Table
I shows the clinical characteristics of these patients.
 | Table IClinical characteristic of the CRC
patients. |
Table I
Clinical characteristic of the CRC
patients.
| Clinical
parameters | Numbers |
|---|
| Cases | 53 |
| Age |
| <65 | 30 |
| ≥65 | 23 |
| Tumor size |
| Small size (<5
cm) | 25 |
| Large size (≥5
cm) | 28 |
| Gender |
| Male | 35 |
| Female | 18 |
| Invasion
levels |
| Mucosa | 4 |
| Submucosa | 3 |
| Muscle | 10 |
| Serosa | 36 |
| TNM stage |
| Stage 1–2 | 34 |
| Stage 3–4 | 19 |
| Lymph node
metastasis |
| Positive | 26 |
| Negative | 27 |
| Grade of
tumors |
| Low grade | 13 |
| Intermediate
grade | 33 |
| High grade | 7 |
| Vascular
invasion |
| Positive | 22 |
| Negative | 31 |
| Perineural
invasion |
| Positive | 17 |
| Negative | 36 |
Cell cultures and irradiation
All the cell lines (FHC, CCL244, HCT116, SW480 and
LOVO) were purchased from Shanghai Institute of Cell Biology
(Shanghai, China) and maintained in DMEM supplemented with 10% FBS
and antibiotics (100 U/ml penicillin G, 100 U/ml streptomycin
sulfates; Gibco, Grand Island, NY, USA) at 37° C in a humidified
atmosphere containing 5% CO2. Cells were exposed to a
single dose of X-ray irradiation from the linear accelerator
(RadSource, Suwanee, GA, USA) at a dose rate of 1.15 Gy/min.
SiRNA construction and transfection
The siRNA sequences targeting human HOTAIR
(si-HOTAIR) or negative control (NC) sequence were designed and
synthesized by Genepharma (Shanghai, China) as shown in Table II. Cells were transfected with
siRNA by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
 | Table IITarget sequences of HOTAIR siRNAs or
negative control (NC) siRNA. |
Table II
Target sequences of HOTAIR siRNAs or
negative control (NC) siRNA.
| siRNA | Target
sequences |
|---|
| si-HOTAIR1 |
| Sense |
5′-GCGCCUUCCUUAUAAGUAUTT-3′ |
| Antisense |
5′-AUACUUAUAAGGAAGGCGCTT-3′ |
| si-HOTAIR2 |
| Sense |
5′-GCACAGAGCAACUCUAUAATT-3′ |
| Antisense |
5′-UUAUAGAGUUGCUCUGUGCTT-3′ |
| si-HOTAIR3 |
| Sense |
5′-GAGGCGCUAAUUAAUUGAUTT-3′ |
| Antisense |
5′-AUCAAUUAAUUAGCGCCUCTT-3′ |
| NC |
| Sense |
5′-UUCUCCGAACGUGUCACGUTT-3′ |
| Antisense |
5′-ACGUGACACGUUCGGAGAATT-3′ |
RNA extraction, reverse transcription and
real-time RT-PCR
Total RNA from tissues and cells was extracted using
TRIzol reagent (Invitrogen). The reverse transcription of mRNAs was
performed using a reverse transcription kit (Thermo-Fisher,
Waltham, MA, USA). Amplification of cDNA template was performed by
real-time RT-PCR using the SYBR Green kit (Tiangen, Beijing,
China). Real-time RT-PCR was performed by the ABI7500 system
(Applied Biosystems, Foster City, CA, USA). GAPDH was used as an
endogenous standard, and HOTAIR values were normalized to GAPDH.
The relative expression of HOTAIR was calculated by the
2−∆∆Ct method. The primer were designed and synthesized
by Genepharma and the primer sequences are listed in Table III.
 | Table IIIPrimer sequences for qRT-PCR
analysis. |
Table III
Primer sequences for qRT-PCR
analysis.
| Gene | Forward
sequence | Reverse
sequence |
|---|
| HOTAIR |
5′-AAACAGAGTCCGTTCAGTGTCA-3′ |
5′-ATTCTTAAATTGGGCCTGGGTC-3′ |
| GAPDH |
5′-CATGAGAAGTATGACAACAGCCT-3′ |
5′-AGTCCTTCCACGATACCAAAGT-3′ |
Cell proliferation assays
The effect of HOTAIR knockdown on cell viability was
monitored by the MTT assay. Cells were seeded in 96-well plates and
cultured for 24, 48 or 72 h. Four hours before termination of
culture, 20 µl of 0.5 mg/ml MTT solution was added into each
well, then the MTT solution was removed and 200 µl DMSO was
added. The supernatant solution was vibrated for 15 min and then
placed in a microplate reader (Bio-Rad, Hercules, CA, USA) to
measure the optical density (OD) value at the wavelength of 490 nm.
The viability index was calculated as the experimental OD value/the
control OD value.
Colony-forming assay
Cell colony formation ability of CRC cells treated
with siRNA transfection was measured by plate colony formation
assay. Cells were seeded into 6-well plates at 300–8,000 cells/well
depending on the dose of radiation. After irradiated with 0, 2, 4,
6 or 8 Gy X-ray irradiation, cells were incubated at 37° C for
10–14 days to allow for colony formation. Then cells were fixed
with 4% paraformaldehyde for 15 min and stained with Giemsa for 20
min. Colony-forming efficiency (CFE) was calculated as follow: CFE
= (number of colonies/number of seeded cells) ×100%. Only viable
colonies containing at least 50 cells were counted.
Cell cycle progression analysis
The detection of CCL244 cell cycle was assessed by
flow cytometry 48 h after siRNA transfection. Cells were harvested
by trypsin digestion, pelleted by centrifugation at 114 × g for 5
min, washed with ice-cold PBS, then fixed with 75% cold ethanol
overnight. The staining solution containing propidium iodide and
DNase-free RNase was added 30 min before detection. Flow cytometric
analysis was performed to determine the fraction of the population
in each phase of the cell cycle using a Coulter flow cytometry
(Beckman-Coulter, Brea, CA, USA).
