Introduction
Chemotherapy is widely used for cancer treatment.
Unfortunately, in a high proportion of cases, cancer cells become
insensitive to the cyctotoxic drugs and are able to proliferate and
metastasize, normally with fatal consequences. How cancer cells
acquire drug resistance remains to be fully understood, although
the effector molecules and pathways involved in the process are
relatively well characterized. Cancer cells can increase the
activity of DNA repair enzymes, metabolize drugs or activate
anti-apoptotic mechanisms (1),
although the most common and best studied mechanism of drug
resistance is the upregulation of membrane transporters (2). ABC transporters are membrane-anchored
proteins that use the energy from ATP hydrolysis to mediate the
export of cytoplasmic or membrane solutes outside of the cell
(3) and have a broad substrate
specificity. This leads to MDR, by which cells become insensitive
to structurally and mechanistically unrelated cytotoxic drugs
(4). In the clinic, resistance due
to P-glycoprotein (ABCB1 or MDR1) has been demonstrated in
leukemias (5) and breast cancer
(6,7).
MicroRNAs (miRs) are a class of 18- to 24-nucleotide
single-stranded non-coding RNAs as negative regulators of gene
expression by triggering translation repression through partial
complementation to 3′-untranslated region (UTR) of target mRNAs
(8). miRs play crucial roles in
multiple biological processes, including cancer (9,10) and
drug resistance. miR-214 confers cell survival and cisplatin
resistance in ovarian cancer cells (11); miR-125b confers the resistance of
breast cancer cells to paclitaxel (12) and miR-221/222 confers tamoxifen
resistance in breast cancer (13).
The miR-106b~25 cluster, consisting of miR-106b,
miR-93 and miR-25, is highly conserved in vertebrates, and is
located in intron 13 of the minichromosome maintenance complex
component 7 (MCM7) oncogene. It may play an important
proto-oncogenic role in cellular transformation and tumorigenesis
by downregulation of several tumor suppressors such as p21, E2F1,
Bim and PTEN (14–16). We have recently reported that the
miR-106b~25 cluster negatively regulates the histone
acetyltransferase EP300, a transcriptional activator of E-cadherin.
This leads to activation of an epithelial-to-mesenchymal transition
(EMT), increase in the ability of cell migration and invasion, and
resistance to doxorubicin and γ-radiation (17).
Here we report that breast cancer cells
overexpressing miR-106b~25 cluster, or in which EP300 or E-cadherin
have been downregulated by RNA interference, have a
P-glycoprotein-independent (transporter independent) MDR phenotype
that involves apoptosis evasion.
Materials and methods
Cells
Minimally transformed mammary epithelial cells
(MTMECs) overexpressing the miR-106b~25 cluster by lentiviral
transfection (MTMEC-miR-106b~25) or expressing a short hairpin
targeting EP300 or E-cadherin (MTMEC-shEP300 and MTMEC-shCDH1,
respectively) mRNAs have been described (17). MTMECs, that are human mammary
epithelial primary cells that have been transformed experimentally
and express TERT, SV40 large T antigen, a constitutively active
form of PI3K, p110α, and oncogenic ras (18), were routinely cultivated on
serum-free HuMEC medium (Life Technologies). The multidrug
resistant cell line NCI/ADR-Res (19) and P-glycoprotein-negative CAL51
cells were maintained in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 1 g/l glucose, 10% foetal calf serum and 4 mM
L-glutamine (Invitrogen) in the presence or absence of 1 μM
doxorubicin, respectively.
Drug resistance clonogenic assay
MTMEC-derived cells were seeded, at least in
duplicate, at a density of 3×105 cells in
25-cm2 culture flasks and exposed to a single dose of
drug for 3 days. Cells were kept in culture for 21 days with
drug-free medium changes every three days. Drug resistant clones
were fixed with 4% paraformaldehyde and stained with 0.2% crystal
violet and counted.
Drug efflux assay
Functional drug efflux assays were performed using
0.1 μM BODIPY-paclitaxel (Invitrogen) in the presence or
absence of 1 μM cyclosporin A (Sigma-Aldrich) essentially as
described (20,21) by flow cytometry in a Becton
Dickinson FACSDiva. Briefly, cells were detached from the culture
dishes with 10 mM EDTA and washed with phenol red-free DMEM
containing 0.1% bovine serum albumin (DMEM-BSA) and
2×106 cells were stained with either 0.1 μM
BODIPY-paclitaxel, 0.1 μM BODIPY-paclitaxel containing 1
μM cyclosporin A to inhibit ABC transporter efflux, or left
untreated to determine auto-fluorescence. After 30 min at 37°C,
cells were washed with DMEM-BSA and dead cells were stained with
TOTO3-iodide (Life Technologies) and gated out.
P-glycoprotein expression assay
Cell surface P-glycoprotein (ABCB1) was determined
by flow cytometry in a Becton Dickinson FACSDiva using the
phycoerythrin-conjugated UIC2 antibody (Immunotech, Marseille,
France) or the corresponding isotype control (Sigma-Aldrich)
essentially as described (22).
Briefly, cells were detached from the culture dishes with 10 mM
EDTA and washed with DMEM-BSA. Antibodies (250 ng) and cells
(5–105) were incubated for 30 min at 37°C in the
presence of 1 μM cyclosporin. Cells were then washed with
DMEM-BSA and dead cells were stained with TOTO3-iodide and gated
out.
Gene expression analysis
For mRNA detection, total RNA (isolated using a
miRCURY RNA isolation kit; Exiqon) was reverse transcribed with
RNase H+ MMLV reverse transcriptase (iScript cDNA
Synthesis kit) and real-time quantitative PCR was performed using
SYBR-Green (Bioline) and ABCB1 specific primers (19) on an ABI Prism 7700 detection system
(PerkinElmer Life Sciences). A comparative threshold cycle was used
to determine the relative gene expression using two normalizers,
RPS6 and RPS14 as previously described (22,23).
Apoptosis
Apoptotic assessment was by detection of active
caspase-9 and -3/-7 using Caspase-Glo assays (Promega) following
the manufacturer's protocol. Caspase activity was normalized to
cell density determined by sulphorhodamine B (Sigma-Aldrich)
staining (24). For cell cycle
analysis cells were stained with propidium iodide after fixation
with ice-cold 70% methanol and the DNA content estimated determined
by flow cytometry essentially as described (23). Annexin V staining was determined by
flow cytometry using an Annexin V-FITC apoptosis detection kit
(BioVision) as described (25).
IC50
The drug concentration necessary to kill 50% of
cells (IC50) was obtained after sulphorhodamine B
(Sigma-Aldrich) staining (24) as
previously described (26).
Statistical analysis
Statistical evaluations were performed by Student's
t-test for paired data, and data were considered significant at a
p-value <0.05.
