Introduction
Although the majority of newly diagnosed bladder
cancer is non-muscle invasive bladder cancer (NMIBC), most patients
with NMIBC relapse after adequate treatment, and some progress to
muscle invasive disease (1,2). Furthermore, the potential of these
tumors for recurrence and progression into muscle invasive disease
is highly unpredictable. The detection of high-risk cases after
transurethral resection (TUR) of the bladder tumor is essential and
depends on efficient prognostic biomarkers for high-risk patients.
Research efforts worldwide have focused on the identification of
clinically helpful tumor markers or potentially valuable
therapeutic targets to improve current diagnostic and management
strategies for patients with bladder cancer (3).
Disease-specific aberrant DNA methylation is
recognized as a hallmark of many cancers, which led to new
opportunities for the understanding, detection, treatment, and
prevention of diseases including bladder cancer (4–12). The
use of DNA meth-ylation as a biomarker has gained increasing
interest in recent years, as aberrant DNA methylation is a major
characteristic of bladder cancer and plays a crucial role in tumor
initiation and progression (7–13).
Although many of the different genetic or epigenetic changes that
lead to aberrant gene expression in bladder cancer have been
identified, the discovery of novel candidate methylation biomarkers
was accelerated by the advent of high-throughput methylation
profiling using normal and malignant cells (14).
NMIBC of similar morphology may behave differently,
and it is difficult to predict clinical outcomes after initial
treatment (1,2). Therefore, novel biomarkers predictive
of disease outcomes, in addition to commonly used
clinicopatho-logical parameters, would be valuable for guiding
appropriate management strategies (7). The aim of the present study was to
identify novel methylation markers predictive of patient outcomes
using microarray analysis of DNA methylation profiles in long-term
follow-up NMIBC samples.
Materials and methods
Subjects and sample collection
A total of 136 human bladder specimens were used for
pyrosequencing (PSQ) analyses, including 8 normal controls (NC) and
128 NMIBC samples (Table I). NMIBC
specimens were obtained from 128 primary NMIBC patients who
underwent TUR for histo-logically diagnosed transitional cell
carcinomas between 1995 and 2010 at our institute. To exclude the
possibility of incomplete resection or confounding factors that may
unduly affect the analyses, patients followed-up for less than 6
months or those that experienced disease relapse within 6 months
were excluded from the study. Samples of normal bladder urothelium
obtained from individuals with benign prostate hyperplasia or
bladder injury were used as controls.
 | Table IBaseline characteristics of the study
subjects. |
Table I
Baseline characteristics of the study
subjects.
| Variables | NC (n=8) | NMIBC (n=128) |
|---|
| Age (years),
mean | 59.0±22.2 | 62.6±14.5 |
| Gender, no. of
patients (%) | | |
| Male | 6 (75.0) | 103 (80.5) |
| Female | 2 (25.0) | 25 (19.5) |
| No. of tumors
(%) | | |
| Single | – | 77 (60.2) |
| Multiple | – | 51 (39.8) |
| Tumor size, no. of
patients (%) | | |
| <3 cm | – | 74 (57.8) |
| ≥3 cm | – | 54 (42.2) |
| Grade, no. of
patients (%) | | |
| G1 | – | 40 (31.3) |
| G2 | – | 75 (58.6) |
| G3 | – | 13 (10.2) |
| T stage, no. of
patients (%) | | |
| Ta | – | 50 (39.1) |
| T1 | – | 78 (60.9) |
| Median RFS, months
(range) | – | 30.6
(6.0–205.3) |
| Recurrence, no. of
patients (%) | | |
| No | – | 74 (57.8) |
| Yes | – | 54 (42.2) |
| Median PFS, months
(range) | – | 54.1
(6.4–205.3) |
| Progression, no. of
patients (%) | – | |
| No | – | 113 (88.3) |
| Yes | – | 15 (11.7) |
All tumors were macro-dissected within 15 min of
surgical resection. Each NMIBC specimen was confirmed by
pathological analysis of a part of the tissue sample (i.e.,
sections were taken from TUR specimens and then snap-frozen in
liquid nitrogen and stored at −80°C). The specimens were provided
by the Chungbuk National University Hospital, a member of the
National Biobank of Korea, which is supported by the Ministry of
Health, Welfare and Family Affairs. The collection and analysis of
all samples was approved by the Chungbuk National University
Hospital Institutional Review Board (GR2010-12-010), and informed
consent was obtained from each subject.
Tumors were staged according to the 2002 TNM
classification and the 1973 WHO grading system (1,2). A
second TUR was performed 2–4 weeks after the initial resection if a
bladder cancer specimen did not include proper muscle, or if a
high-grade tumor was detected. Patients with intermediate- or
high-risk NMIBC received one cycle of intravesical therapy. Each
patient was followed-up and managed according to the standard
recommendations (1,2). Recurrence was defined as the
recurrence of primary NMIBC at a lower or equivalent pathologic
stage (Ta/T1), and progression was defined as muscular invasion
(TNM stage T2 or higher) or metastatic disease.
DNA methylation profiling
Microarray methylation data of 24 human bladder
specimens (NMIBC=18, NC=6) previously published by our group were
used (15). Methylation patterns
were assayed using the genome-wide Illumina Infinium
HumanMethylation27 BeadChip array (Illumina Inc., San Diego, CA,
USA), which enables interrogation of 27,578 CpG dinucleotides
covering 14,495 genes. Methylation assays were carried out
according to the manufacturer's protocol. Bisulfite conversion of
genomic DNA was carried out using the EZ DNA Methylation kit (Zymo
Research, Orange, CA, USA). Fluorescence signals corresponding to
C- or T-nucleotides were measured, and the data were used to assign
a quantitative measure of methylation level (β-value). Each
methylation data point represents the fluorescent signal from the
methylated (M) and unmethylated (U) alleles. Background intensity
was computed from a set of negative controls and subtracted from
each analytic data point. The ratio of the fluorescent signals from
the 2 alleles was then computed. The β-value value represents a
quantitative measure of the DNA methylation level of specific CpG
islands, and ranges from 0 (completely unmethylated) to 1
(completely methylated).