Wound healing assay
CRC cells transfected with siRNA were seeded into
6-well plates. Then the confluent cell monolayers were scratched
using a 200-µl pipette tip, followed by removal of the
supernatant and addition of fresh culture solution. The wound
healing was detected using an inverted microscope (Olympus, Tokyo,
Japan). The analysis of the cell migration assay was performed
using the ImageJ software (National Institutes of Health, Bethesda,
MD, USA).
Cell invasion assay
Cells (5×104) transfected with siRNA were
seeded into the upper part of a Transwell® chamber
(Corning Incorporated, Corning, NY, USA), which was pre-coated with
50 µl matrigel for 1 h, and 500 µl DMEM medium
containing 10% FBS was added into the lower part of the chamber.
Cells were cultured at 37° C with 5% CO2 for 24 h to
allow the cells to migrate through the matrigel. The remaining
cells on the upper surface were removed with a cotton swab
moistened with PBS. Penetrated cells on the lower surface were
fixed in ice-cold methanol for 30 min followed by staining with 2%
crystal violet in methanol for 15 min and counted under an inverted
microscope (Olympus, Tokyo, Japan). The cell number represented
migration activity.
Cell apoptosis analysis
Cells were transfected with siRNA 24 h prior to
treatment with sham or 6 Gy X-ray irradiation. The apoptosis rate
of CRC cells was detected by flow cytometry. Cells
(5×105) were washed with cold PBS at 179 × g for 5 min
and then resuspended in 1X Annexin V binding buffer. The solution
(500 µl) was transferred to a culture tube and Annexin
V-FITC and PI, 5 µl each, was added. The solution was mixed
and then incubated for 10 min at room temperature in the dark. Flow
cytometric analysis was performed after 30 min of staining.
Western blotting
Cells were transfected with siRNA 24 h prior to
treatment with sham or 6 Gy X-ray irradiation. The cells were
washed with PBS and then harvested in lysis buffer with 1 mM PMSF
on ice. The proteins extracted from supernatants were separated on
10% polyacrylamide gels and subsequently transferred onto PVDF
membranes. For immunoblotting analysis, PVDF membranes were