Results
Overexpression of the miR-106b~25 cluster
leads to a MDR phenotype
We have recently reported that experimental
upregulation of the miR-106b~25 cluster in MTMEC cells leads to the
acquisition of doxorubicin and γ-radiation resistance (17). We asked whether upregulation of this
miR cluster would lead to resistance to other structurally
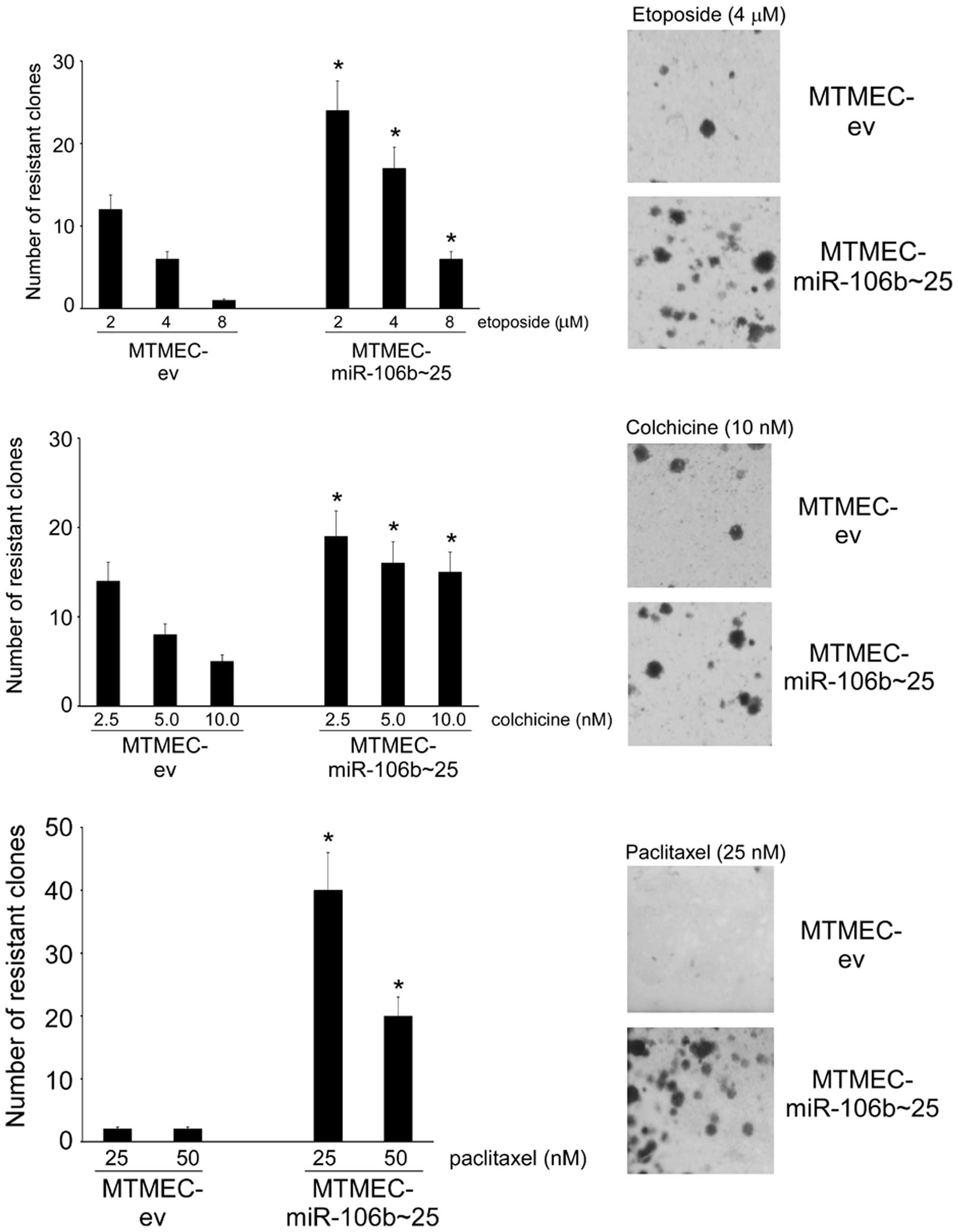
unrelated drugs. First, we tested etoposide, that, as doxorubicin,
is a toposiomerase II inhibitor (27). Indeed, MTMEC cells overexpressing
the miR-106b~25 cluster generated a higher number of
etoposide-resistant clones than control cells transfected only with
the empty vector (Fig. 1). Next we
asked whether overexpression of the miR-106b~25 cluster would also
influence the generation of resistance to other drugs with
different modes of action. For this we selected two microtubule
interfering agents, colchicine and paclitaxel. Long-term clonogenic
assays indicated that indeed MTMEC cells overexpressing miR-106b~25
cluster generated more colchicine- and paclitaxel-resistant clones
than control cells transfected only with empty vector (Fig. 1).
Doxorubicin, at low to moderate concentrations, and
γ-irradiation trigger a senescent phenotype, very similar to the
well-characterized replicative senescence, often termed drug (or
therapy)-induced senescence (28).
Experimental upregulation of miR-106b~25 cluster in MTMEC cells
allows cells to bypass senescence and to proliferate after
doxorubicin or γ-irradiation treatment, becoming resistant
(17). However, other drugs, such
as the taxanes paclitaxel and docetaxel, exert their cytotoxic
effect by kinetic suppression of micro-tubules that block cells in
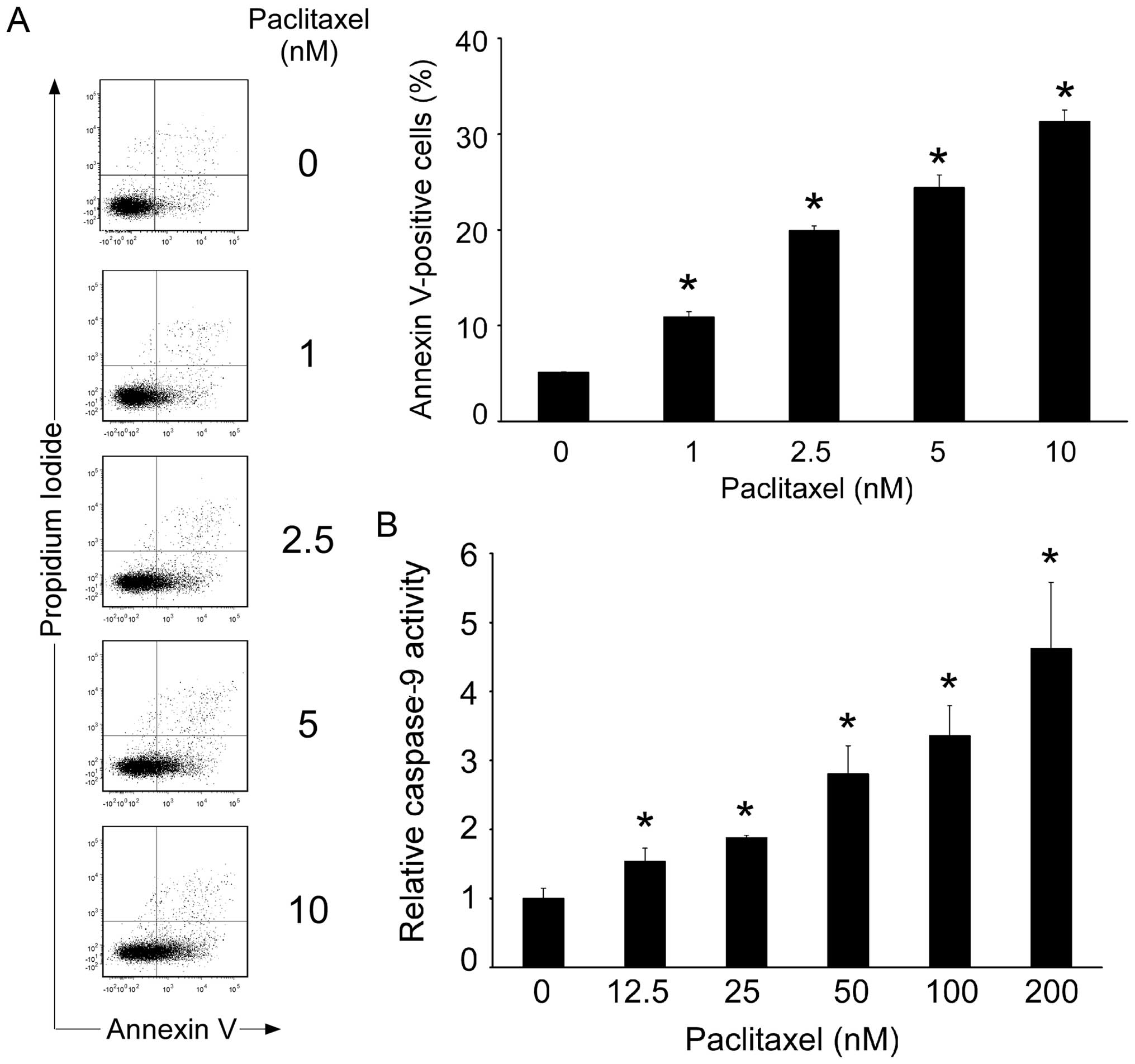
the G2/M phase of the cell cycle and trigger apoptosis (29). When MTMEC cells were treated with
increasing concentrations of paclitaxel there was a dose-dependent
increase in Annexin V staining and activation of caspase-9
(Fig. 2). This confirms that
paclitaxel, as expected, acts on breast cancer cells triggering an
apoptotic programme.
