Introduction
Head and neck squamous cell carcinoma (HNSCC) is a
group of common malignant cancers located in the oral cavity,
hypopharynx, oropharynx and larynx (1). Annually, HNSCCs account for more than
550,000 cases of cancer worldwide (2). Research suggests that tobacco use and
alcohol consumption are two major risk factors for HNSCC. Human
papillomaviruse (HPV) infection, particularly high-risk HPVs, are
closely associated with HNSCCs, particularly carcinomas of the lip
and oral cavity (3,4). The tumor-suppressor p53, which plays
essential roles in many different types of human malignant tumors,
was also found to be associated with the occurrence of HNSCC
(5). Various types of mutations in
the p53-encoding gene (TP53) have been identified in HNSCC
tissues (6). In addition, various
single-nucleotide polymorphisms (SNPs) in TP53, such as the
polymorphism in codon 72 (SNP72), significantly influence the age
at onset of various types of HNSCCs (7).
Prion protein (PrP) is a copper binding
glycoprotein, which is highly conserved throughout evolution
(8). PrP exists in many types of
tissues, but is mostly concentrated in the central nervous system
(CNS). PrP is directly involved in prion diseases or transmissible
spongiform encephalopathies (TSEs) that are a group of fatal
neurodegenerative diseases, such as human Creutzfeldt-Jakob disease
(CJD), scrapie and bovine spongiform encephalopathy. In these
cases, normal cellular PrP (PrPC) is converted into a
proteinase-resistant and pathological isoform (PrPSc)
(9). As a cellular membrane
protein, normal PrP is expressed in the neurons and glia of the
brain and spinal cord, as well as in several peripheral tissues and
in leukocytes (10). The efficient
transcription of PrP is detectable in the brains of mice and
chickens beginning early in embryogenesis, and its level increases
as development proceeds (11).
Although its wide expression in many cell types indicate a more
cosmopolitan biological function, the exact cellular function
remains undefined. Evidence suggests that PrPC is
important for synaptic activity, cell adhesion and recognition,
ligand uptake, transmembrane signaling and neuroprotection
(12). The biological function of
PrP in peripheral tissues has been rarely addressed.
In the past decade, the association of PrP with
human malignant tumors has attracted great attention. PrP has been
found to be overexpressed in a variety of cancers including
gastric, pancreatic and breast cancers, osteosarcoma and melanoma
(13–15). Various studies have even proposed
that overexpression of PrP is closely associated with the poor
prognosis of pancreatic and breast cancers, highlighting that it
may affect the growth and invasiveness of cancers (16). PrP expression is also believed to
play an important role in the acquisition of multidrug-resistant
(MDR) gastric cancer (17).
However, the expression of PrP and its role in HNSCCs remain
undescribed.
In the present study, PrP expression in tumor
tissues from 92 pathologically diagnosed HNSCC cases were screened
with PrP-specific immunohistochemical assays. We found that 55.43%
(51/92) of the tested carcinomas were PrP-positive. The positive
rate and the staining intensity of PrP were closely related with
the pathological degree of the tumors. Additionally, PrP-positive
rates also showed correlations with the anatomic site and the
clinical grade of the carcinomas. Further evaluation of the
associations of PrP expression with p53 abnormalities and HPV
infection in malignant tissues revealed that PrP-positive staining
was more frequently detected in the tissues with p53-positive
accumulation and the wild-type TP53 gene.
Materials and methods
Specimens
The paraffin-embedded specimens of HNSCCs used in
our previous studies (7,18) were employed in the present study,
except one laryngocarcinoma case without further available tissue.
Totally, 92 HNSCCs were recruited, including 63 laryngocarcinoma, 5
oropharyngeal, 15 hypopharynx and 9 lip carcinomas. All patients
were hospitalized at the Department of Head and Neck Surgery,
Peking University Cancer Hospital and Institute, from 2006 to 2011.
The demographic, clinical and pathological information of these
patients was previously described (18). Six surgically removed tissues from
the patients with laryngocarcinoma were stored at −80°C.
Immunohistochemistry (IHC)
Paraffin sections (5-µm in thickness) were
deparaffinized in xylene for 5 min twice and gradually routinely
rehydrated. The sections were quenched for endogenous peroxidases
in 3% H2O2 in methanol for 15 min, and
pretreated with enzyme digestion antigen retrieval for 1 min. After
blocking in 1% normal goat serum, the sections were incubated
overnight at 4°C with 1:500-diluted PrP-specific mAbs, including
3F4 (MAB1562; Millipore), 6D11 (sc-58581), 7D9 (sc-58582) (both
from Santa Cruz Biotechnology) and 8H4 (ab-61409; Abcam). The
sections were then incubated for 60 min with 1:1,000-diluted
HRP-conjugated goat anti-mouse secondary antibody (Vector Labs,
USA), and visualized by incubation with 3,3-diaminobenzidine
tetrahydrochloride (DAB). The slices were dehydrated and mounted in
Permount. Photomicrographs were captured with a DP70 digital camera
mounted on a BX5 microscope (Olympus Optical, Japan).
Determination of the degree of
PrP-positive staining
The strategy for determination of the degree of
PrP-positive staining in the tested tissues was based on a protocol
described elsewhere (19). Briefly,
five fields under microscopy were randomly selected for each slice.
The score was given based on the ratio of positively stained cells
and gradation of the stained color. A total of <10% positive
cells was recorded as 0, 10–25% as 1, >25–50% as 2, >50–75%
as 3, and >75% as 4. No positive staining was scored as 0, light
brown signals in cells was scored as 1, brown signals in cells was
scored as 2, deep brown signals was scored as 3. The score of the
percentage of positive cells and the score of the gradation of the
stained color was multiplied and the final result was given based
on the product: 0–1 as negative (−), 2–4 as weakly positive (+),
5–8 as positive (++), 9–12 as strongly positive (+++).