incubated with specific antibodies recognizing target proteins at
4° C overnight. After washing with TBST for three times, the
membranes were then incubated with corresponding HRP conjugated
secondary antibody (1:1000) for 1 h at room temperature. Finally
the membranes were analyzed by ECL detection system and visualized
by FluroChem MI imaging system (Alpha Innotech Corp., Santa Clara,
CA, USA). The primary antibodies were MMP2, MMP9 (1:1000; Cell
Signaling Technology, Boston, MA, USA), Bax and Bcl-2 (1:1000;
Santa Cruz Biotechnology, Santa Cruz, CA, USA). Protein GAPDH
(1:1000; Sigma-Aldrich, St. Louis, MO, USA) was used as loading
controls.
Statistical analysis
Statistical analysis was performed using SPSS 19.0.
Data are presented as mean ± SEM. The Student's t-test was used to
measure the difference between two groups. The differences among
three or more groups were tested for significance using one-way
analysis of variance. P<0.05 was considered to be statistically
significant.
Results
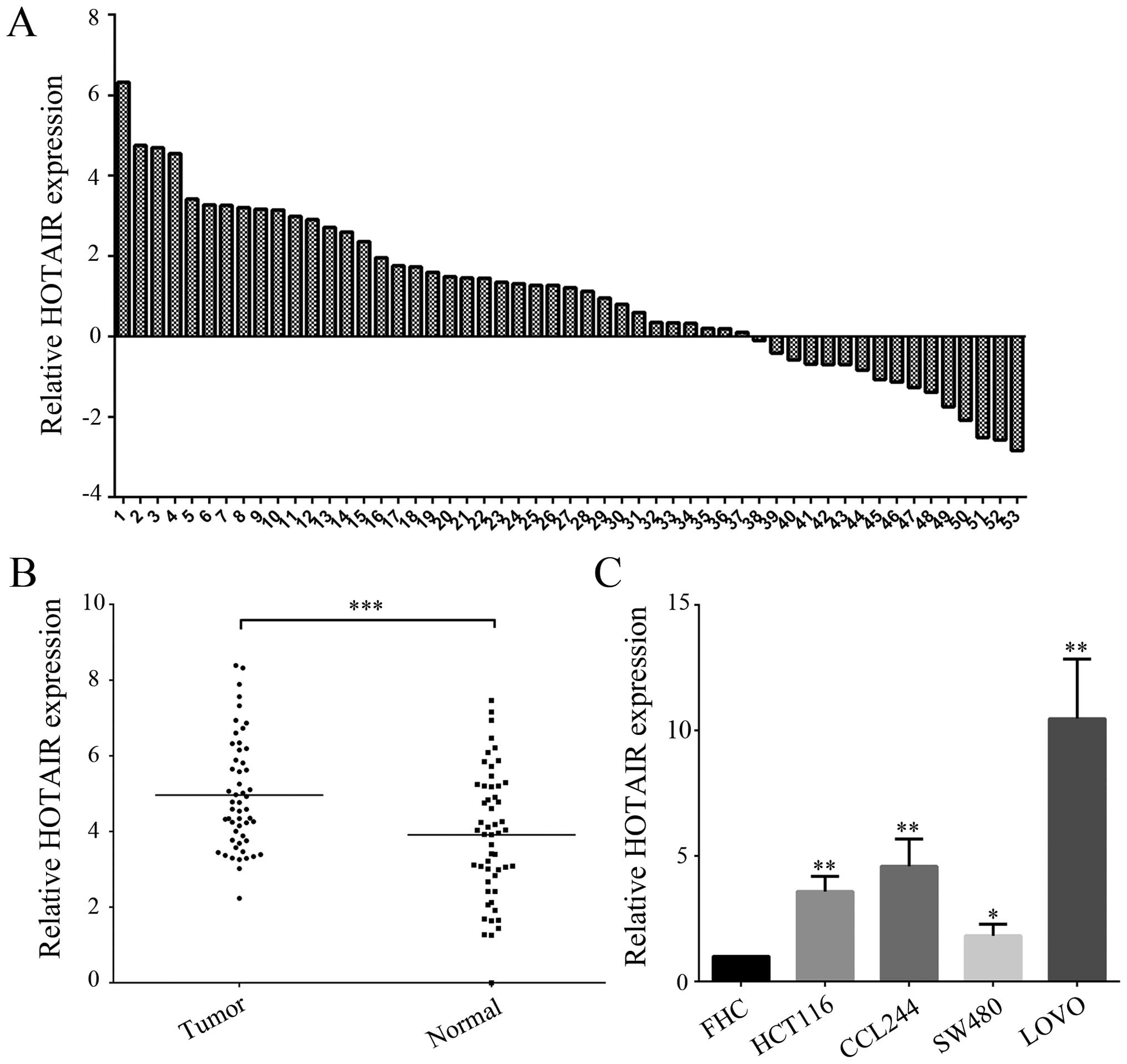
Overexpression of HOTAIR in human
CRC
The expression level of HOTAIR in 53 pairs of human
CRC and matched adjacent normal tissues was detected by real-time
RT-PCR. Fig. 1A and B showed the
overexpression of HOTAIR in 53 CRC tissues compared to their
non-tumorous counterparts. Furthermore, the expression level of
HOTAIR in 5 cell lines including one normal colonic mucosal cell
line (FHC) and 4 CRC cell lines (HCT116, CCL244, SW480, LOVO) was
also determined by real-time RT-PCR. Our results showed that the
expression of HOTAIR in HCT116, CCL244, SW480 and LOVO CRC cells
was higher than that in FHC cells (Fig.
1C). These results indicated that HOTAIR was highly expressed
in CRC tumor tissues and cell lines. Our previous study has
demonstrated that the ascending order of these four CRC cell lines
in terms of radiosensitivity was CCL244, SW480, LOVO and HCT116
(17). Therefore, CCL244 cell line
was selected to perform the following experiments.
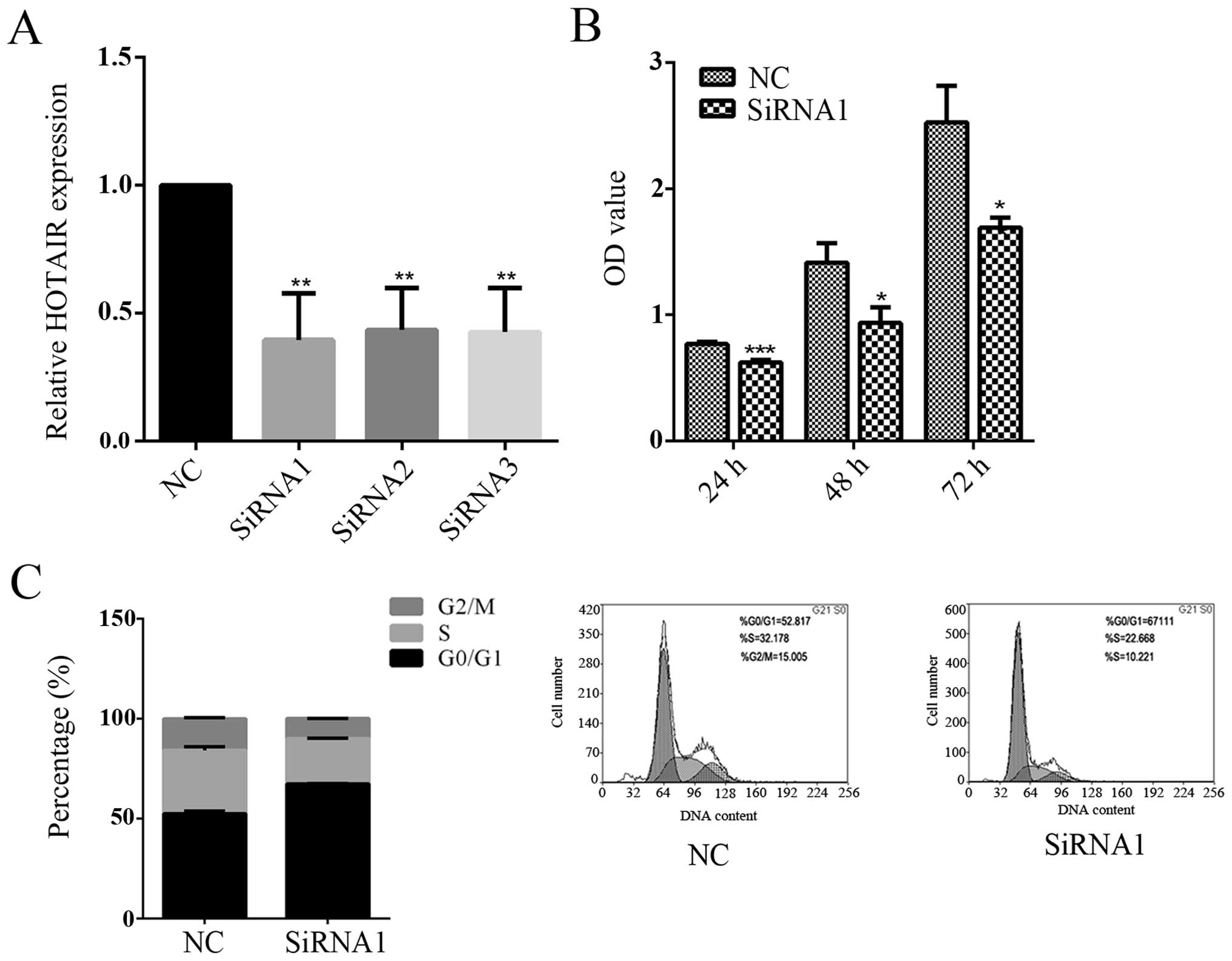
HOTAIR silencing inhibits the
proliferation of CRC cells
CCL244 cells were transfected with siRNA against
HOTAIR to further explore the role of HOTAIR in proliferation.