Thus, upregulation of miR-106b~25 cluster confers
cells the ability to generate resistance to a variety of
structurally and mechanistically different drugs, a hallmark of MDR
(4).
Downregulation of EP300 leads to the MDR
phenotype
The three miRs in the miR-106b~25 cluster bind
EP300 3′-UTR mRNA leading to its downregulation, decreasing
E-cadherin levels and activating an EMT accompanied by resistance
to doxorubicin and γ-irradiation in MTMECs (17). As resistance to doxorubicin and
γ-irradiation can be mimicked by experimental downregulation of
EP300, we asked whether the same would apply to paclitaxel.
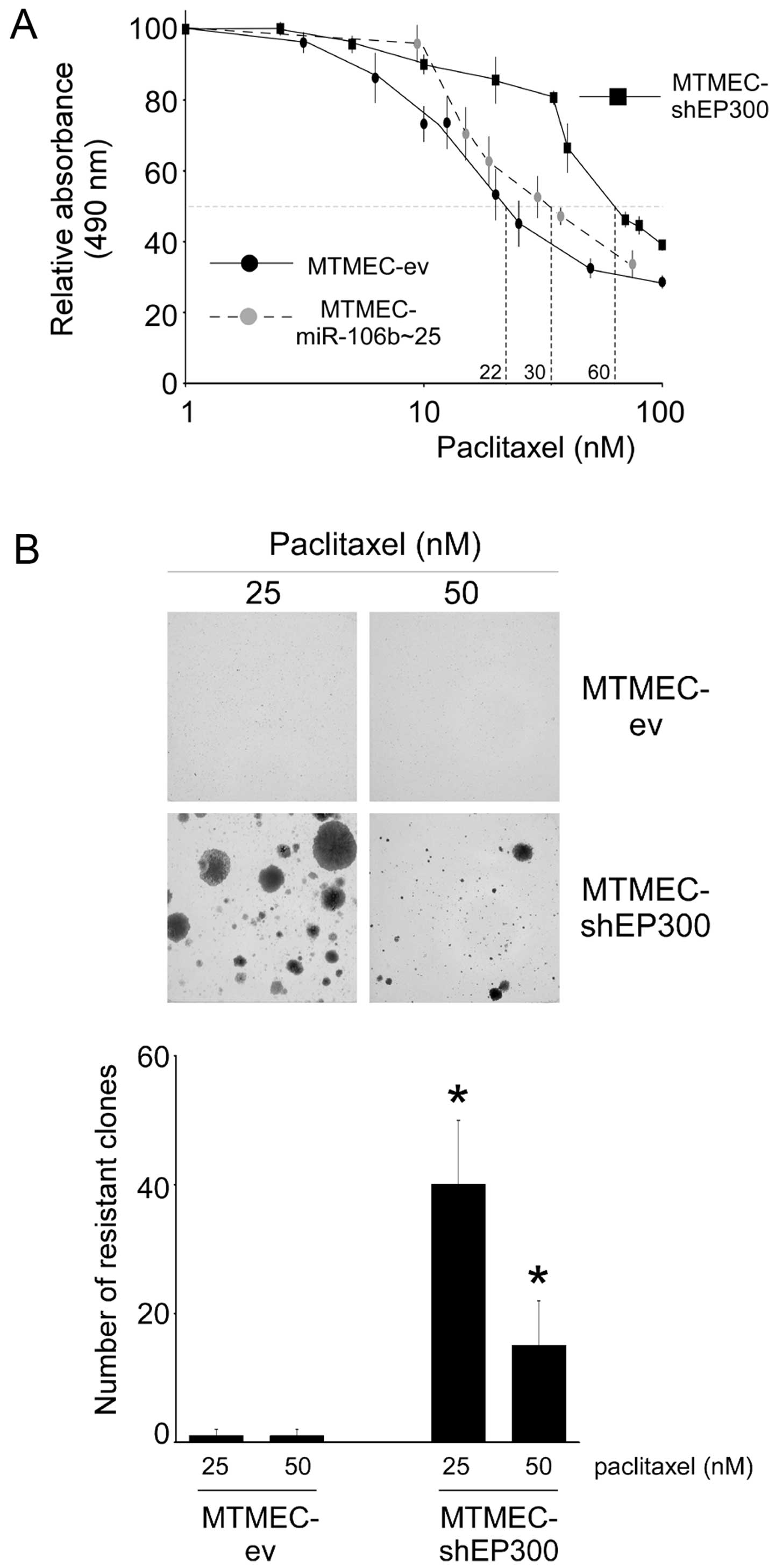
Short-term drug sensitivity assays indicated that MTMECs
overexpressing miR-106b~25 were slightly more resistant to
paclitaxel than control cells (IC50 values of 30 and 22
nM, respectively). In MTMECs with EP300 downregulated by RNA
interference the paclitaxel IC50 had increased to 60 nM
(~3-fold; Fig. 3A). Long-term
generation of paclitaxel resistance was also affected. When
MTMEC-shEP300 cells were treated with paclitaxel, a large number of
drug-resistant proliferating clones were generated whereas this was
not the case with control cells (Fig.
3B and C). Thus, downregulation of EP300 leads to a decrease in
paclitaxel sensitivity and generation of paclitaxel-resistant
cells.
The above, and our previously published data,
indicate that downregulation of EP300 leads to the MDR
phenotype.