PSQ analysis
The DNA methylation status of candidate
NMIBC-specific hypermethylated CpG sites was assessed by PSQ using
PyroMark Q96 ID (Qiagen, Valencia, CA, USA) according to the
manufacturer's instructions. PSQ primers were designed to encompass
the CpG sites assayed on the Illumina Infinium array. The primer
sequences and amplification conditions are described in Table II.
 | Table IIPrimers used for pyrosequencing
analysis. |
Table II
Primers used for pyrosequencing
analysis.
| Genes | Forward
(5′-3′) | Reverse
(5′-3′) | Sequencing primer
(5′-3′) | Annealing Tm
(°C) | Amplicon location
relative to TSS | Sequence to
analyze | Product size
(bp) |
|---|
| BARHL2 | GGTTTTTTTTATTG |
(Biotin)-ACAAATTACCACTTCCCAATTA |
AAAATAAAATAAATATTAAATAATG | 56 | (−106)–(−94) | TTGTYGTYGTTT | 184 |
| TTATTGTTATATGA | | | | | | |
| RSPH9 | GTTAAGGATGAGG |
(Biotin)-ACAACCTCCTACTATCTCT |
AGGATGAGGTTTTAGAAGAGAAT | 52 | 117–147 |
TYGATTATAGYGGTA | 146 |
| TTTTAGAAGAGAA | | | | |
GTYGYGTTTAATTAG | |
| RAB37 |
(Biotin)-TGAATGAAATAA |
AACCAAAATTCTAAAATCCTATC |
CTTAAAACTCCAAAATACTAAC | 52 | (−348) –
(−327) |
GTYGGAGGGTAAAAACR | 111 |
|
GTAGGGATTATTAGT | | | | |
CAAAATTTAAATCCRACT | |
Bladder cancer cell lines, culture, and
drug treatment
The human bladder cancer cell lines T24 (KCLB 30004)
and J82 (KCLB 30001) were purchased from the Korean Cell Line Bank
(Seoul, Korea). All cells were maintained in RPMI-1640 medium
supplemented with 10% FBS (both from Gibco-BRL, Grand Island, NY,
USA) and L-glutamine, and cells were incubated in a humidified
atmosphere with 5% CO2 at 37°C. Both cell lines were
cultured until confluent and then treated with PBS or 0.3
μmol/l 5-Aza-CdR (both from Sigma-Aldrich, St. Louis, MO,
USA). The medium was changed the following day, and cells were
counted and harvested on days 3 and 7, and then once per week.
Statistical analysis
DNA methylation data were normalized using quantile
normalization in the R language environment (version 2.10.0,
available at http://www.r-project.org/). The detailed analytical
methods have been described previously (16,17).
To detect NMIBC-specific candidate methylation markers, genes whose
methylation levels differed between NMIBC and NC by a β-value
(∆β-values) >0.5 were selected. To validate the genes identified
in this study, our ∆β-values were compared with those of two
microarray data sets obtained from Western populations (9,13): i),
32 bladder tissues (NMIBC =26, NC =6); and ii), 70 bladder tissues
(NMIBC =64, NC =6). Continuous variables between groups were
compared using a two-sample t-test or ANOVA trend analysis using
polynomial contrasts. To minimize bias against arbitrary cut-off
points, median values were applied to divide patients into
subgroups (hypomethylation or hypermeth-ylation), and the survival
function of candidate genes was evaluated. The Kaplan-Meier method
was used to estimate the time-to-recurrence or progression based on
methylation status, and differences were assessed using log-rank
statistics. For the multivariate Cox proportional hazards
regression models, the prognostic value of methylation status was
evaluated separately and adjusted for well-known
clinicopathological (gender, age, tumor size, tumor number,
intravesical therapy, grade, and stage) factors. Statistical
analysis was performed using IBM SPSS Statistics ver. 21.0 (IBM
Co., Armonk, NY, USA). P<0.05 was considered statistically
significant.
Results
Baseline characteristics
The baseline characteristics of the NC and NMIBC
patients are presented in Table I.
Mean recurrence-free survival and progression-free survival in
patients with NMIBC was 44.1±39.1 months (median, 30.6; range,
6.0–205.3) and 60.8±40.7 months (median, 54.1; range, 6.4–205.3),
respectively.
Identification of differentially
methylated and expressed genes in NMIBC and NC
Our previously published genome-wide methylation
profiles obtained from 18 NMIBC patients were analyzed and compared
with those from 6 NC (15). The
complete sets of microarray data derived from the human bladder
tissues are available online (http://www.ncbi.nlm.nih.gov/geo/) under the data
series accession number GSE37817. To select NMIBC-specific
methylation markers, a highly stringent selection criterion
(∆β-value >0.5) was applied. We identified 25 unique CpG island
loci in 23 genes that were hypermethylated in NMIBC compared with
NC, and these are listed in Table
III. The validity of our candidate genes as methylation markers
for NMIBC was evaluated using an independent set of Infinium
microarray methylation data derived from two Western populations
(9,13). Only partial validation was possible
because of the limited data provided in both studies. The ∆β-values
obtained in the present study were comparable to those of other
studies (Table III). Of the
candidate markers identified, genes in the top fifth ∆β-value were
selected for evaluation of their clinical relevance in the
validation cohort.