Preparation of tissue homogenates
The tissue samples of HNSCC and the brain samples of
normal and scrapie-infected mice were homogenized in 10% lysis
buffer (w/v, 100 mM NaCl, 10 mM EDTA, 0.5% Nonidet P-40, 0.5%
sodium deoxycholate, 10 mM Tris, pH 7.5) according to a protocol
described elsewhere (20). Then,
tissue debris was removed with low speed centrifugation at 2,000 ×
g for 10 min and the supernatants were collected for further
study.
Ethics statement
Written consent for further investigation and
publication was obtained from the patients or the patients'
relatives, respectively. Usage of the stored human samples in the
present study was approved by the Ethics Committees of Peking
University Cancer Hospital and the Institute and National Institute
for Viral Disease Prevention and Control, China CDC.
Statistical analysis
Statistical analysis was performed using Chi-square
and Fisher's exact tests for correlations between groups for HPV
infection, SNP72 type, p53 mutation and IHC. Mann-Whitney method
was used for the relationship between SNP72 and age. A probability
value of <0.05 was considered to indicate a statistically
significant result. All statistical analyses were performed by SPSS
20 (IBM, USA).
Results
PrP expression is detectable in more than
half of the carcinoma tissues
PrP protein is widely expressed in CNS. To ascertain
the expression of PrP in HNSCC tissues, several commercial PrP
monoclonal antibodies were selected and screened for the potential
usage in the IHC assays: mAbs 3F4, 6D11, 7D9 and 8H4. Using the
sections from the same paraffin-embedded tissues of 3 different
patients, the immunoreactivities of the mAbs were comparably
evaluated under the same experimental conditions. mAb 3F4 produced
the most significant reactivity in the IHC assays, with clear brown
positive staining in the cytoplasm and membrane of the carcinoma
cells. Therefore, PrP-specific mAb 3F4 was used in the IHC assays
in the subsequent tests.
Totally, 92 slices were stained with mAb 3F4
immunofluorescently, and 51 cases (55.43%) were PrP-positive. Based
on the judgment protocol described above, 41 (44.57%) cases were
weakly positive for PrP (+), 9 (9.78%) were PrP positive (++) and
only one case was strongly positive for PrP (+++). As shown in
Fig. 1, the PrP-specific staining
was concentrated in the carcinoma cells, mostly distributed in the
cytoplasm. In some areas, the strongly positive staining formed
large positive masses. PrP-positive staining was rarely observed in
the regions of morphologically normal tissues.
Among the 92 tested HNSCC specimens, 63 were located
in the larynx, 5 in the oropharynx, 15 in the hypopharynx and 9 in
the lip. PrP expression was positive in 44.44% (4/9) of the lip
carcinomas, 60% (3/5) of the oropharyngeal carcinomas, 46.67%
(7/15) of the hypopharynx carcinomas and 58.73% (37/63) of the
laryngeal carcinomas (Table I).
Statistical analysis showed significantly higher PrP expression in
the cancers of the oropharynx and larynx.
 | Table IPrP-positive staining in the HNSCC
cases. |
Table I
PrP-positive staining in the HNSCC
cases.
| Site | No. of cases | PrP
| Positive rate
(%) |
|---|
| − n (%) | + n (%) | ++ n (%) | +++ n (%) |
|---|
| Lip | 9 | 5 (55.56) | 2 (22.22) | 2 (22.22) | 0 (0.00) | 44.44 |
| Oropharynx | 5 | 2 (40.00) | 2 (40.00) | 0 (0.000) | 1 (20.00) | 60.00a |
| Hypopharynx | 15 | 8 (53.33) | 5 (33.33) | 2 (13.33) | 0 (0.00) | 46.67b |
| Larynx | 63 | 26 (41.27) | 32 (50.79) | 5 (7.94) | 0 (0.00) | 58.73c |
| Total | 92 | 41 (44.57) | 41 (44.57) | 9 (9.78) | 1 (1.09) | 55.43 |
Correlation between PrP expression and
the clinical degrees of the HNSCCs
To test the potential correlation between
PrP-positive staining and the clinical grade at the time of
surgical operation, 92 of the HNSCCs were grouped according to
their clinical degrees. The PrP-positive rates of clinical degree
I, II, III and IV cases were 44.44 (12/27), 62.86 (22/35), 56
(14/25) and 60% (3/5), respectively (Table II). Statistical assays revealed
significantly low PrP-positive rates in the group of clinical
degree I than that in II (P=0.0001), III (P=0.0199) and IV
(P=0.0015). There was no statistical difference in the PrP-positive
rate among the tumors of clinical degree II, III and IV. The
distributions of the PrP-positive carcinomas based on the
intensities were quite comparable. Tumor tissues in later clinical
degree had higher PrP-positive rates.
 | Table IIPrP-positive staining in HNSCC based
on the clinical degrees. |
Table II
PrP-positive staining in HNSCC based
on the clinical degrees.