Three different siRNA vectors were designed to knockdown the coding
gene of HOTAIR and the silencing efficiency of these siRNA vectors
was detected by real-time RT-PCR. The results revealed that the
silencing efficiency of siRNA1, siRNA2 and siRNA3 was respectively
39.6, 43.5 and 42.6% (Fig. 2A).
Therefore, siRNA1 was selected to perform the following
experiments. MTT assay showed that knockdown of HOTAIR caused 18.8%
(P=0.0008), 33.9% (P=0.0403) and 33.1% (P=0.0242) reduction in the
cell viability at 24, 48 and 72 h after siRNA transfection compared
with negative control (NC) siRNA-transfected cells (Fig. 2B). Furthermore, cell cycle
progression analysis demonstrated that the population of cells in
G0/G1 phase was increased and the percentage of cells in S phase
was significantly decreased in HOTAIR knockdown cells (Fig. 2C). Taken together, these results
revealed that HOTAIR knock-down obviously inhibited CCL244 cell
proliferation through blocking cells in G0/G1 phase.
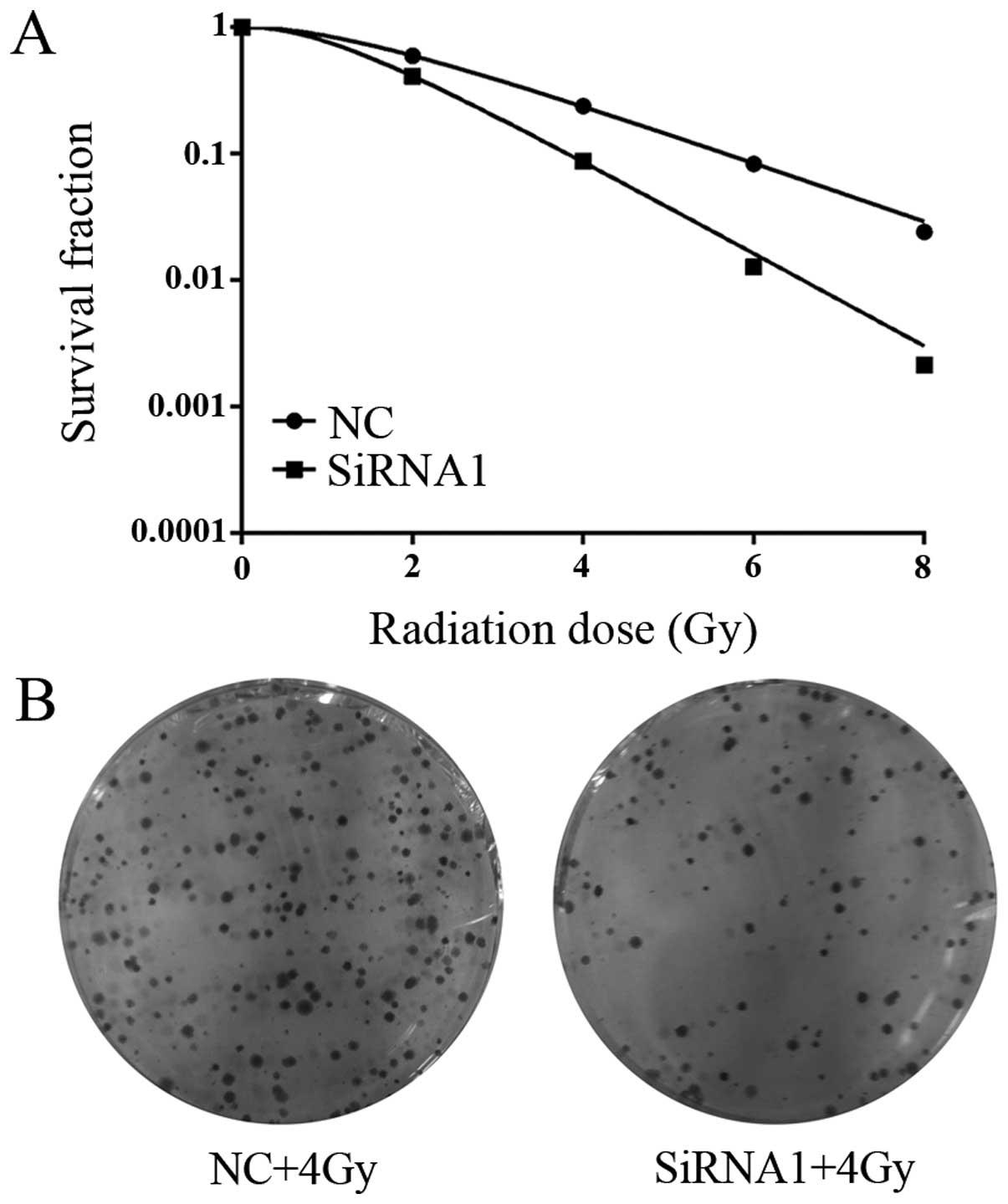
Knockdown of HOTAIR increases the
radiosensitivity of CCL244 cells
Clonogenic survival assay was performed to explore
the influence of HOTAIR on radiosensitivity of CCL244 CRC cells.