ABC transporters are not responsible for
the MDR phenotype of cells either overexpressing the miR-106b~25
cluster or with EP300 downregulated
Upregulation of ABCB1 (P-glycoprotein) is the main
mechanism by which cells become multidrug resistant. To test
whether the multidrug resistant phenotype of MTMEC-miR-106b~25 and
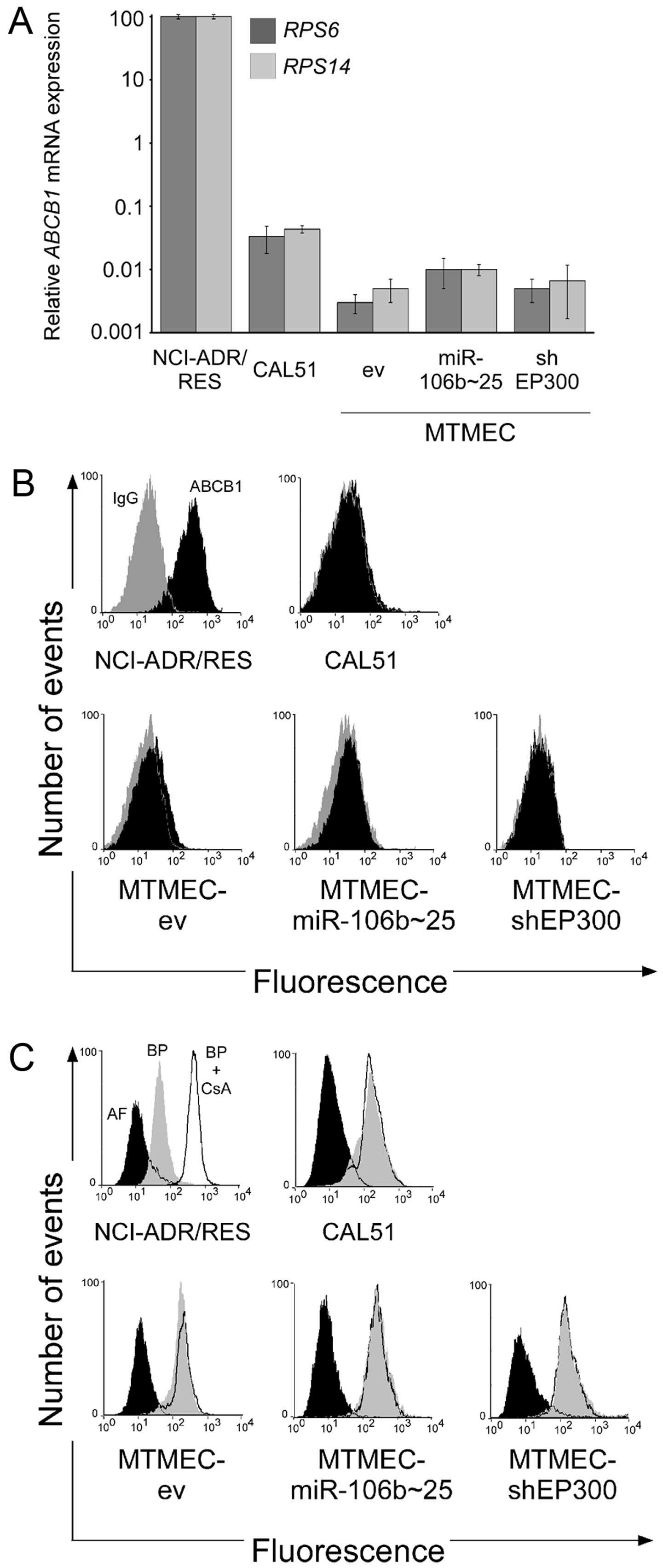
MTMEC-shEP300 cells was due to ABCB1, we tested first ABCB1
mRNA levels by RT-QPCR including NCI-ADR/RES and CAL51 as positive
and negative controls, respectively. ABCB1 mRNA levels in
CAL51 were between 1,000 and 10,000 lower than those found in
ABCB1-positive NCI-ADR/RES cells (Fig.
4A). Importantly, ABCB1 mRNA expression in all MTMECs
was one order of magnitude lower than in CAL51 cells (Fig. 4A). As we have previously
demonstrated that ABCB1 mRNA and protein levels do not
correlate (22), we determined
functional ABCB1 by flow cytometry using UIC2 antibody (23) and NCI-ADR/RES and CAL51 cell lines
as positive and negative controls, respectively. None of the MTMECs
showed an increase in fluorescence after ABCB1-specific antibody
binding with respect to the isotype IgG control (Fig. 4B). These results indicate that MDR
in MTMECs is not due to upregulation of ABCB1.
As several other transporters can be responsible for
multi-drug resistance (30), we
asked whether MTMECs were able to efflux paclitaxel out of the
cell. For this, we took advantage of BODIPY-paclitaxel, a
drug-derivative with a fluorescent dye used to determine efflux
pump activity (23). Importantly,
all MTMECs, as well as the negative cell line CAL51, were unable to
efflux BODIPY-paclitaxel (Fig. 4C).
In contrast, NCI-ADR/RES cells showed, as expected, efflux activity
that was inhibited by cyclosporin A (Fig. 4C).
In summary, the multidrug resistance phenotype of
MTMECs, either overexpressing miR-106b~25 or with EP300
downregulated, is not due to transporter activity.
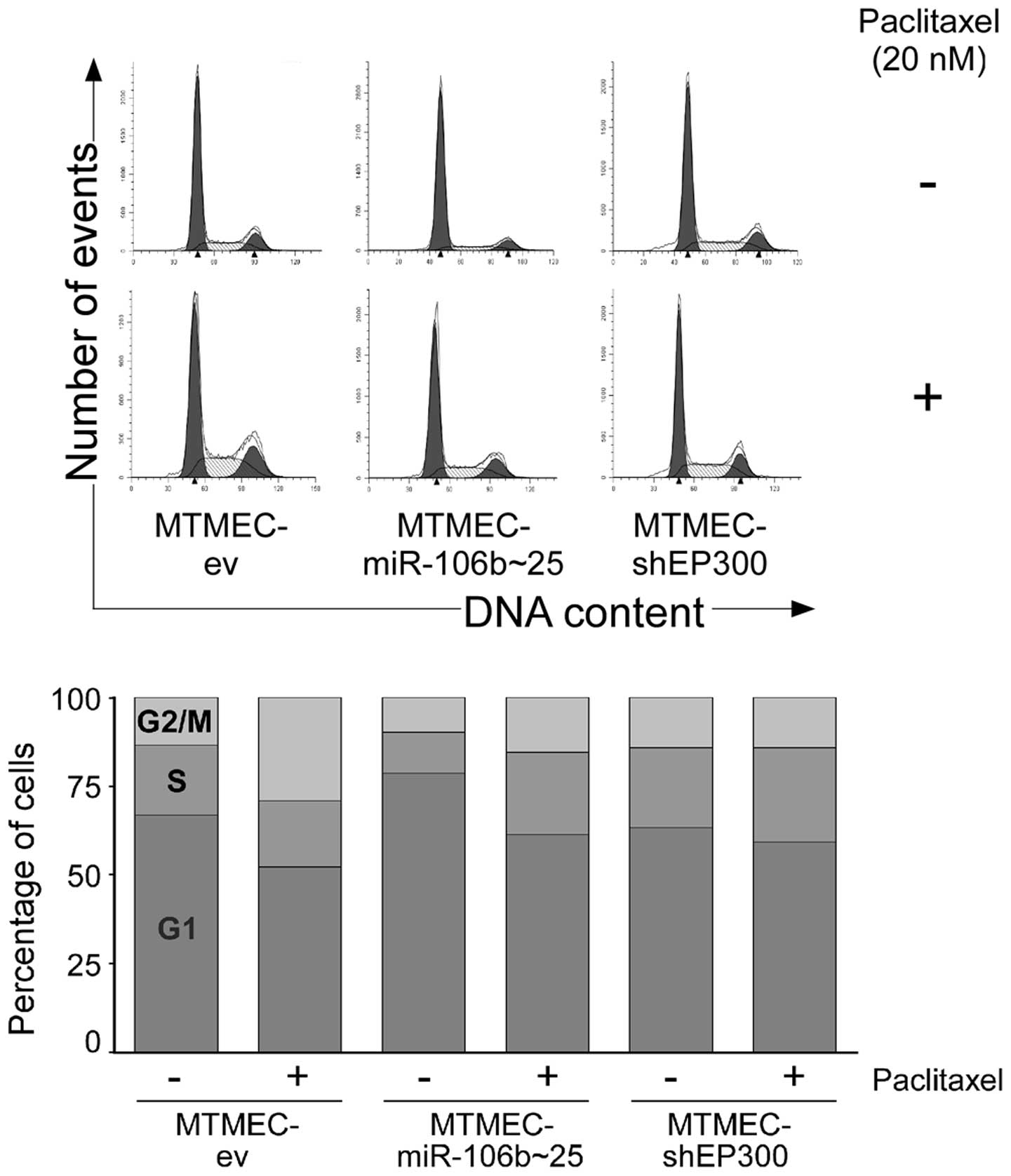
Paclitaxel alterations in the cell cycle
are abolished in MTMEC-miR-106b~25 and MTMEC-shEP300 cells
As taxanes block cells in the G2/M phase of the cell
cycle (29), we asked whether MDR
MTMECs have altered cell cycle profiles. After treating cells with
20 nM paclitaxel for 48 h MTMEC-ev control cells showed, as
expected, a decrease in the percentage of cells at the G1 phase of
the cell cycle and an increase in G2/M (from 67 to 52% in G1 and
from 13 to 29% in G2/M). The percentage of cells in S was
practically the same (Fig. 5).