| Clinical
degree | No. of cases | PrP
| Positive rate
(%) |
|---|
| − n (%) | + n (%) | ++ n (%) | +++ n (%) |
|---|
| I | 27 | 15 (55.56) | 9 (33.33) | 3 (11.11) | 0 (0.000) | 44.44 |
| II | 35 | 13 (37.14) | 19 (54.29) | 3 (8.57) | 0 (0.000) | 62.86a |
| III | 25 | 11 (44.00) | 11 (44.00) | 3 (12.00) | 0 (0.000) | 56.00b |
| IV | 5 | 2 (40.00) | 2 (40.00) | 0 (0.000) | 1 (20.00) | 60.00c |
| Total | 92 | 41 (44.57) | 41 (44.57) | 9 (9.78) | 1 (1.09) | 55.43 |
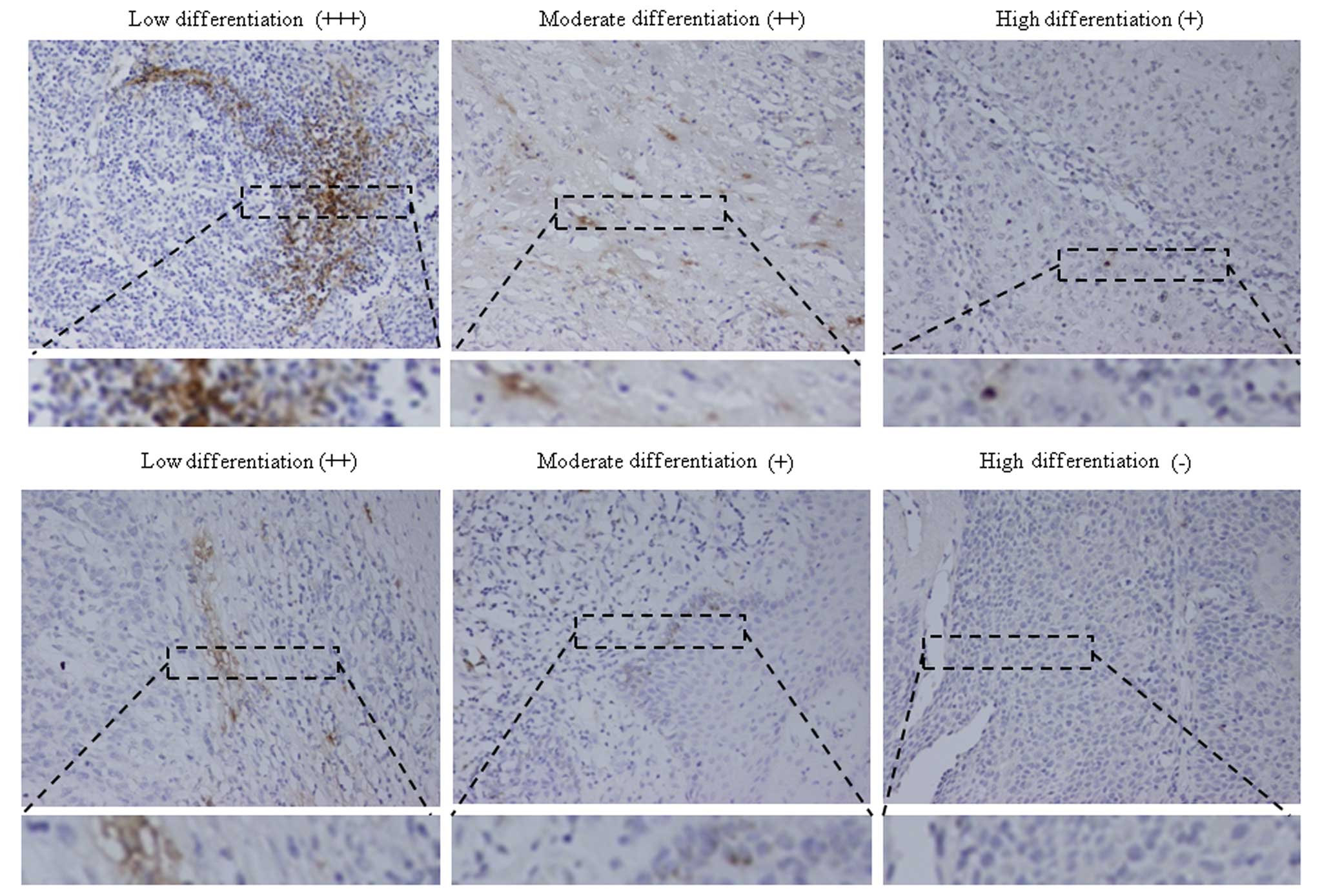
Correlation between PrP expression and
the pathological grade of the HNSCCs
To ascertain the possible relationship between PrP
expression and pathological differentiation, the tested HNSCCs were
divided into groups of low, moderately and highly differentiated
carcinomas. The PrP-positive rates were 30.77% (8/26) in the highly
differentiated, 60.78% (31/51) in the moderately differentiated and
80% (12/15) in the lowly differentiated HNSCCs, respectively
(Table III). Statistical analysis
obviously showed significance among the three groups and between
each group (P<0.0001). Further analysis identified that 8 out of
9 PrP-positively stained (++) cases were distributed in the groups
of moderately and lowly differentiated carcinomas, and one strongly
PrP-positive (+++) case was poorly differentiated carcinoma. This
strongly indicates a close association of PrP expression with
poorly differentiated HNSCCs.
 | Table IIIPrP-positive staining in HNSCC based
on the pathological degrees. |
Table III
PrP-positive staining in HNSCC based
on the pathological degrees.