 | Table IIIComparison of β-value differences
between tumors and normal controls with Infinium DNA methylation
array data. |
Table III
Comparison of β-value differences
between tumors and normal controls with Infinium DNA methylation
array data.
| Target ID | Symbol | Annotation | Our study
| Reinert et
al (9)
| Ibragimova et
al (13)
| CpG island
locationb |
|---|
| Normal | Tumor | ∆β-valuea | ∆β-valuea | ∆β-valuea |
|---|
| cg02983451 | KLF11 | Kruppel-like factor
11 | 0.13 | 0.75 | 0.62 | 0.42 | 0.58 |
2:10099704-10102471 |
| cg17241310 | BARHL2 | BarH-like homeobox
2 | 0.12 | 0.7 | 0.57 | 0.46 | N/A |
1:90954376-90957242 |
| cg05899618 | GDF7 | Growth
differentiation factor 7 | 0.12 | 0.69 | 0.57 | 0.39 | N/A |
2:20728362-20731136 |
| cg04600618 | RSPH9 | Radial spoke head 9
homolog | 0.14 | 0.71 | 0.57 | 0.44 | 0.59 |
6:43720593-43721485 |
| cg12448933 | RAB37 | RAB37, member RAS
oncogene family | 0.2 | 0.76 | 0.56 | 0.49 | N/A |
17:70178644-70179471 |
| cg12374721 | PRAC | Prostate cancer
susceptibility candidate | 0.15 | 0.71 | 0.56 | 0.48 | N/A |
17:44154292-44154687 |
| cg04396791 | SHANK2 | SH3 and multiple
ankyrin repeat domains 2 | 0.25 | 0.81 | 0.56 | 0.29 | N/A |
11:70185287-70186268 |
| cg24199834 | POU4F2 | POU class 4
homeobox 2 | 0.14 | 0.68 | 0.54 | 0.47 | 0.61 |
4:147778503-147781596 |
| cg25802093 | SPAG6 | Sperm associated
antigen 6 | 0.21 | 0.75 | 0.54 | 0.52 | N/A |
10:22673932-22675060 |
| cg08536841 | IZUMO1 | Izumo sperm-egg
fusion 1 | 0.21 | 0.73 | 0.53 | 0.4 | N/A |
19:53942196-53942451 |
| cg00117172 | RUNX3 | Runt-related
transcription factor 3 | 0.08 | 0.61 | 0.53 | 0.42 | 0.46 |
1:25127692-25131906 |
| cg21790626 | ZNF154 | Zinc finger protein
154 | 0.09 | 0.61 | 0.53 | 0.52 | 0.59 |
19:62911404-62912681 |
| cg23563234 | PCDHGB7 | Protocadherin γ
subfamily B, 7 | 0.16 | 0.68 | 0.53 | 0.45 | N/A |
5:140777221-140777959 |
| cg08260959 |
HIST1H4F | Ηistone cluster 1,
H4f | 0.09 | 0.61 | 0.52 | 0.45 | 0.58 |
6:26348554-26349101 |
| cg21475402 | BCAN | Βrevican | 0.18 | 0.7 | 0.52 | N/A | N/A |
1:154878094-154879230 |
| cg05159188 |
HIST1H4F | Histone cluster 1,
H4f | 0.11 | 0.63 | 0.52 | 0.46 | 0.48 |
6:26348554-26349101 |
| cg14456683 | ZIC1 | Zic family member
1 | 0.21 | 0.73 | 0.51 | 0.52 | N/A |
3:148609249-148612068 |
| cg12874092 | VIM | Vimentin | 0.04 | 0.55 | 0.51 | 0.42 | 0.43 |
10:17310042-17312147 |
| cg08668790 | ZNF154 | Zinc finger protein
154 | 0.11 | 0.62 | 0.51 | 0.5 | 0.61 |
19:62911404-62912681 |
| cg23129478 | ST8SIA5 | ST8
α-N-acetyl-neuraminide α-2,8-sialyltransferase 5 | 0.09 | 0.6 | 0.51 | 0.37 | 0.47 |
18:42589800-42592268 |
| cg19352038 | PAX3 | Paired box 3 | 0.25 | 0.76 | 0.51 | 0.42 | N/A |
2:222872611-222873277 |
| cg07778029 | HOXA9 | Homeobox A9 | 0.06 | 0.57 | 0.51 | 0.5 | 0.46 |
7:27170287-27173690 |
| cg03975694 | ZNF540 | Zinc finger protein
540 | 0.18 | 0.68 | 0.51 | 0.39 | N/A |
19:42733846-42734771 |
| cg01295203 | PRDM14 | PR domain
containing 14 | 0.14 | 0.65 | 0.5 | 0.48 | 0.55 |
8:71144163-71147771 |
| cg12111714 | ATP8A2 | ATPase,
aminophospholipid transporter, class I, type 8A, member 2 | 0.32 | 0.82 | 0.5 | 0.42 | N/A |
13:24940557-24941659 |
PSQ analysis
To verify the clinical relevance of candidate
methylation markers, PSQ analysis was performed using
bisul-fite-modified genomic DNA obtained from 136 human bladder
specimens (NMIBC=128, NC=8). PSQ analysis was technically possible
in three out of five candidate genes [BarH-like homeobox 2
(BARHL2), radial spoke head 9 homolog (RSPH9), and
member RAS oncogene family (RAB37)], and these genes were
analyzed by PSQ in the present study.