The dose-survival curves of CCL244 cells were obtained according to
the multi-target single hit model. Results showed that
HOTAIR-silencing caused a significant dose-dependent
radiosensitization in CCL244 cells (Fig. 3). The mean lethal dose (D0) for
cells treated with radiation alone was 1.85 Gy, while cells treated
with irradiation plus HOTAIR silencing exhibited a D0 of 1.18 Gy.
The quasi-threshold dose (Dq) in radiation alone and radiation plus
HOTAIR silencing group was 1.44 Gy and 1.12 Gy, respectively. The
sensitizer enhancement ratio (SER) was 1.57 for cells treated with
radiation plus HOTAIR silencing, compared to cells treated with
radiation alone. Therefore, these results demonstrated that
downregulation of HOTAIR significantly increased the
radiosensitivity of CCL244 cells.
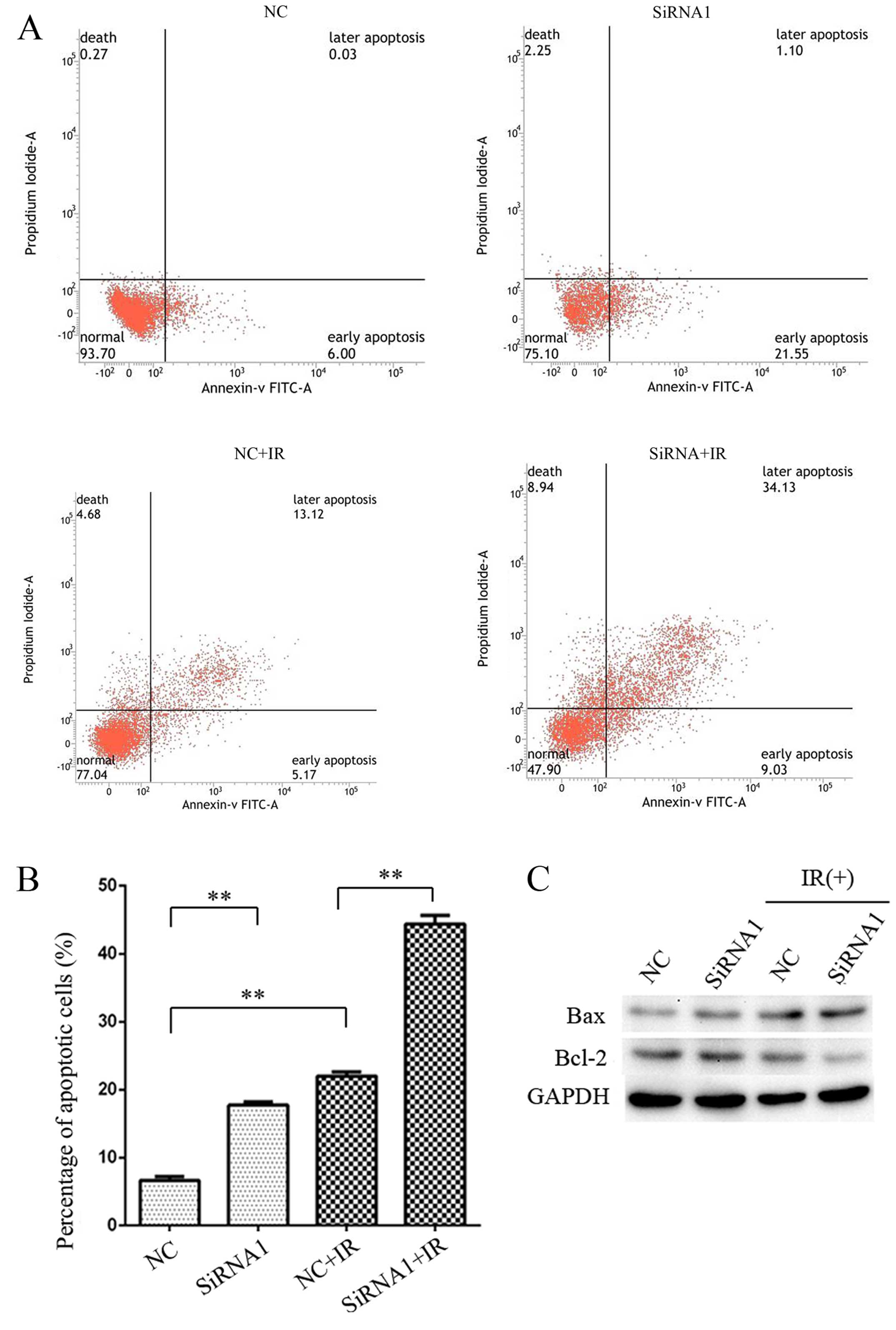
HOTAIR silencing increases
radiation-induced apoptosis in CCL244 cells and regulated the
expression of apoptosis-related proteins
To further investigate the role of HOTAIR in
radiation-induced apoptosis, flow cytometry was used to detect the
percentage of apoptotic cells in HOTAIR silenced CCL244 cells with
or without irradiation. As shown in Fig. 4A and 4B, 6 Gy X ray irradiation induced
apoptosis in CCL244 cells (NC 6.63% vs. NC+IR 21.99%, P=0.0033).
HOTAIR silencing further enhanced the apoptosis response to 6 Gy X
ray (NC+IR 21.99% vs. siRNA1+IR 44.41%, P=0.0039). Furthermore,
HOTAIR silencing CCL244 cells combined with irradiation showed
downregulation of anti-apoptotic protein Bcl-2 and upregulation of
pro-apoptotic protein Bax (Fig.
4C). These results demonstrated that HOTAIR knockdown
significantly increased radiation-induced apoptosis in CCL244
cells.