| Pathological
degree | No. of cases | PrP
| Positive rate
(%) |
|---|
| − n (%) | + n (%) | ++ n (%) | +++ n (%) |
|---|
| Highly
differentiated | 26 | 18 (69.23) | 7 (26.92) | 1 (3.85) | 0 (0.00) | 30.77 |
| Moderately
differentiated | 51 | 20 (39.22) | 26 (50.98) | 5 (9.80) | 0 (0.00) | 60.78a |
| Lowly
differentiated | 15 | 3 (20.00) | 8 (53.33) | 3 (20.00) | 1 (6.67) | 80.00b |
| Total | 92 | 41 (44.57) | 41 (44.57) | 9 (9.78) | 1 (1.09) | 55.43 |
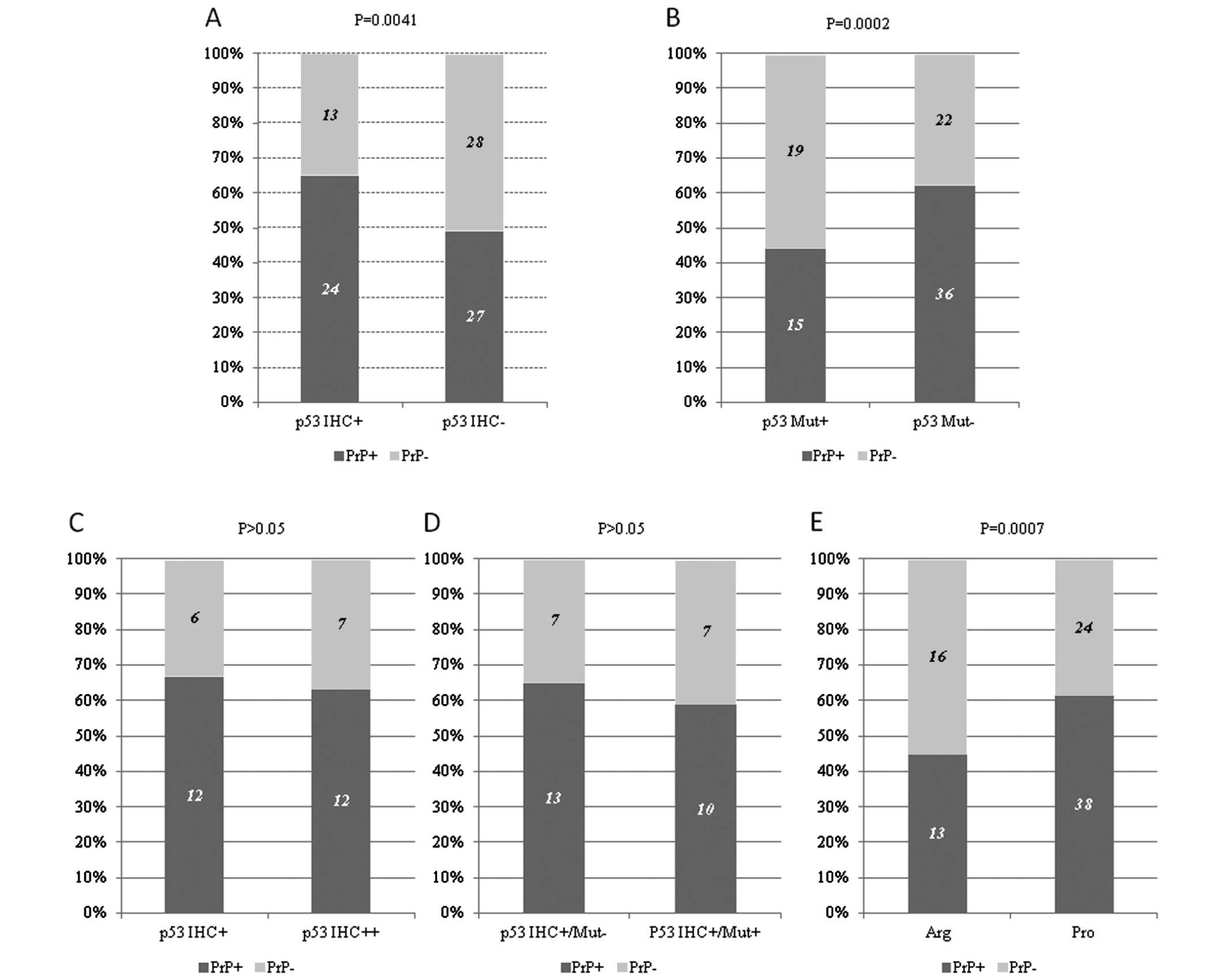
Correlation between the PrP expression
and abnormalities of p53 in the HNSCCs
Our previous study proposed that among all 92 HNSCC
cases, 37 cases (40.22%) were p53-positive and 55 cases (59.78%)
were p53-negative. Sequencing analysis of the TP53 gene
revealed that 34 (36.96%) cases contained various mutations
(7). Analysis of the possible
relationship between PrP and p53 staining in IHC identified that
the rate of PrP-positive staining was 64.86% (24/37) in the
p53-positive patients, while the rate was 49.09% (27/55) in the
p53-negative patients, showing statistical difference (P=0.0041)
(Fig. 2A). All 6 HNSCC cases
showing p53 strong positive p53 staining was also PrP-positive. In
contrary, positive staining for PrP was detected in 44.11% (15/34)
of the patients containing mutations in TP53 and 62.07%
(36/58) of those without a mutation in TP53, showing high
significance (P=0.0002) (Fig. 2B).
This indicates that the patients with p53-positive cancer cells and
the patients with the wild-type TP53 gene had a higher
possibility to be PrP-positive.
To ascertain the possible linkage of PrP-positive
staining with the intensities of the p53-positive staining in the
IHC assays, 37 p53-positive cases in IHC were grouped as weakly
positive (p53 IHC+) (18 cases) and strongly positive (p53 IHC++)
(19 cases). The positive rates of PrP expression in the groups of
p53 IHC+ and p53 IHC++ were 66.67% (12/18) and 63.15% (12/19),
respectively, without statistical difference in the PrP-positive
rate (P>0.05) (Fig. 2C).
Furthermore, the cases with PrP-positive expression in the 37 cases
with p53-positivity in IHC were analyzed according to whether they
had mutations in their TP53 gene. Out of 17 patients with
mutations in TP53, 58.82% (10/17) were PrP-positive, while
out of 20 patients with wild-type TP53, 65.00% (13/20) were
PrP-positive (Fig. 2D). Although
the PrP-positive rate in the patients with p53-positive staining
and mutated TP53 was slightly lower than in the patients
with p53-positive staining and wild-type TP53, statistical
analysis did not achieve significance (P>0.05). This highlights
that either the intensity of p53-positive staining or the status of
TP53 did not notably influence the PrP expression in the
group of p53-positive HNSCCs.