Association between methylation levels
and clinicopatho-logical variables
As shown in Table
IV, the methylation levels of candidate genes were
significantly higher in NMIBC patient samples than in normal
samples (P<0.001). To evaluate the relationship between
methylation patterns and clinicopathological factors, methylation
levels were examined in correlation with well-known prognostic
factors such as tumor number, tumor size, and tumor grade and
stage. The results showed that high levels of methylation of
BARHL2 and RSPH9 were significantly associated with
tumor size, grade, and stage.
 | Table IVAssociation between methylation
markers and clinicopathological characteristics. |
Table IV
Association between methylation
markers and clinicopathological characteristics.
| Variables | BARHL2
| RSPH9
| RAB37
|
|---|
| Methylation level
(%) | P-value | Methylation level
(%) | P-value | Methylation level
(%) | P-value |
|---|
| Normal vs.
cancer | | <0.001a | | <0.001a | | <0.001a |
| Normal | 17.1±3.5 | | 18.0±6.6 | | 24.4±1.0 | |
| Cancer | 53.8±24.0 | | 52.6±25.9 | | 42.6±18.2 | |
| No. of tumors | | 0.043a | | 0.030a | | 0.496a |
| Single | 50.5±25.3 | | 48.6±25.6 | | 43.5±18.4 | |
| Multiple | 58.9±21.2 | | 58.7±25.4 | | 41.2±18.0 | |
| Tumor size
(cm) | | 0.461a | | 0.196a | | 0.386a |
| <3 | 52.4±25.2 | | 55.1±25.0 | | 41.4±17.8 | |
| ≥3 | 55.7±22.4 | | 49.1±26.0 | | 44.2±18.7 | |
| Grade | | 0.002b | | 0.001b | | 0.753b |
| G1 | 46.7±23.4 | | 45.6±22.8 | | 45.7±23.4 | |
| G2 | 54.7±23.0 | | 52.1±27.0 | | 39.6±17.6 | |
| G3 | 70.8±23.5 | | 76.6±11.5 | | 49.6±20.2 | |
| T stage | | 0.004a | | 0.042a | | 0.216a |
| Ta | 46.2±24.1 | | 47.7±23.1 | | 45.1±20.0 | |
| T1 | 58.7±22.7 | | 57.8±27.2 | | 41.0±17.1 | |
| Recurrence | | 0.048a | | 0.005a | | 0.154a |
| No | 50.5±24.4 | | 47.1±24.3 | | 40.5±16.1 | |
| Yes | 58.7±22.7 | | 60.1±26.4 | | 45.4±20.5 | |
| Progression | | 0.022a | | 0.002a | | 0.554a |
| No | 52.1±24.2 | | 50.1±25.1 | | 43.0±25.1 | |
| Yes | 67.1±18.6 | | 71.5±24.4 | | 40.0±23.5 | |
Methylation status as a predictor of
prognosis
The methylation levels of BARHL2 and
RSPH9 were significantly higher in the poor prognosis group
(recurrence or progression) than in the favorable prognosis group
(Table IV). Stratification of
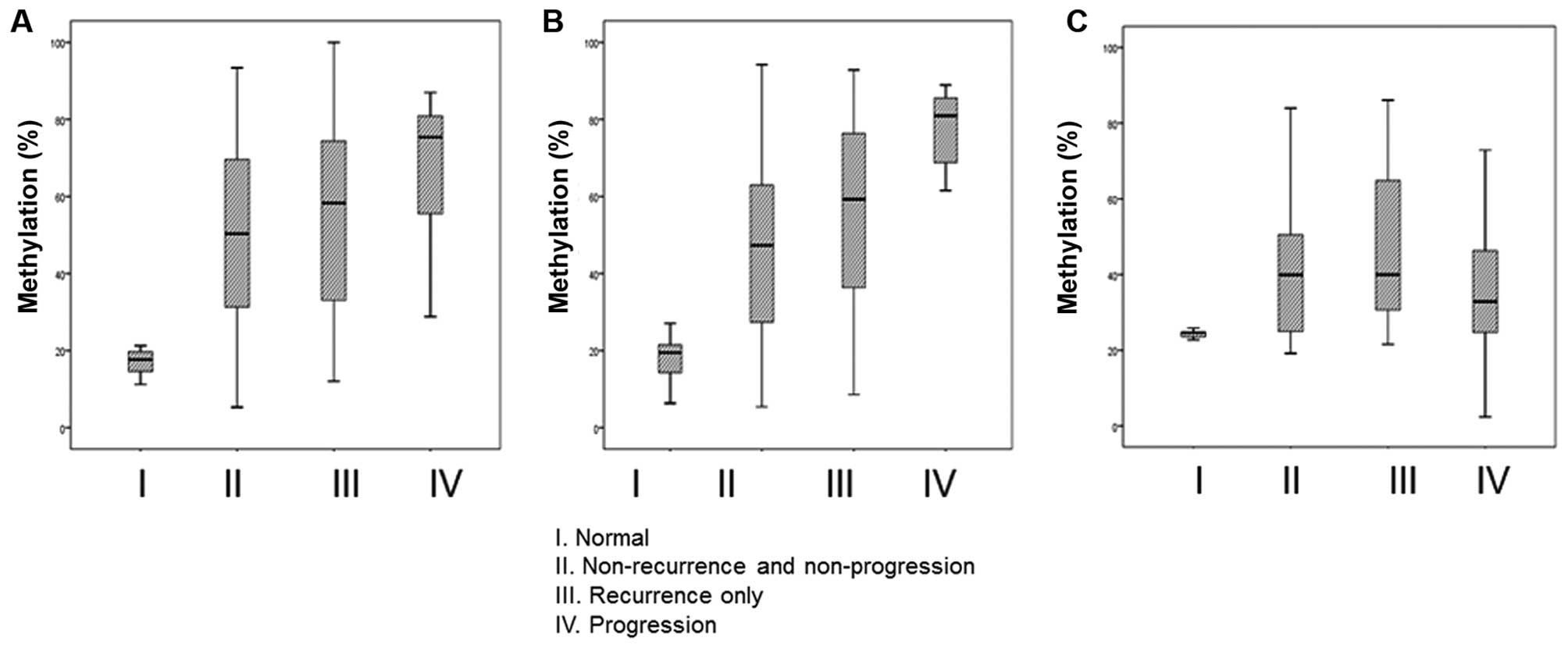
NMIBC patients into three prognostic groups (non-recurrence and
non-progression, recurrence only, and progression) showed that the
methylation of BARHL2 and RSPH9 was positively
correlated with poor prognosis (Fig.