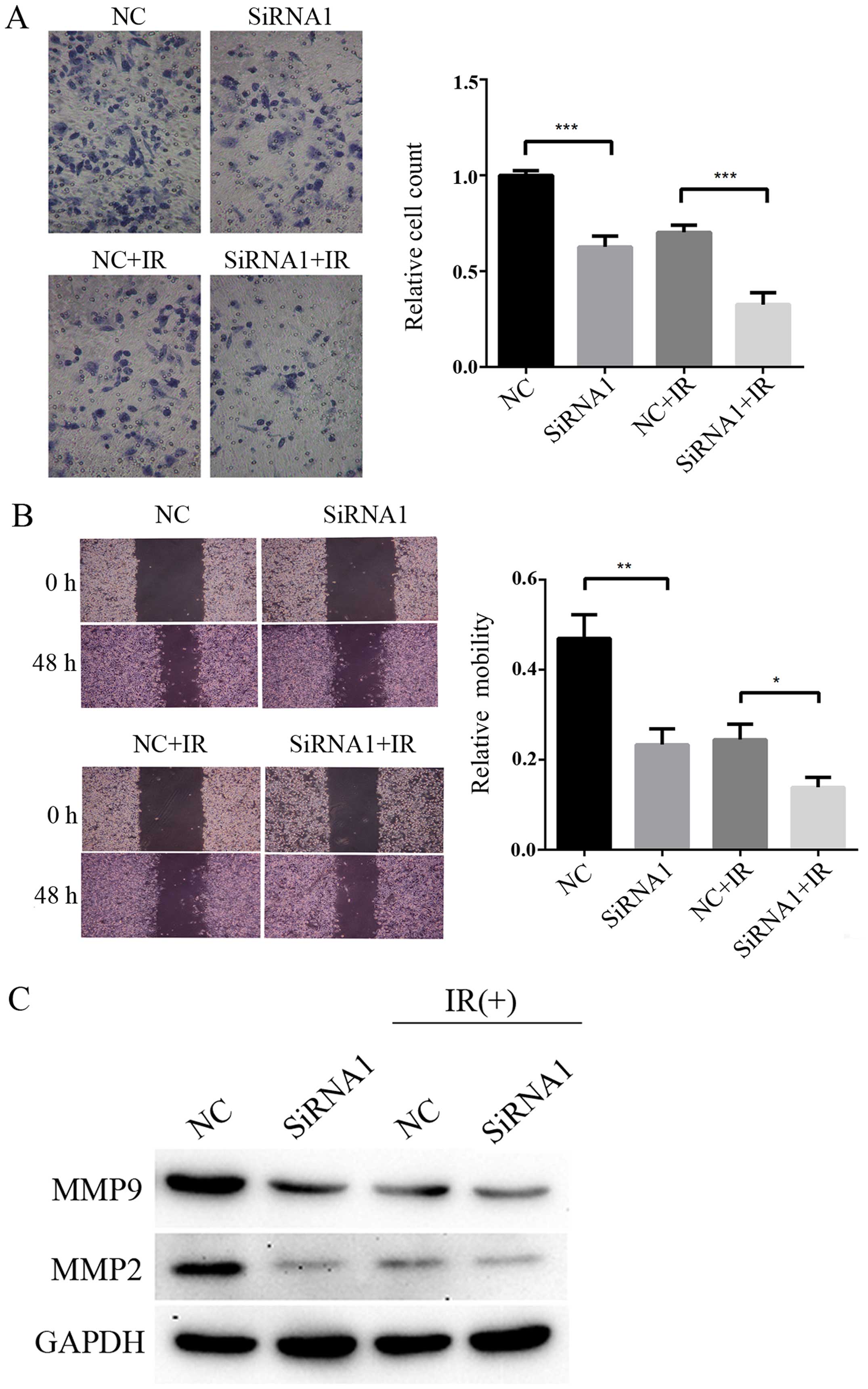
Knockdown of HOTAIR inhibits the invasion
and migration of CCL244 cells after irradiation
Metastasis and invasion are the most important
prognostic factors in patients with CRC. Transwell matrigel
invasion and wound healing assays were performed to further explore
the participation of HOTAIR in cell metastasis. As shown in
Fig. 5A, knockdown of HOTAIR
inhibited the invasiveness of CCL244 cells compared with negative
control (NC) CCL244 cells (P=0.0005). HOTAIR silencing CCL244 cells
combined with 6 Gy X-ray irradiation showed obvious inhibition in
invasiveness compared with cells treated with 6 Gy X-ray radiation
alone (P=0.0008). As shown in Fig.
5B, knockdown of HOTAIR inhibited the migration of CCL244 cells
compared with (NC) CCL244 cells (P=0.0029). HOTAIR silencing CCL244
cells combined with 6 Gy X-ray irradiation showed obvious
inhibition in migration compared with cells treated with 6 Gy X-ray
radiation alone (P=0.0108). As a family of extracellular matrix
degrading enzymes, matrix metalloproteinases (MMPs) are expressed
in various stages of CRC and participate in tumor invasion and
metastasis (18). The expression
level of some MMPs was also detected by western blotting. As shown
in Fig. 5C, HOTAIR silencing CCL244
cells combined with irradiation showed downregulation of MMP2 and
MMP9. Therefore, these results indicated that HOTAIR silencing
significantly inhibited the invasion and migration of CCL244 cells
after irradiation.
Discussion
Long non-coding RNAs (lncRNAs) are emerging as key
regulators of diverse cellular processes (19,20).
As the first found lncRNA, HOTAIR is co-expressed with the HOXC
gene cluster to regulate various genes through reverse
transcription (21,22). Numerous evidence has shown that
dysregulation of HOTAIR was closely associated with tumor
metastasis and poor prognosis (23–26).
However, the function role of HOTAIR in CRC proliferation,
invasion, radiation-induced apoptosis and radiosensitivity is still
being explored.
It is reported that HOTAIR is highly expressed in
various types of cancers such as primary breast tumors, hepato
cellular carcinoma, and gastrointestinal stromal tumors (27–29).