However, the percentage of cells in G2/M after paclitaxel treatment
increased only by 5% in MTMEC-miR-106b~25 cells (from 10 to 15%)
and was practically the same in MTMEC-shEP300 cells (from 14 to
15%). Thus, the cell cycle effects of paclitaxel are abolished in
MDR MTMECs.
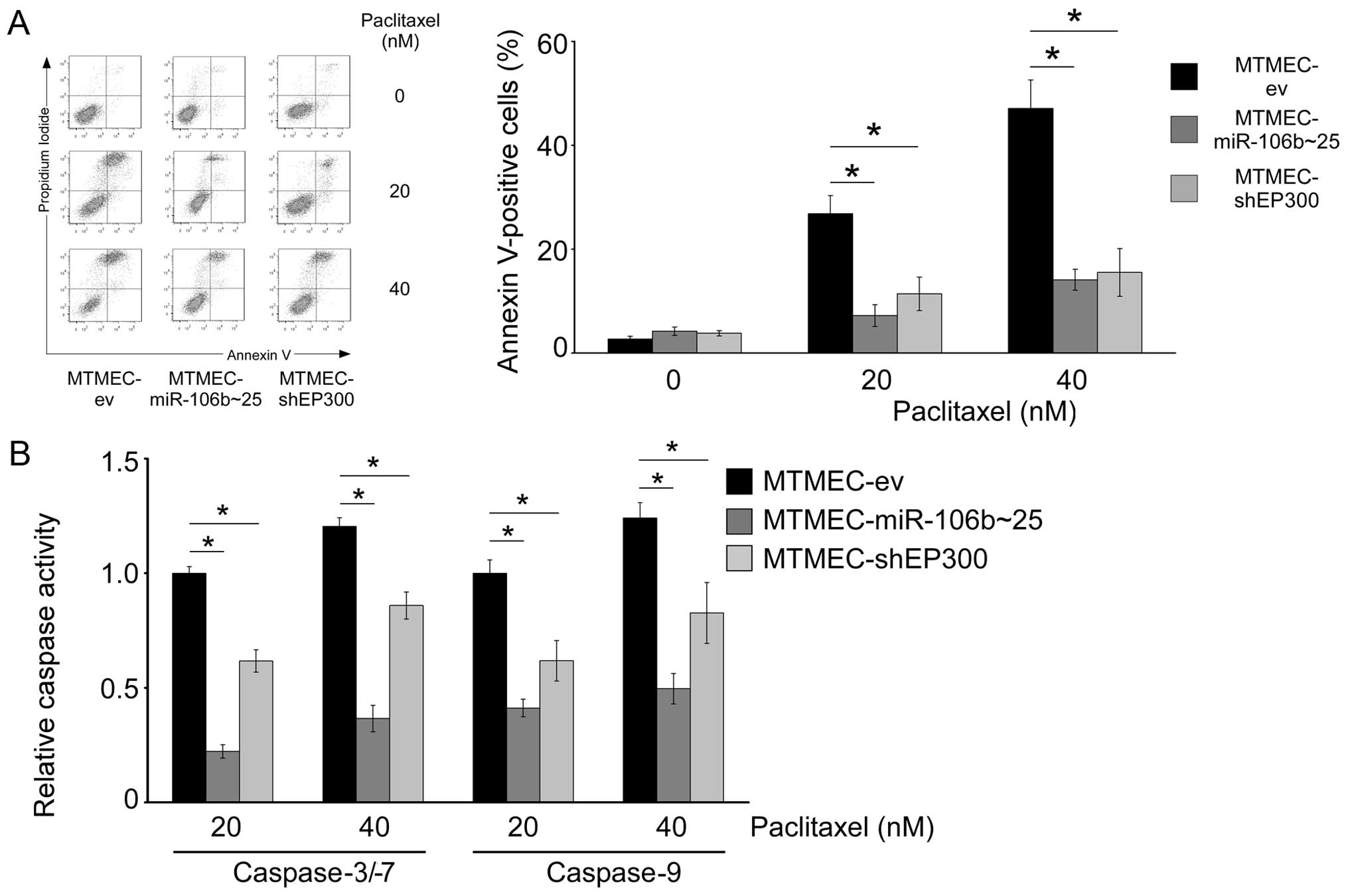
MDR in cells overexpressing miR-106b~25
cluster, or with downregulation of EP300, is due to apoptosis
evasion
Apoptosis evasion is another mechanism by which
cancer cells may become resistant to a variety of drugs such as
doxorubicin, etoposide or paclitaxel (31). In order to test whether apoptosis
was involved in the MDR phenotype of MTMEC-miR-106b~25 and
MTMEC-shEP300 cells, we determined the percentage of dead cells
after paclitaxel treatment (20 and 40 nM for 48 h) by flow
cytometry after staining with Annexin V and propidium iodide. As
expected, there was a dose-dependent effect on cell death in
MTMEC-ev control cells (25% at 20 nM and 45% at 40 nM). However,
MTMEC-miR-106b~25 and MTMEC-shEP300 cells showed only 10 and 15%
cell death after treatment with 20 and 40 nM paclitaxel,
respectively (Fig. 6A).
As paclitaxel triggers activation of caspase-9 in
MTMECs (Fig. 2B), we asked whether
MDR MTMEC-miR-106b~25 and MTMEC-shEP300 cells responded to
paclitaxel treatment with lower activation of both initiator and
executioner caspases. Caspase-9 activation after treatment with
paclitaxel (20 and 40 nM for 72 h) was lower in MTMEC-miR-106b~25
and MTMEC-shEP300 cells than in MTMEC-ev control cells (Fig. 6B). Interestingly, activation in
MTMEC-miR-106b~25 cells was lower than in MTMEC-shEP300 cells.