1). To further determine the relevance of candidate gene
methylation status as a predictive indicator, the methylation
levels of each gene were dichoto-mized (hypomethylation or
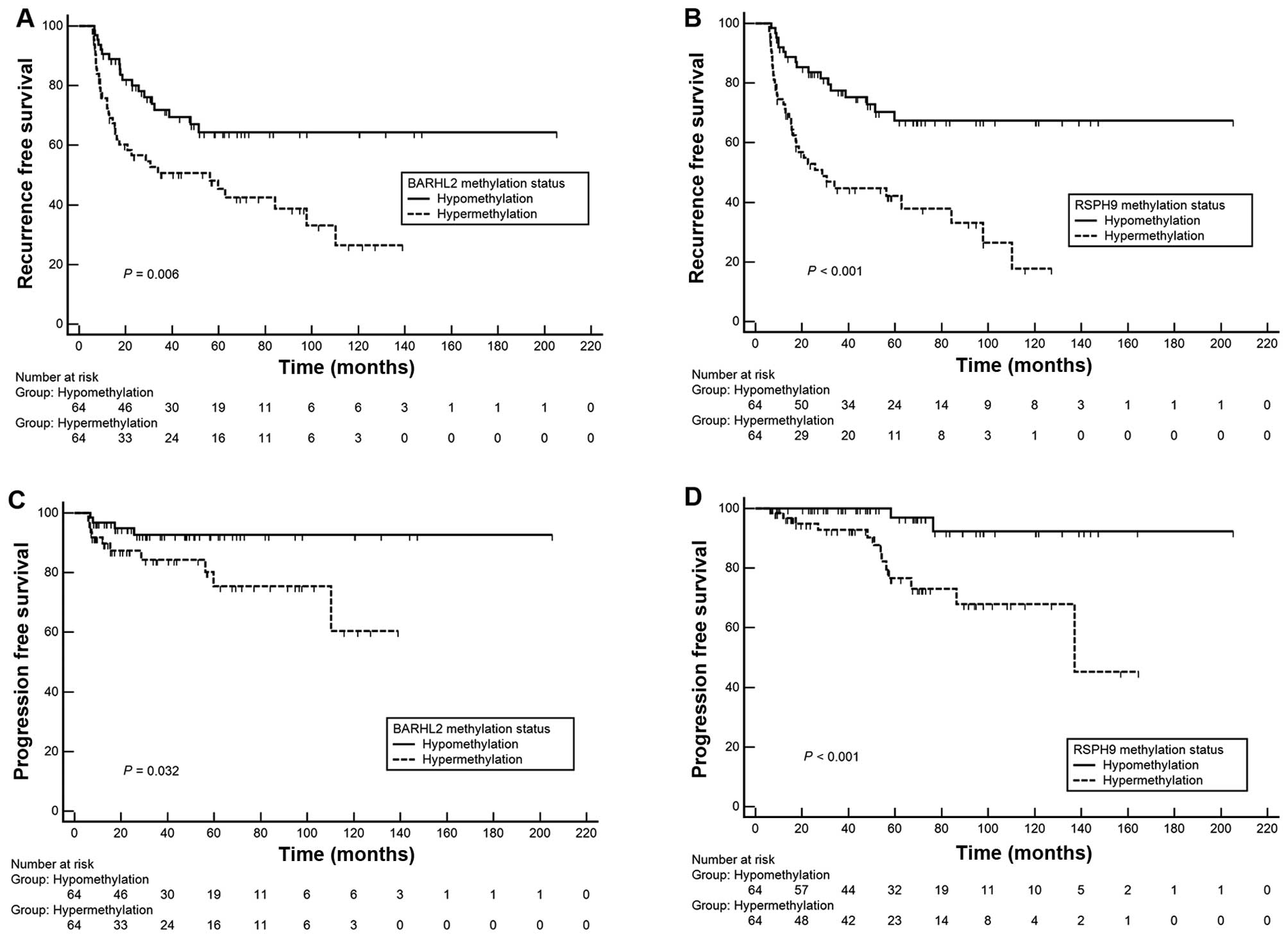
hypermethylation) using median cut-off points. Kaplan-Meier
estimates identified significant differences in time-to-recurrence
or progression according to methylation status of BARHL2 and
RSPH9 (Fig. 2, log-rank
test, each P<0.05). In univariate and multivariate Cox
regression analyses, RSPH9 methylation status was an
independent predictor of recurrence [hazard ratio (HR), 3.02;
P=0.001] and progression (HR, 8.25; P=0.028, Table V) in primary NMIBC patients.