Single-nucleotide polymorphisms (SNPs) in codon 72
(SNP72) of TP53 include arginine (Arg)/Arg, Pro/Pro and
Pro/Arg. Among the 91 HNSCCs with data for p53 SNP72, 29 cases were
Arg/Arg homozygote whose phenotype is nominated as Arg, 62 were
Pro/Pro homozygote or Pro/Arg heterozygote whose phenotype is
termed as Pro (7). The
distributions of PrP-positive rates in these two groups were
obviously different; the PrP-positive rate in the group of Pro/Pro
homozygotes or Pro/Arg heterozygotes (61.29%, 38/62) was
significantly higher than those of the Arg/Arg homozygote (44.83%,
13/29), indicating a close correlation between PrP expression and
p53 SNP72 (Fig. 2E).
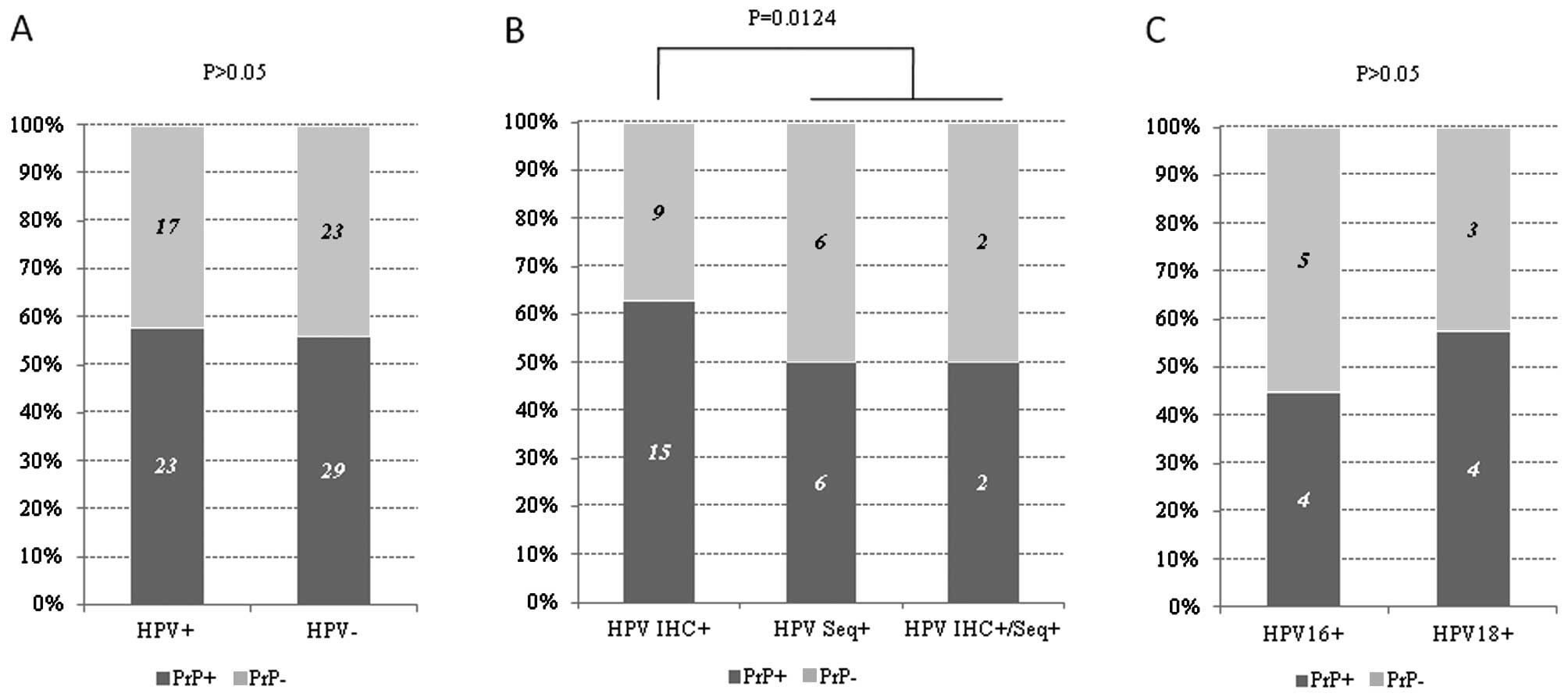
Correlation between PrP expression and
HPV infection in HNSCCs
Among the 92 HNSCCs, HPV-related sequences and/or
proteins were detectable in 40 cases. The positive rates of PrP
immunostaining were 57.50% (23/40) and 55.76% (29/52) in the
HPV-positive and -negative cases, respectively. No statistical
difference in PrP expression was found between the HPV+
and HPV− groups (P>0.05) (Fig. 3A). Among the 40 HPV-positive HNSCCs,
12 were HPV sequence positive only, and 24 were HPV early (E6/E7)
protein positive only, and 4 were HPV sequence and protein positive
both (18). Obvious expression of
PrP was detected in half of the patients in the HPV sequence
positive (HPV Seq+) and HPV sequence and protein
positive (HPV IHC+/Seq+) groups, but in 62.5% of the
patients showing HPV protein positivity (HPV IHC+),
showing statistical difference (P<0.05) (Fig. 3B). Additionally, 4 out of 9 HPV16
sequence-positive cases and 4 out of 7 HPV18 sequence-positive
cases were PrP-positive, without statistical significance (Fig. 3C). These data indicate that HPV
infection does not influence the expression of PrP in malignant
cells.
Discussion
In the present study, we screened the PrP expression
in malignant tissues of HNSCCs with IHC. PrP-positive staining was
observable in 55.43% of the tested HNSCCs. The positive rate and
the intensity of PrP in the tumor tissues were closely associated
with the pathological grade of the carcinomas. Meanwhile, the
PrP-positive rate was correlated with the anatomic site and
clinical degree of the HNSCCs. Additionally, the HNSCCs with
p53-positivity in IHC and the Pro/Pro and Pro/Arg genotypes in
SNP27 of p53 showed a higher probability for positive expression of
PrP in their cells. PrP expression in the HNSCCs were not related
with HPV infections.