Similar results were obtained when the activity of executioner
caspases-3/-7 was determined, including the lowest activation in
MTMEC-miR-106b~25 cells.
Thus, overexpression of miR-106b~25, or
downregulation of EP300 (a direct target of the three miRs in the
cluster), leads to a transporter-independent MDR phenotype
involving apoptosis evasion.
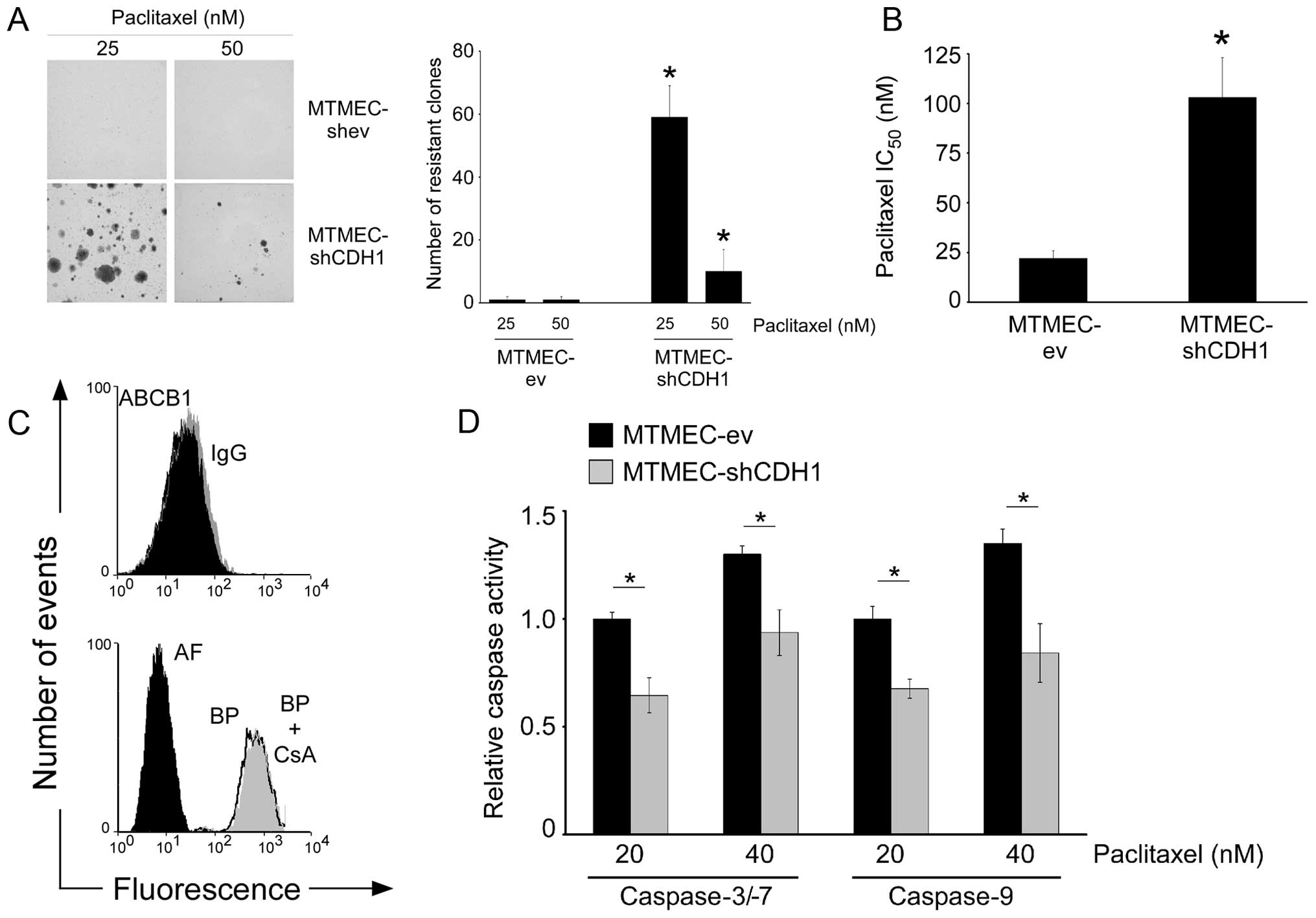
Downregulation of E-cadherin leads to the
MDR phenotype which is transporter-independent
We have previously determined that doxorubicin
resistance in MTMECs due to overexpression of miR-106b~25 cluster,
or to downregulation of EP300, can be mimicked by experimentally
downregulating E-cadherin expression (17). As the three miRs in the miR-106b~25
cluster target EP300 downregulating its expression and EP300 is a
transcriptional activator of E-cadherin, we asked whether the
experimental downregulation of E-cadherin would also lead to
paclitaxel resistance. Indeed, MTMEC-shCDH1 cells led to a slightly
higher number of paclitaxel resistant clones (Fig. 7A) than MTMEC-miR-106b~25 (Fig. 1) and MTMEC-shEP300 cells (Fig. 3B). Short-term paclitaxel sensitivity
was also lower in E-cadherin knockdown cells (IC50 ~100
nM; Fig. 7B) than in those with the
miR-106b~25 upregulated (IC50 ~30 nM) or EP300
downregulated (IC50 ~60 nM; Fig. 3A). This indicates that MTMEC-shCDH1
cells have an MDR phenotype despite not having overexpression of
ABCB1 and not transporting paclitaxel out of the cells (Fig. 7C). However, activation of caspase-9
and caspase-3/-7 in response to paclitaxel was lower in cells with
E-cadherin knocked down (Fig. 7D).
Thus, transporter-independent MDR can be acquired by downregulation
of E-cadherin via apoptosis evasion.
Discussion
Here we report that the axis EP300→E-cadherin, which
is controlled by the miR-106b~25 cluster, regulates paclitaxel
resistance in breast cancer cells by apoptosis evasion. This
pathway also determines the resistance to DNA damaging agents, such
as doxorubicin or γ-radiation, bypassing therapy-induced
senescence, hallmarks of MDR. This phenotype was independent of
membrane pumps, but involved apoptosis evasion. Thus,
transporter-independent MDR can by generated by modulation of the
miR-106b~25 cluster→EP300→E-cadherin pathway.
Resistance to chemotherapeutics used in cancer
therapy remains one of the main hurdles to overcome for the
successful treatment of this disease. Although in many cases the
initial response to chemotherapy is positive, in a high proportion
of cases, and after a disease-free period, resistant cells give
rise to secondary tumors normally at distant sites following
metastasis, normally with fatal consequences (32). The use of combination therapies aims
to overcome resistance to single agents. However, drug resistant
cancer cells develop many times acquiring decreased sensitivity to
a broad spectrum of structurally and functionally different drugs,
a phenomenon early recognized and termed MDR (33). During tumor progression, epithelial
cells lose polarity and acquire characteristics of mesenchymal
cells including the capacity to invade surrounding tissues. This
EMT is normally accompanied by an increase in the stem cell
population, the so-termed cancer stem cells, and acquisition of
drug resistance (34). We have
recently demonstrated that a cluster of three miRs (miR-106b,
miR-93 and miR-25) negatively regulates the expression of EP300, a
histone acetyltransferase that transcriptionally activates
E-cadherin, leading to an increase in motility and invasion and
doxorubicin and γ-irradiation resistance in breast cancer cells
(17). Loss of E-cadherin
constitutes one of the hallmarks of the EMT process and negative
regulators of E-cadherin are well studied (35,36).