The present study examined the expression level of HOTAIR in CRC
tissues and matched normal tissues. Results showed that the HOTAIR
level was significantly upregulated in CRC cancer tissues compared
with adjacent normal tissues. Furthermore, CRC cell lines showed
high expression level of HOTAIR compared with normal colonic
mucosal cell line. The above evidence reveals the important role of
HOTAIR in CRC tumorigenesis.
To further explore the role of HOTAIR in CRC, we
investigated the effects of HOTAIR knockdown on CRC cell
proliferation. MTT assay revealed that RNAi-mediated suppression of
HOTAIR led to a significant inhibition in CCL244 cell viability.
Furthermore, HOTAIR silencing results in a significant inhibition
in the proliferation through blocking cells in G0/G1 phase.
Therefore, HOTAIR knockdown can inhibit the proliferation in CRC
and HOTAIR represents a new promising target and a prognosis marker
for CRC therapy.
It was demonstrated that HOTAIR is a negative
prognostic factor and HOTAIR silencing results in inhibition of
cell invasion in pancreatic tumors (30). Xu et al found that HOTAIR
promoted epithelial-mesenchymal transition (EMT) through regulating
snail expression thus contributing to gastric cancer metastasis
(31). Furthermore, HOTAIR may
promote tumor aggressiveness through the upregulation of VEGF and
MMP-9 and EMT-related genes (32).
Additionally, it is reported that HOTAIR promotes tumor cell
invasion and metastasis by recruiting EZH2 and repressing
E-cadherin in squamous cell carcinoma (33). We performed Transwell invasion and
wound healing assays to further explore the influence of HOTAIR
knockdown and irradiation treatment on cell migration and invasion.
Results showed that irradiation treatment caused an obvious
inhibition in cell invasion and HOTAIR silencing further enhanced
the inhibition response to irradiation. Furthermore, western
blotting was performed to detect the metastasis-related proteins of
MMPs to support the role of HOTAIR in CRC metastasis mechanically.
Similarly, the present study demonstrated that HOTAIR silencing
plus irradiation treatment significantly suppressed the expression
of MMP2 and MMP9, indicating the promoting effect of HOTAIR in
cancer metastasis. This phenomenon indicates that HOTAIR may
function as an oncogene in CRC tumorigenesis.
Apoptotic cell death is involved in almost every
mode of cell death. Radiation induced cell death is closely
associated with apoptosis. Dysregulation of apoptotic genes such as
caspase-3, Bax, and Bcl-2 may play a key role in radiation induced
cell death (34). HOTAIR has been
found to promote the proliferation of serous ovarian cancer cells
through the regulation of cell cycle arrest and apoptosis (35). Recently, it was reported that
calycosin and genistein induce apoptosis by inactivation of
HOTAIR/p-Akt signaling pathway in human breast cancer MCF-7 cells
(36). Consistent with these
studies, we also observed a significant increase in the apoptotic
cell population following HOTAIR silencing plus irradiation
treatment. Furthermore, HOTAIR silenced CRC cells plus irradiation
treatment showed a significant increase in pro-apoptotic protein
Bax levels and decrease in anti-apoptotic protein Bcl-2 levels. In
conclusion, irradiation treatment induced apoptosis in CRC CCL244
cells and HOTAIR silencing further enhanced the apoptotic response
to X ray irradiation.
Few studies have addressed the role of HOTAIR in
cancer cell radiosensitivity. Wang et al found that Curcumin
enhances the radiosensitivity in nasopharyngeal carcinoma cells
involving the reversal of differentially expressed long non-coding
RNAs (37). Several experiments
were performed to further evaluate the role of HOTAIR in CCL244 CRC
cell radio sensitivity in our study. Flow cytometry showed that
knockdown of HOTAIR significantly increased the radiation-induced
apoptosis in CCL244 cells. Furthermore, the SER of HOTAIR knockdown
was 1.57 in the clone formation assay. Therefore, our finding
indicates that HOTAIR knockdown sensitized CCL244 cells to
irradiation. However, the underlying mechanism needs further
exploration.
In summary, the present study showed that HOTAIR was
overexpressed in CRC and HOTAIR silencing inhibited the
proliferation and metastasis and enhanced the radio sensitivity in
CCL244 cells. These results indicate that targeting HOTAIR may
serve as a potentially novel approach for CRC diagnosis and
therapy.
Acknowledgments
This study was partially supported by the National
Natural Science Foundation of China (grant no. 81172348 and
81301933), Health Research Projects in Jiangsu Province (H201313),
the projects of Suzhou City Technology Bureau (SYSD2013090) and the
Priority Academic Program Development of Jiangsu Higher Education
Institutions (PAPD).
References
|
1
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Karsa LV, Lignini TA, Patnick J, Lambert R
and Sauvaget C: The dimensions of the CRC problem. Best Pract Res
Clin Gastroenterol. 24:381–396. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karim-Kos HE, de Vries E, Soerjomataram I,
Lemmens V, Siesling S and Coebergh JW: Recent trends of cancer in
Europe: A combined approach of incidence, survival and mortality
for 17 cancer sites since the 1990s. Eur J Cancer. 44:1345–1389.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lacombe J, Azria D, Mange A and Solassol
J: Proteomic approaches to identify biomarkers predictive of
radiotherapy outcomes. Expert Rev Proteomics. 10:33–42. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nie L, Wu HJ, Hsu JM, Chang SS, Labaff AM,
Li CW, Wang Y, Hsu JL and Hung MC: Long non-coding RNAs: versatile
master regulators of gene expression and crucial players in cancer.
Am J Transl Res. 4:127–150. 2012.PubMed/NCBI
|
|
6
|
Hajjari M, Khoshnevisan A and Shin YK:
Molecular function and regulation of long non-coding RNAs:
Paradigms with potential roles in cancer. Tumour Biol.
35:10645–10663. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li G, Zhang H, Wan X, Yang X, Zhu C, Wang
A, He L, Miao R, Chen S and Zhao H: Long noncoding RNA plays a key
role in metastasis and prognosis of hepatocellular carcinoma.