Apart from its indispensible and unique role in
human and animal prion diseases, a disparate significance of PrP in
human malignant tumors has attracted great attention. Numerous
lines of evidence support the potential involvement of PrP in
different types of human cancers, in which PrP is overexpressed,
such as colorectal (21), gastric
(22,23) breast (24,25),
pancreatic (26,27) and prostrate (28). In addition, PrP expression is
observable in many tumor cell lines derived from the above
malignant tumors or from other cancers, such as melanoma and
hepatocarcinoma (16,27). However, the PrP status in cancer
derived from squamous cells appears to be rarely described, except
a report of oral squamous cell carcinomas in a Chinese journal
(29). Our data in the present
study strongly indicate a distinct involvement of PrP protein in
the cancer biology of squamous cell carcinoma, at least HNSCCs.
Despite the diversities of the PrP-positive rates in
different types of cancers from different studies, a close
correlation between PrP expression and pathological and clinical
grades of cancers has been well documented in almost all studies
(16,27). The frequency of PrP expression was
found to be ~40% in patients with pancreatic ductal cell
adenocarcinoma, but only 13% in pancreatic intraepithelial
neoplasia (PIN)-3 cases and none in PIN-2 and -1 cases (27). Higher PrP-positive rates and
stronger PrP staining are also noted in breast and gastric cancers
(24,30). More importantly, PrP expression
appears to be associated with poor prognosis in pancreatic and
breast cancers (24), which
highlights a contributory role of PrP in tumor progression. In line
with the above observations, our data confirmed that the positive
rates and staining intensities of PrP correlate well with the
progression of HNSCCs, particularly the pathological grade of the
tumors, which emphasizes again a common phenomenon in different
human malignant tumors. Further prospective analysis of the
relationship between the survival times and PrP expression in
HNSCCs will help to define the prognostic value of PrP IHC
assays.
Disruption of the p53 network favours cell survival
and tumor progression. Abnormalities of p53, either mutations in
TP53 or overexpression, are frequently observed in a number
of human cancers, including HNSCCs, which is likely to be
associated with increased susceptibility to cancer development. Our
data in the present study illustrated a positive correlation of PrP
expression with p53- positive IHC but a negative correlation with
mutated TP53 in the HNSCCs. The exact biological meanings of
such a pattern remain unclear. Recently, the abnormal accumulation
of either WT or mutant p53 in the cytoplasm and nucleus were
described in human cancers, such as neuroblastoma, retinoblastoma,
breast and colon cancers (31–34).
Notably, the aggregation of p53 could be amyloid-like, which
highlights a possibility that p53 amyloid formation may participate
in the malignant process (35).
SNP72 in TP53 is able to influence the
function of p53, in which TP53 encoding Arg is more
effective than that encoding Pro at inducing apoptosis and
preventing cells from neoplastic development (7). The significance of the SNP72
polymorphism in association with the susceptibility of several
types of cancers and the survival of cancer patients have been
proposed (36). Our previous study
found that HNSCC patients with a Pro polymorphism showed average
younger onset-age and had higher percentages of strong
p53-positivity in IHC (7).
Coincidental with the clinical and pathological features of SNP72,
we revealed that the patients with a Pro polymorphism in SNP72 had
a significantly higher PrP-positive rate in the present study. This
may indicate a possible cooperative effect of PrP and p53 in cancer
biology.
HPV infections are frequently detected in HNSCCs
(3,37). Among the tested 92 HNSCCs in the
present study, 40 cases showed HPV positivity, either HPV sequences
or proteins. We did not observe a notable difference in PrP
expression between HPV-positive and HPV-negative groups, implying
that HPV infection has little effect on PrP expression in HNSCCs.
HPV infection in the 92 HNSCCs showed an anatomic increasing trend
from inside to outside; more HPV-positive cases were noted in the
lip and oropharynx than in the hypopharynx and larynx (18). However, PrP expression in the HNSCCs
did not reveal anatomic-dependent alteration. As the limited
numbers of other HNSCCs besides laryngeal cancers, a final
conclusion of the relationship between HPV infection and PrP
expression in HNSCCs requires large score assays.
Cellular PrP appears to be involved in many
essential biological processes, such as cell adhesion, neurite
outgrowth, synaptic transmission, oxidative stress, anti-apoptosis
and neuroprotection, which are closely associated with cellular
survival, proliferation and differentiation (38,39).
Therefore, it is reasonable to speculate whether aberrant PrP
function may contribute to tumorigenesis. However, numerous studies
including ours demonstrated that the PrP proteins in cancer cells
are an aglycosyl form of PrP or so-called 'pro-prion', which
accumulate mainly in the cytoplasm but do not anchor on the cell
membrane (40). In this situation,
it is hard to simply attribute the role of PrP in tumorigenesis to
its aberrantly enhanced biological effects. In fact, accumulation
of aglycosyl PrP (not PrPSc) in the cytoplasm usually
reduces the viability of many different types of cultured cell
lines. Recently, many potential mechanisms have been proposed to
explain the possible role of PrP in cancer cells, such as
activation of the PI3K/Akt signaling pathway to upregulate cyclin D
in gastric cancer cells (41),
chemotherapy drug-induced PrP interaction with P-glycoprotein
(P-gp; ATP-dependent drug-efflux pumps ABCB1) in drug-resistant
MCF7 breast cancer cells, enhanced doxorubicin resistance via the
ERK1/2 signaling pathway in MDA-MB-435 breast cancer cells
(42), the fatal attraction between
pro-PrP to filamin A in melanoma and pancreatic cancer cells
(26). Nevertheless, a more
comprehensive knowledge of PrPs may facilitate the understanding of
the pathogenesis and the development of more effective tools for
diagnosis, prognosis, therapy and prevention not only for TSEs, but
also for a number of cancers.