Loss of functional E-cadherin renders cells more resistant to
paclitaxel (37) and downregulation
of EP300 (both experimental and in drug resistant lines) is
associated with doxorubicin and cisplatin resistance in bladder
cancer cells (38,39) and metastatic properties in
pancreatic cancer (40). Here we
demonstrate that minimally transformed mammary epithelial cells in
which the miR-106b~25 cluster is upregulated, or in which either
EP300 or E-cadherin have been downregulated by RNA interference
(17) are able to generate
paclitaxel resistance and are thus MDR.
There are several effectors of MDR, although the
most common is upregulation of drug transporters such as ABCB1
(41). MDR has also been associated
with E-cadherin loss (42,43). However, none of the MDR cells in
this study upregulate ABCB1 mRNA or functional ABCB1 at the
cell surface. Other ABC transporters, such as ABCG2, which is
frequently found upregulated in cancer stem cells (44), are not responsible for the MDR
phenotype as are not able to efflux a fluorescent derivative of
paclitaxel. Thus, MDR controlled by the miR-106b~25 cluster via
downregulation of EP300 and E-cadherin is
transporter-independent.
Paclitaxel, as well as docetaxel, the other taxane
currently used in the clinic, is a microtubule-stabilizing agent
that interferes with spindle microtubule dynamics causing cell
cycle arrest and apoptosis (29).
Although some studies indicate a mode of action via the extrinsic
apoptotic pathway (45,46), the mitochondrial pathway is clearly
involved as caspase-9 activation has been unequivocally
demonstrated (47). In addition,
paclitaxel has also been shown to activate caspase-2 (48). MTMECs have p53 inactivated due to
expression of SV40 large T (18).
Thus, the caspase activation observed following paclitaxel
treatment must be p53-independent. Although p53 is normally
associated with the cytochrome c release from the
mitochondria and activation of caspase-9, this can also occur in a
p53-independent manner (49). Our
data also indicates activation of caspase-9, although we cannot
rule out the involvement of a non-mitochondrial pathway. However,
although the initiator signals triggering apoptosis may not have
been fully elucidated yet, these ultimately converge into
executioner caspases, such as caspase-3/-7. Indeed, overexpression
of caspase-3 restores sensitivity for drug-induced apoptosis in
breast cancer cells with acquired resistance to epirubicin,
etoposide and paclitaxel (31).
Importantly, we show here that upregulation of miR-106b~25 cluster,
or downregulation of either EP300 or E-cadherin, leads to a lower
activation of caspase-3/-7 than in control cells, upon paclitaxel
treatment.
The MDR phenotype can be fully mimicked by
experimental downregulation of E-cadherin in MTMECs. However, we
cannot rule out the possibility that either the miRs in the
miR-106b~25 cluster, or EP300, or both, act on downstream molecules
that regulate themselves drug resistance. miRs act repressing the
expression of hundred of targets and EP300, which as a
transcriptional co-activator, can also affect expression of many
genes. This offer the prospect of finding novel molecules and
regulatory pathways controlling transporter-independent MDR.
Abbreviations:
|
UTR
|
3′-untranslated region
|
|
ABC
|
ATP-binding cassette
|
|
DMEM-BSA
|
DMEM containing 0.1% bovine serum
albumin
|
|
IC50
|
drug concentration necessary to kill
50% of cells
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
miR
|
microRNA
|
|
MTMECs
|
minimally transformed mammary
epithelial breast cancer cells
|
|
MDR
|
multidrug resistant
|
Acknowledgments
We thank the China Scholarship Council (Y.H.),
Commonwealth Scholarship Comission (M.A.), Cancer Research UK China
Programme (Y.Z., C.C., E.Y.), Chinese National Natural Sciences
Foundation (81402480 to Y.H.), the Science and Technology
Foundation of Tianjin Municipal Health Bureau (2014KZ078 to Y.H.)
for their support.
References
|
1
|
Raguz S and Yagüe E: Resistance to
chemotherapy: New treatments and novel insights into an old
problem. Br J Cancer. 99:387–391. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gillet JP and Gottesman MM: Overcoming
multidrug resistance in cancer: 35 years after the discovery of
ABCB1. Drug Resist Updat. 15:2–4. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gottesman MM, Fojo T and Bates SE:
Multidrug resistance in cancer: Role of ATP-dependent transporters.
Nat Rev Cancer. 2:48–58. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Steinbach D and Legrand O: ABC
transporters and drug resistance in leukemia: Was P-gp nothing but
the first head of the Hydra? Leukemia. 21:1172–1176. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Trock BJ, Leonessa F and Clarke R:
Multidrug resistance in breast cancer: A meta-analysis of
MDR1/gp170 expression and its possible functional significance. J
Natl Cancer Inst. 89:917–931. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Clarke R, Leonessa F and Trock B:
Multidrug resistance/P-glycoprotein and breast cancer: Review and
meta-analysis. Semin Oncol. 32(Suppl 7): S9–S15. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Negrini M and Calin GA: Breast cancer
metastasis: A microRNA story. Breast Cancer Res. 10:2032008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jang JS, Jeon HS, Sun Z, Aubry MC, Tang H,
Park CH, Rakhshan F, Schultz DA, Kolbert CP, Lupu R, et al:
Increased miR-708 expression in NSCLC and its association with poor
survival in lung adenocarcinoma from never smokers. Clin Cancer
Res. 18:3658–3667. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang H, Kong W, He L, Zhao JJ, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al:
MicroRNA expression profiling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi
Y, Xiong W, Li G, Lu J, Fodstad O, et al: MicroRNA-125b confers the
resistance of breast cancer cells to paclitaxel through suppression
of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J
Biol Chem. 285:21496–21507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Miller TE, Ghoshal K, Ramaswamy B, Roy S,
Datta J, Shapiro CL, Jacob S and Majumder S: MicroRNA-221/222
confers tamoxifen resistance in breast cancer by targeting p27Kip1.
J Biol Chem. 283:29897–29903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Poliseno L, Salmena L, Riccardi L, Fornari
A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A and Fedele G:
Identification of the miR-106b~25 microRNA cluster as a
proto-oncogenic PTEN-targeting intron that cooperates with its host
gene MCM7 in transformation. Sci Signal. 3:ra292010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Petrocca F, Visone R, Onelli MR, Shah MH,
Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini
M, et al: E2F1-regulated microRNAs impair TGFbeta-dependent
cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell.
13:272–286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Acosta JC and Gil J: Senescence: A new
weapon for cancer therapy. Trends Cell Biol. 22:211–219. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou Y, Hu Y, Yang M, Jat P, Li K,
Lombardo Y, Xiong D, Coombes RC, Raguz S and Yagüe E: The
miR-106b~25 cluster promotes bypass of doxorubicin-induced
senescence and increase in motility and invasion by targeting the
E-cadherin transcriptional activator EP300. Cell Death Differ.
21:462–474. 2014. View Article : Google Scholar :
|
|
18
|
Zhao JJ, Gjoerup OV, Subramanian RR, Cheng
Y, Chen W, Roberts TM and Hahn WC: Human mammary epithelial cell
transformation through the activation of phosphatidylinositol
3-kinase. Cancer Cell. 3:483–495. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Unsworth H, Raguz S, Edwards HJ, Higgins
CF and Yagüe E: mRNA escape from stress granule sequestration is
dictated by localization to the endoplasmic reticulum. FASEB J.