However, differences in BARHL2 methylation did not reach
statistical significance for the prediction of prognosis
(P>0.05).
 | Table VMultivariate Cox regression analysis
of disease outcomes according to RHPH9 methylation in
non-muscle invasive bladder cancer (n=128). |
Table V
Multivariate Cox regression analysis
of disease outcomes according to RHPH9 methylation in
non-muscle invasive bladder cancer (n=128).
| Variables | Recurrence
| Progression
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (<66 vs. ≥66
years) | 0.99
(0.57–1.76) | 0.973 | 2.31
(0.69–7.73) | 0.175 |
| Gender (male vs.
female) | 0.56
(0.25–1.26) | 0.158 | 0.18
(0.20–1.55) | 0.118 |
| No. of tumors
(single vs. multiple) | 1.14
(0.62–2.08) | 0.672 | 6.91
(1.70–28.10) | 0.007 |
| Tumor size (<3
vs. ≥3 cm) | 1.27
(0.69–2.23) | 0.158 | 2.92
(0.85–9.96) | 0.088 |
| Stage (Ta vs.
T1) | 0.69
(0.33–1.43) | 0.322 | 1.17
(0.12–11.87) | 0.896 |
| Grade | | | | 0.021 |
| G1 | 1 | – | 1 | – |
| G2 | 1.37
(0.65–2.90) | 0.408 | 3.04
(0.23–40.06) | 0.399 |
| G3 | 1.20
(0.41–3.53) | 0.744 | 17.49
(1.11–276.29) | 0.042 |
| Intravesical
therapy (no vs. yes) | 1.41
(0.70–2.83) | 0.331 | 0.84
(0.14–4.91) | 0.835 |
| RSPH9
(hypomethylation vs. hypermethylation) | 3.02
(1.61–5.67) | 0.001 | 8.25
(1.26–54.09) | 0.028 |
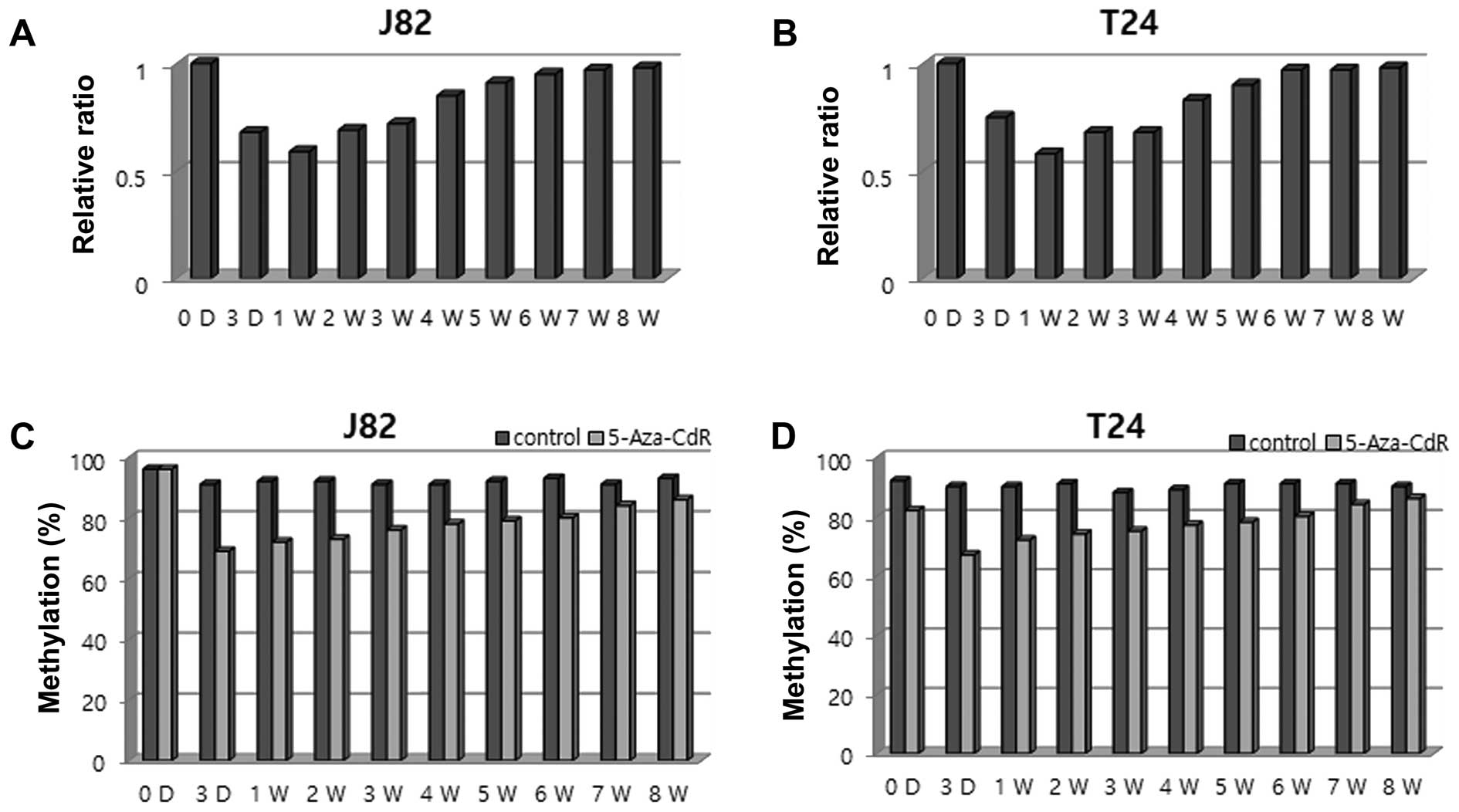
Reversibility of RSPH9 methylation with
AZA treatment
To investigate the potential reversibility of
RSPH9 methylation, cells from two bladder cancer lines (T24
and J82) were treated with 0.3 μM 5-Aza-CdR for 24 h, and
sequential changes in cell number and RSPH9 methylation
level were evaluated in each cell line. Compared to the untreated
cells, a decrease in cell count was detected on day 1 after
5-Aza-CdR treatment, reaching a maximal level on day 3 in the J82
and T24 cells, followed by a gradual increase in cell number
starting 1 week after 5-Aza-CdR treatment, reaching constant values
after 6 weeks in both cell lines. Consistent with the cell-count
changes, the methylation level of RSPH9 decreased on day 3
and progressively increased 1 week after treatment in the J82 and
T24 cell lines (Fig. 3).