Acknowledgments
The present study was supported by the Chinese
National Natural Science Foundation Grants (81301429 and 81572048),
the China Mega-Project for Infectious Disease (2011ZX10004-101 and
2012ZX10004215), and the SKLID Development Grant (2012SKLID102 and
2015SJLID503).
References
|
1
|
Inglehart RC, Scanlon CS and D'Silva NJ:
Reviewing and reconsidering invasion assays in head and neck
cancer. Oral Oncol. 50:1137–1143. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Machiels JP, Lambrecht M, Hanin FX, Duprez
T, Gregoire V, Schmitz S and Hamoir M: Advances in the management
of squamous cell carcinoma of the head and neck. F1000Prime Rep.
6:442014. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kaba G, Dzudzor B, Gyasi RK, Asmah RH,
Brown CA, Kudzi W and Wiredu EK: Human papillomavirus genotypes in
a subset of head and neck squamous cell carcinoma. West Afr J Med.
33:121–124. 2014.PubMed/NCBI
|
|
4
|
Woods R Sr, O'Regan EM, Kennedy S, Martin
C, O'Leary JJ and Timon C: Role of human papillomavirus in
oropharyngeal squamous cell carcinoma: A review. World J Clin
Cases. 2:172–193. 2014.PubMed/NCBI
|
|
5
|
Dang M, Lysack JT, Wu T, Matthews TW,
Chandarana SP, Brockton NT, Bose P, Bansal G, Cheng H, Mitchell JR,
et al: MRI texture analysis predicts p53 status in head and neck
squamous cell carcinoma. AJNR Am J Neuroradiol. 36:166–170. 2015.
View Article : Google Scholar
|
|
6
|
Maruyama H, Yasui T, Ishikawa-Fujiwara T,
Morii E, Yamamoto Y, Yoshii T, Takenaka Y, Nakahara S, Todo T,
Hongyo T, et al: Human papillomavirus and p53 mutations in head and
neck squamous cell carcinoma among Japanese population. Cancer Sci.
105:409–417. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi Q, Xiao K, Wei W, Zhang BY, Chen C, Xu
Y, Chen LN, Song YT, Ma X, Zhang NS, et al: Associations of TP53
mutations, codon 72 polymorphism and human papillomavirus in head
and neck squamous cell carcinoma patients. Oncol Rep. 30:2811–2819.
2013.PubMed/NCBI
|
|
8
|
Colby DW and Prusiner SB: Prions. Cold
Spring Harb Perspect Biol. 3:a0068332011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liberski PP: Historical overview of prion
diseases: A view from afar. Folia Neuropathol. 50:1–12.
2012.PubMed/NCBI
|
|
10
|
Gasperini L and Legname G: Prion protein
and aging. Front Cell Dev Biol. 2:442014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Llorens F, Ferrer I and del Río JA: Gene
expression resulting from PrPC ablation and
PrPC overexpression in murine and cellular models. Mol
Neurobiol. 49:413–423. 2014. View Article : Google Scholar
|
|
12
|
Yusa S, Oliveira-Martins JB,
Sugita-Konishi Y and Kikuchi Y: Cellular prion protein: From
physiology to pathology. Viruses. 4:3109–3131. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He M, Wang L, Pu J, Yang Q, Li G and Hao
J: Proliferin-related protein overexpression in SGC-7901 gastric
cancer cells inhibits in vitro cell growth and tumorigenesis in
nude mice. Oncol Rep. 29:2243–2248. 2013.PubMed/NCBI
|
|
14
|
Sy MS, Altekruse SF, Li C, Lynch CF,
Goodman MT, Hernandez BY, Zhou L, Saber MS, Hewitt SM and Xin W:
Association of prion protein expression with pancreatic
adenocarcinoma survival in the SEER residual tissue repository.
Cancer Biomark. 10:251–258. 2011–2012.
|
|
15
|
Farah J, Sayah R, Martinetti F, Donadille
L, Lacoste V, Hérault J, Delacroix S, Nauraye C, Vabre I, Lee C, et
al: Secondary neutron doses in proton therapy treatments of ocular
melanoma and craniopharyngioma. Radiat Prot Dosimetry. 161:363–367.
2014. View Article : Google Scholar
|
|
16
|
Antony H, Wiegmans AP, Wei MQ, Chernoff
YO, Khanna KK and Munn AL: Potential roles for prions and
protein-only inheritance in cancer. Cancer Metastasis Rev. 31:1–19.
2012. View Article : Google Scholar :
|
|
17
|
Hinton C, Antony H, Hashimi SM, Munn A and
Wei MQ: Significance of prion and prion-like proteins in cancer
development, progression and multi-drug resistance. Curr Cancer
Drug Targets. 13:895–904. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei W, Shi Q, Guo F, Zhang BY, Chen C,
Zhang NS and Dong XP: The distribution of human papillomavirus in
tissues from patients with head and neck squamous cell carcinoma.
Oncol Rep. 28:1750–1756. 2012.PubMed/NCBI
|
|
19
|
Gao JM, Gao C, Han J, Zhou XB, Xiao XL,
Zhang J, Chen L, Zhang BY, Hong T and Dong XP: Dynamic analyses of
PrP and PrPSc in brain tissues of golden hamsters
infected with scrapie strain 263K revealed various PrP forms.
Biomed environ Sci. 17:8–20. 2004.PubMed/NCBI
|
|
20
|
Zhang J, Chen L, Zhang BY, Han J, Xiao XL,
Tian HY, Li BL, Gao C, Gao JM, Zhou XB, et al: Comparison study on
clinical and neuropathological characteristics of hamsters
inoculated with scrapie strain 263K in different challenging
pathways. Biomed Environ Sci. 17:65–78. 2004.PubMed/NCBI
|
|
21
|
Antonacopoulou AG, Palli M, Marousi S,
Dimitrakopoulos FI, Kyriakopoulou U, Tsamandas AC, Scopa CD,
Papavassiliou AG and Kalofonos HP: Prion protein expression and the
M129V polymorphism of the PRNP gene in patients with colorectal
cancer. Mol Carcinog. 49:693–699. 2010.PubMed/NCBI
|
|
22
|
Duhayon S, Hoet P, Van Maele-Fabry G and
Lison D: Carcinogenic potential of formaldehyde in occupational
settings: A critical assessment and possible impact on occupational
exposure levels. Int Arch Occup Environ Health. 81:695–710. 2008.