24:3370–3380. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee JS, Paull K, Alvarez M, Hose C, Monks
A, Grever M, Fojo AT and Bates SE: Rhodamine efflux patterns
predict P-glycoprotein substrates in the National Cancer Institute
drug screen. Mol Pharmacol. 46:627–638. 1994.PubMed/NCBI
|
|
21
|
Yagüe E, Arance A, Kubitza L, O'Hare M,
Jat P, Ogilvie CM, Hart IR, Higgins CF and Raguz S: Ability to
acquire drug resistance arises early during the tumorigenesis
process. Cancer Res. 67:1130–1137. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yague E, Armesilla AL, Harrison G, Elliott
J, Sardini A, Higgins CF and Raguz S: P-glycoprotein (MDR1)
expression in leukemic cells is regulated at two distinct steps,
mRNA stabilization and translational initiation. J Biol Chem.
278:10344–10352. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Raguz S, Adams C, Masrour N, Rasul S,
Papoutsoglou P, Hu Y, Cazzanelli G, Zhou Y, Patel N, Coombes C, et
al: Loss of O6-methylguanine-DNA methyltransferase
confers collateral sensitivity to carmustine in topoisomerase
II-mediated doxo-rubicin resistant triple negative breast cancer
cells. Biochem Pharmacol. 85:186–196. 2013. View Article : Google Scholar
|
|
24
|
Vichai V and Kirtikara K: Sulforhodamine B
colorimetric assay for cytotoxicity screening. Nat Protoc.
1:1112–1116. 2006. View Article : Google Scholar
|
|
25
|
Hu Y, Cheng X, Li S, Zhou Y, Wang J, Cheng
T, Yang M and Xiong D: Inhibition of sorcin reverses multidrug
resistance of K562/A02 cells and MCF-7/A02 cells via regulating
apoptosis-related proteins. Cancer Chemother Pharmacol. 72:789–798.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rasul S, Balasubramanian R, Filipović A,
Slade MJ, Yagüe E and Coombes RC: Inhibition of gamma-secretase
induces G2/M arrest and triggers apoptosis in breast cancer cells.
Br J Cancer. 100:1879–1888. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nitiss JL: Targeting DNA topoisomerase II
in cancer chemotherapy. Nat Rev Cancer. 9:338–350. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ewald JA, Desotelle JA, Wilding G and
Jarrard DF: Therapy-induced senescence in cancer. J Natl Cancer
Inst. 102:1536–1546. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McGrogan BT, Gilmartin B, Carney DN and
McCann A: Taxanes, microtubules and chemoresistant breast cancer.
Biochim Biophys Acta. 1785:96–132. 2008.
|
|
30
|
Wu CP, Hsieh CH and Wu YS: The emergence
of drug transporter-mediated multidrug resistance to cancer
chemotherapy. Mol Pharm. 8:1996–2011. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Friedrich K, Wieder T, Von Haefen C,
Radetzki S, Jänicke R, Schulze-Osthoff K, Dörken B and Daniel PT:
Overexpression of caspase-3 restores sensitivity for drug-induced
apoptosis in breast cancer cell lines with acquired drug
resistance. Oncogene. 20:2749–2760. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Holohan C, Van Schaeybroeck S, Longley DB
and Johnston PG: Cancer drug resistance: An evolving paradigm. Nat
Rev Cancer. 13:714–726. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kathawala RJ, Gupta P, Ashby CR Jr and
Chen ZS: The modulation of ABC transporter-mediated multidrug
resistance in cancer: A review of the past decade. Drug Resist
Updat. 18:1–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
May CD, Sphyris N, Evans KW, Werden SJ,
Guo W and Mani SA: Epithelial-mesenchymal transition and cancer
stem cells: A dangerously dynamic duo in breast cancer progression.
Breast Cancer Res. 13:2022011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ferreira P, Oliveira MJ, Beraldi E, Mateus
AR, Nakajima T, Gleave M, Yokota J, Carneiro F, Huntsman D, Seruca
R, et al: Loss of functional E-cadherin renders cells more
resistant to the apoptotic agent taxol in vitro. Exp Cell Res.
310:99–104. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shiota M, Yokomizo A, Kashiwagi E, Tada Y,
Inokuchi J, Tatsugami K, Kuroiwa K, Uchiumi T, Seki N and Naito S:
Foxo3a expression and acetylation regulate cancer cell growth and
sensitivity to cisplatin. Cancer Sci. 101:1177–1185. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Takeuchi A, Shiota M, Tatsugami K,
Yokomizo A, Tanaka S, Kuroiwa K, Eto M and Naito S: p300 mediates
cellular resistance to doxorubicin in bladder cancer. Mol Med Rep.
5:173–176. 2012.
|
|
40
|
Mees ST, Mardin WA, Wendel C, Baeumer N,
Willscher E, Senninger N, Schleicher C, Colombo-Benkmann M and
Haier J: EP300 - a miRNA-regulated metastasis suppressor gene in
ductal adenocarcinomas of the pancreas. Int J Cancer. 126:114–124.
2010. View Article : Google Scholar
|
|
41
|
Callaghan R, Luk F and Bebawy M:
Inhibition of the multidrug resistance P-glycoprotein: Time for a
change of strategy? Drug Metab Dispos. 42:623–631. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chekhun VF, Lukyanova NY, Kovalchuk O,
Tryndyak VP and Pogribny IP: Epigenetic profiling of
multidrug-resistant human MCF-7 breast adenocarcinoma cells reveals
novel hyper- and hypomethylated targets. Mol Cancer Ther.
6:1089–1098. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lu L, Zhou D, Jiang X, Song K, Li K and
Ding W: Loss of E-cadherin in multidrug resistant breast cancer
cell line MCF-7/Adr: Possible implication in the enhanced invasive
ability. Eur Rev Med Pharmacol Sci. 16:1271–1279. 2012.PubMed/NCBI
|
|
44
|
Doyle L and Ross DD: Multidrug resistance
mediated by the breast cancer resistance protein BCRP (ABCG2).
Oncogene. 22:7340–7358. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sprowl JA, Reed K, Armstrong SR, Lanner C,
Guo B, Kalatskaya I, Stein L, Hembruff SL, Tam A and Parissenti AM:
Alterations in tumor necrosis factor signaling pathways are
associated with cytotoxicity and resistance to taxanes: A study in
isogenic resistant tumor cells. Breast Cancer Res. 14:R22012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Park SJ, Wu CH, Gordon JD, Zhong X, Emami
A and Safa AR: Taxol induces caspase-10-dependent apoptosis. J Biol
Chem. 279:51057–51067. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Janssen K, Pohlmann S, Jänicke RU,
Schulze-Osthoff K and Fischer U: Apaf-1 and caspase-9 deficiency
prevents apoptosis in a Bax-controlled pathway and promotes
clonogenic survival during paclitaxel treatment. Blood.
110:3662–3672. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jelínek M, Balušíková K, Kopperová D,
Nĕmcová-Fürstová V, Šrámek J, Fidlerová J, Zanardi I, Ojima I and
Kovář J: Caspase-2 is involved in cell death induction by taxanes
in breast cancer cells. Cancer Cell Int. 13:422013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yamakawa N, Takahashi A, Mori E, Imai Y,
Furusawa Y, Ohnishi K, Kirita T and Ohnishi T: High LET radiation
enhances apoptosis in mutated p53 cancer cells through caspase-9
activation. Cancer Sci. 99:1455–1460. 2008. View Article : Google Scholar : PubMed/NCBI
|