Discussion
Similar to other human cancers, bladder cancer is a
molecular disease driven by multiple genetic, epigenetic, and
environmental factors (6,7). DNA methylation, which is the most
common and best characterized epigenetic change in bladder cancer,
inactivates tumor-suppressor genes and may be used as potential
biomarkers (6,7). In the present study, we used
microarray-based profiling to discover novel epigenetic markers
relevant to NMIBC. Among the candidate meth-ylation markers
identified, the RSPH9 methylation pattern showed close
associations with aggressive NMIBC characteristics, including
advanced stage and high tumor grade. The methylation status of
RSPH9 was identified as an independent predictive indicator
of prognosis.
The evolution of classic single-gene DNA methylation
detection assays to genome-wide microarray-based analyses enabled a
better understanding of the role of DNA methylation in cancer
(6,14). However, the identification of bona
fide candidate methylation markers of clinical relevance requires
appropriate selection criteria, validation with external data sets,
confirmation of the methylation status using highly targeted
locus-specific assays of human tissues, and comparison with
clinicopathological parameters or disease outcomes (4,18).
Considering these criteria, the results of the present study are
promising. Candidate methylation markers were selected by
genome-wide microarray profiling, and their relevance was validated
with two microarray data sets obtained from Western populations
with NMIBC (9,13). Furthermore, the association between
the methylation status of RSPH9 and prognostic outcomes was
verified in long-term follow-up NMIBC patients. The results
suggested that the novel methylation marker identified is specific
to NMIBC and appropriate for predicting prognosis.
Little information is available concerning the
function of the RSPH9 gene (19–21).
It is located on chromosome 6p21.1 and encodes radial spoke head
protein 9. Previous studies suggested that RSPH9 is mutated
in primary ciliary dyski-nesia patients with microtubule defects
(19–21). To the best of our knowledge, the
present study is the first to identify RSPH9 as a
cancer-related methylation marker. Despite the prognostic
significance of RSPH9 in NMIBC identified in the present
study, these findings do not indicate that it plays a crucial role
in bladder tumor initiation or progression. The lack of a clear
association between these candidate markers and bladder cancer is a
limitation of the present study, and this issue will be addressed
in future studies. However, the objective of the present study was
the identification of disease markers; we focused on the
association between methylation changes of specific methylation
markers and disease phenotype rather than analyzing the effect of
methylation status on gene transcription and function (4).
DNA methylation is a reversible modification, which
makes it a potential therapeutic target. Drugs that target
epigenetic alterations, such as DNA methylation inhibitors, restore
the activity of genes by targeting aberrant hetero-chromatic
regions, ultimately leading to the reactivation of tumor-suppressor
genes and/or other genes that are crucial for normal cellular
function (5). In the present study,
we showed that the methylation level of RSPH9 in human
bladder cancer cell lines decreased in response to 5-Aza-CdR
treatment and increased progressively in its absence. The
reversibility of RSPH9 methylation shown in the present
study confirms its value as a candidate therapeutic target.
Large-scale validation studies using human samples, as well as
functional analyses and a gene ontologic approach to the study of
RSPH9, may provide additional knowledge about its biological
mechanism and clinical relevance.
From a clinical point of view, the most promising
applications for epigenetic markers are early detection, prediction
of response to treatment, and indication of disease prognosis. The
results presented herein are promising because the candidate
methylation markers were selected from a genome-wide analysis and
validated in a relatively large number of human tissue samples
obtained from long-term follow-up patients. In addition, the
selected methylation markers are independent predictors of disease
outcome. An accurate prediction of prognosis made using these
candidate methylation markers would aid clinicians in terms of
patient counseling, determining the frequency and extent of
monitoring, and whether more aggressive therapy is needed. However,
despite these promising results, further validation studies are
necessary to reduce false prediction rates and achieve reliable
clinical relevance. It may also lead to new therapies that target
specific molecular defects, thereby significantly lowering the
morbidity associated with NMIBC.
In conclusion, our findings suggest that the novel
methylation marker RSPH9 is an independent indicator of
prognosis in NMIBC patients. This prognostic marker may constitute
a promising tool for assessing the recurrence and progression of
NMIBC and may facilitate the design of individualized therapeutic
modalities.
Abbreviations:
|
BARHL2
|
BarH-like homeobox 2
|
|
HR
|
hazard ratio
|
|
NC
|
normal controls
|
|
NMIBC
|
non-muscle invasive bladder cancer
|
|
PSQ
|
pyrosequencing
|
|
RAB37
|
member RAS oncogene family
|
|
RSPH9
|
radial spoke head 9 homolog
|
|
TUR
|
transurethral resection
|
Acknowledgments
The present study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
(NRF) funded by the Ministry of Education (2011-0023308) and
Ministry of Science, ICT and Future Planning (NRF-2014R1A2A1
A09006983). All samples derived from the National Biobank of Korea
were obtained with informed consent under institutional review
board-approved protocols.