Biomed Res Int. 2014:7805212014.PubMed/NCBI
|
|
8
|
Sarkar SP and Adshead G: Whose DNA is it
anyway? European court, junk DNA, and the problem with prediction.
J Am Acad Psychiatry Law. 38:247–250. 2010.PubMed/NCBI
|
|
9
|
Popov N and Gil J: Epigenetic regulation
of the INK4b-ARF-INK4a locus: In sickness and in health.
Epigenetics. 5:685–690. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kotake Y, Nakagawa T, Kitagawa K, Suzuki
S, Liu N, Kitagawa M and Xiong Y: Long non-coding RNA ANRIL is
required for the PRC2 recruitment to and silencing of p15(INK4B)
tumor suppressor gene. Oncogene. 30:1956–1962. 2011. View Article : Google Scholar :
|
|
11
|
Wojcik SE, Rossi S, Shimizu M, Nicoloso
MS, Cimmino A, Alder H, Herlea V, Rassenti LZ, Rai KR, Kipps TJ, et
al: Non-codingRNA sequence variations in human chronic lymphocytic
leukemia and colorectal cancer. Carcinogenesis. 31:208–215. 2010.
View Article : Google Scholar :
|
|
12
|
Calin GA, Liu CG, Ferracin M, Hyslop T,
Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et
al: Ultraconserved regions encoding ncRNAs are altered in human
leukemias and carcinomas. Cancer Cell. 12:215–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F,
Yu W, Wang X, Zhang L, Yu J, et al: Long noncoding RNA HOTAIR
involvement in cancer. Tumour Biol. 35:9531–9538. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cai B, Song XQ, Cai JP and Zhang S:
HOTAIR: A cancer-related long non-coding RNA. Neoplasma.
61:379–391. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu
ZL, Zhou GZ, Cao G, Jin L, Xie HW, et al: Upregulation of the long
non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma
metastasis and poor prognosis. Mol Carcinog. 52:908–915. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu Y, Liu J, Zheng Y, You L, Kuang D and
Liu T: Suppressed expression of long non-coding RNA HOTAIR inhibits
proliferation and tumourigenicity of renal carcinoma cells. Tumour
Biol. 35:11887–11894. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang XD, Xu XH, Zhang SY, Wu Y, Xing CG,
Ru G, Xu HT and Cao JP: Role of miR-100 in the radioresistance of
colorectal cancer cells. Am J Cancer Res. 5:545–559.
2015.PubMed/NCBI
|
|
18
|
Wagenaar-Miller RA, Gorden L and Matrisian
LM: Matrix metalloproteinases in colorectal cancer: Is it worth
talking about? Cancer Metastasis Rev. 23:119–135. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu L, Zhu G, Zhang C, Deng Q, Katsaros D,
Mayne ST, Risch HA, Mu L, Canuto EM, Gregori G, et al: Association
of large noncoding RNA HOTAIR expression and its downstream
intergenic CpG island methylation with survival in breast cancer.
Breast Cancer Res Treat. 136:875–883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nie Y, Liu X, Qu S, Song E, Zou H and Gong
C: Long non-coding RNA HOTAIR is an independent prognostic marker
for nasopharyngeal carcinoma progression and survival. Cancer Sci.
104:458–464. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Geng YJ, Xie SL, Li Q, Ma J and Wang GY:
Large intervening non-coding RNA HOTAIR is associated with
hepatocellular carcinoma progression. J Int Med Res. 39:2119–2128.
2011. View Article : Google Scholar
|
|
28
|
Niinuma T, Suzuki H, Nojima M, Nosho K,
Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki
Y, et al: Upregulation of miR-196a and HOTAIR drive malignant
character in gastrointestinal stromal tumors. Cancer Res.
72:1126–1136. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ishibashi M, Kogo R, Shibata K, Sawada G,
Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et
al: Clinical significance of the expression of long non-coding RNA
HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 29:946–950.
2013.PubMed/NCBI
|
|
30
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar
|
|
31
|
Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ,
Huang L, Yu PF and Cheng XD: Knockdown of long non-coding RNA
HOTAIR suppresses tumor invasion and reverses
epithelial-mesenchymal transition in gastric cancer. Int J Biol
Sci. 9:587–597. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim
SW and Kim YT: Long non-coding RNA HOTAIR is associated with human
cervical cancer progression. Int J Oncol. 46:521–530. 2015.
|
|
33
|
Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren
X, Wei F, Yu W, Liu T, Wang X, et al: Long non-coding RNA HOTAIR
promotes tumor cell invasion and metastasis by recruiting EZH2 and
repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol.
46:2586–2594. 2015.PubMed/NCBI
|
|
34
|
Shinomiya N: New concepts in
radiation-induced apoptosis: 'premitotic apoptosis' and
'postmitotic apoptosis'. J Cell Mol Med. 5:240–253. 2001.
View Article : Google Scholar
|
|
35
|
Qiu JJ, Wang Y, Ding JX, Jin HY, Yang G
and Hua KQ: The long non-coding RNA HOTAIR promotes the
proliferation of serous ovarian cancer cells through the regulation
of cell cycle arrest and apoptosis. Exp Cell Res. 333:238–248.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen J, Lin C, Yong W, Ye Y and Huang Z:
Calycosin and genistein induce apoptosis by inactivation of
HOTAIR/p-Akt signaling pathway in human breast cancer MCF-7 cells.
Cell Physiol Biochem. 35:722–728. 2015.PubMed/NCBI
|
|
37
|
Wang Q, Fan H, Liu Y, Yin Z, Cai H, Liu J,
Wang Z, Shao M, Sun X, Diao J, et al: Curcumin enhances the
radiosensitivity in nasopharyngeal carcinoma cells involving the
reversal of differentially expressed long non-coding RNAs. Int J
Oncol. 44:858–864. 2014.PubMed/NCBI
|