View Article : Google Scholar
|
|
23
|
Pan Y, Zhao L, Liang J, Liu J, Shi Y, Liu
N, Zhang G, Jin H, Gao J, Xie H, et al: Cellular prion protein
promotes invasion and metastasis of gastric cancer. FASEB J.
20:1886–1888. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Meslin F, Conforti R, Mazouni C, Morel N,
Tomasic G, Drusch F, Yacoub M, Sabourin JC, Grassi J, Delaloge S,
et al: Efficacy of adjuvant chemotherapy according to Prion protein
expression in patients with estrogen receptor-negative breast
cancer. Ann Oncol. 18:1793–1798. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang J, Pan YL, Ning XX, Sun LJ, Lan M,
Hong L, Du JP, Liu N, Liu CJ, Qiao TD, et al: Overexpression of
PrPC and its antiapoptosis function in gastric cancer.
Tumour Biol. 27:84–91. 2006. View Article : Google Scholar
|
|
26
|
Li C, Yu S, Nakamura F, Yin S, Xu J,
Petrolla AA, Singh N, Tartakoff A, Abbott DW, Xin W, et al: Binding
of pro-prion to filamin A disrupts cytoskeleton and correlates with
poor prognosis in pancreatic cancer. J Clin Invest. 119:2725–2736.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sy MS, Li C, Yu S and Xin W: The fatal
attraction between pro-prion and filamin A: Prion as a marker in
human cancers. Biomarkers Med. 4:453–464. 2010. View Article : Google Scholar
|
|
28
|
Sauer H, Dagdanova A, Hescheler J and
Wartenberg M: Redox-regulation of intrinsic prion expression in
multicellular prostate tumor spheroids. Free Radic Biol Med.
27:1276–1283. 1999. View Article : Google Scholar
|
|
29
|
Zhang J, Zeng Y, Zheng J and Xu J:
Expression of Prion protein and its clinical significance in oral
squamous cells carcinoma and oral leukoplakia. Zhonghua Kou Qiang
Yi Xue Za Zhi. 48:752–754. 2013.In Chinese.
|
|
30
|
Wang JH, Du JP, Zhang YH, Zhao XJ, Fan RY,
Wang ZH, Wu ZT and Han Y: Dynamic changes and surveillance function
of prion protein expression in gastric cancer drug resistance.
World J Gastroenterol. 17:3986–3993. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Courtney R and Ranganathan S: Simultaneous
adrenocortical carcinoma and neuroblastoma in an infant with a
novel germline p53 mutation. J Pediatr Hematol Oncol. 37:215–218.
2015. View Article : Google Scholar
|
|
32
|
Seema R, Parul S, Nita K and Kamlesh:
High-risk histomorphological features in retinoblastoma and their
association with p53 expression: An Indian experience. Indian J
Ophthalmol. 62:1069–1071. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li ZD, Wang K, Yang XW, Zhuang ZG, Wang JJ
and Tong XW: Expression of aryl hydrocarbon receptor in relation to
p53 status and clinicopathological parameters in breast cancer. Int
J Clin Exp Pathol. 7:7931–7937. 2014.
|
|
34
|
Read ML, Seed RI, Modasia B, Kwan PP,
Sharma N, Smith VE, Watkins RJ, Bansal S, Gagliano T, Stratford AL,
et al: The proto-oncogene PBF binds p53 and is associated with
prognostic features in colorectal cancer. Mol Carcinog. Nov
18–2014.Epub ahead of print. PubMed/NCBI
|
|
35
|
Silva JL, De Moura Gallo CV, Costa DC and
Rangel LP: Prion-like aggregation of mutant p53 in cancer. Trends
Biochem Sci. 39:260–267. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Al-Qasem A, Toulimat M, Tulbah A, Elkum N,
Al-Tweigeri T and Aboussekhra A: The p53 codon 72 polymorphism is
associated with risk and early onset of breast cancer among Saudi
women. Oncol Lett. 3:875–878. 2012.PubMed/NCBI
|
|
37
|
Zaravinos A: An updated overview of
HPV-associated head and neck carcinomas. Oncotarget. 5:3956–3969.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Petit CS, Besnier L, Morel E, Rousset M
and Thenet S: Roles of the cellular prion protein in the regulation
of cell-cell junctions and barrier function. Tissue Barriers.
1:e243772013. View Article : Google Scholar
|
|
39
|
Roucou X: Regulation of PrPC
signaling and processing by dimerization. Front Cell Dev Biol.
2:572014. View Article : Google Scholar
|
|
40
|
Martin-Lannerée S, Hirsch TZ,
Hernandez-Rapp J, Halliez S, Vilotte JL, Launay JM and
Mouillet-Richard S: PrPC from stem cells to cancer.
Front Cell Dev Biol. 2:552014.
|
|
41
|
Cheng Y, Li Y, Liu D, Zhang R and Zhang J:
miR-137 effects on gastric carcinogenesis are mediated by targeting
Cox-2-activated PI3K/AKT signaling pathway. FEBS Lett.
588:3274–3281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xue P, Yang X, Liu Y, Xiong C and Ruan J:
A novel compound RY10-4 downregulates P-glycoprotein expression and
reverses multidrug-resistant phenotype in human breast cancer
MCF-7/ADR cells. Biomed Pharmacother. 68:1049–1056. 2014.
View Article : Google Scholar : PubMed/NCBI
|