References
|
1
|
Babjuk M, Burger M, Zigeuner R, Shariat
SF, van Rhijn BW, Compérat E, Sylvester RJ, Kaasinen E, Böhle A,
Palou Redorta j, et al European Association of Urology: EAU
guidelines on non-muscle-invasive urothelial carcinoma of the
bladder: Update 2013. Eur Urol. 64:639–653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kamat AM, Hegarty PK, Gee JR, Clark PE,
Svatek RS, Hegarty N, Shariat SF, Xylinas E, Schmitz-Dräger BJ,
Lotan Y, et al: International Consultation on Urologic Disease -
European Association of Urology Consultation on Bladder Cancer
2012: ICUD-EAU International Consultation on Bladder Cancer 2012:
Screening, diagnosis, and molecular markers. Eur Urol. 63:4–15.
2013. View Article : Google Scholar
|
|
3
|
Cheng L, Davison DD, Adams J,
Lopez-Beltran A, Wang L, Montironi R and Zhang S: Biomarkers in
bladder cancer: Translational and clinical implications. Crit Rev
Oncol Hematol. 89:73–111. 2014. View Article : Google Scholar
|
|
4
|
Ushijima T: Detection and interpretation
of altered methylation patterns in cancer cells. Nat Rev Cancer.
5:223–231. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome - biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Besaratinia A, Cockburn M and Tommasi S:
Alterations of DNA methylome in human bladder cancer. Epigenetics.
8:1013–1022. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim WJ and Kim YJ: Epigenetic biomarkers
in urothelial bladder cancer. Expert Rev Mol Diagn. 9:259–269.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim EJ, Kim YJ, Jeong P, Ha YS, Bae SC and
Kim WJ: Methy-lation of the RUNX3 promoter as a potential
prognostic marker for bladder tumor. J Urol. 180:1141–1145. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reinert T, Modin C, Castano FM, Lamy P,
Wojdacz TK, Hansen LL, Wiuf C, Borre M, Dyrskjøt L and Orntoft TF:
Comprehensive genome methylation analysis in bladder cancer:
Identification and validation of novel methylated genes and
application of these as urinary tumor markers. Clin Cancer Res.
17:5582–5592. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sánchez-Carbayo M: Hypermethylation in
bladder cancer: Biological pathways and translational applications.
Tumour Biol. 33:347–361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kandimalla R, van Tilborg AA and Zwarthoff
EC: DNA meth-ylation-based biomarkers in bladder cancer. Nat Rev
Urol. 10:327–335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vinci S, Giannarini G, Selli C, Kuncova J,
Villari D, Valent F and Orlando C: Quantitative methylation
analysis of BCL2, hTERT, and DAPK promoters in urine sediment for
the detection of non-muscle-invasive urothelial carcinoma of the
bladder: A prospective, two-center validation study. Urol Oncol.
29:150–156. 2011. View Article : Google Scholar
|
|
13
|
Ibragimova I, Dulaimi E, Slifker MJ, Chen
DY, Uzzo RG and Cairns P: A global profile of gene promoter
methylation in treatment-naïve urothelial cancer. Epigenetics.
9:760–773. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kalari S and Pfeifer GP: Identification of
driver and passenger DNA methylation in cancer by epigenomic
analysis. Adv Genet. 70:277–308. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim YJ, Yoon HY, Kim JS, Kang HW, Min BD,
Kim SK, Ha YS, Kim IY, Ryu KH, Lee SC, et al: HOXA9, ISL1 and
ALDH1A3 methylation patterns as prognostic markers for nonmuscle
invasive bladder cancer: Array-based DNA methylation and expression
profiling. Int J Cancer. 133:1135–1142. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee JS, Leem SH, Lee SY, Kim SC, Park ES,
Kim SB, Kim SK, Kim YJ, Kim WJ and Chu IS: Expression signature of
E2F1 and its associated genes predict superficial to invasive
progression of bladder tumors. J Clin Oncol. 28:2660–2667. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS,
Jeong P, Kim MJ, Yun SJ, Lee KM, Moon SK, et al: Predictive value
of progression-related gene classifier in primary non-muscle
invasive bladder cancer. Mol Cancer. 9:32010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Olkhov-Mitsel E and Bapat B: Strategies
for discovery and validation of methylated and hydroxymethylated
DNA biomarkers. Cancer Med. 1:237–260. 2012. View Article : Google Scholar
|
|
19
|
Castleman VH, Romio L, Chodhari R, Hirst
RA, de Castro SC, Parker KA, Ybot-Gonzalez P, Emes RD, Wilson SW,
Wallis C, et al: Mutations in radial spoke head protein genes RSPH9
and RSPH4A cause primary ciliary dyskinesia with
central-micro-tubular-pair abnormalities. Am J Hum Genet.
84:197–209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Onoufriadis A, Shoemark A, Schmidts M,
Patel M, Jimenez G, Liu H, Thomas B, Dixon M, Hirst RA, Rutman A,
et al: Targeted NGS gene panel identifies mutations in RSPH1
causing primary ciliary dyskinesia and a common mechanism for
ciliary central pair agenesis due to radial spoke defects. Hum Mol
Genet. 23:3362–3374. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ziętkiewicz E, Bukowy-Bieryłło Z, Voelkel
K, Klimek B, Dmeńska H, Pogorzelski A, Sulikowska-Rowińska A,
Rutkiewicz E and Witt M: Mutations in radial spoke head genes and
ultrastructural cilia defects in East-European cohort of primary
ciliary dyskinesia patients. PLoS One. 7:e336672012. View Article : Google Scholar
|