Introduction
Colorectal cancer (CRC) is the third most common
cancer worldwide (1) and also the
third leading cause of cancer-related death in Taiwan (2). The pathogenesis of colon cancer is
unclear, but it is believed that family history, genetic
alteration, diet and lifestyle changes are likely involved. The
current cytotoxic drugs in the treatment of CRC include
5-fluorouracil (5-FU) alone or in combination with capecitabine,
irinotecan and/or oxaliplatin (3).
Although 5-FU has been used as the first-line treatment for colon
cancer, the response rate is <20% and patients who have
responded may become resistant (4).
In addition, serious side effects and complications (e.g., fatigue,
pain, diarrhea, nausea, vomiting, and hair loss) can add extra
burden to patients during treatment (5). Thus, it is necessary to seek a novel
antitumor agent including phytochemical compounds in the treatment
of CRC.
The dried root of Astragalus membranaceus
(Fischer) Bge. var. mongolicus (Bge.) Hsiao (AM) is commonly
used in the treatment of common cold, diarrhea, fatigue, and
anorexia (6). The active
constituents of AM include saponins, flavonoids and polysaccharides
(7). Pharmacological studies on AM
have demonstrated its effect on anti-oxidation, anti-inflammation
and enhancement of immune system response (8). There are growing evidence that AM may
be a potential anti-tumorigenic agent. For instance, AM was able to
suppress hepatocarcinogenesis in rats (9). Formononetin, a type of isoflavonoid
isolated from AM, presented anti-angiogenic effect in colon cancer
cells in vitro and in vivo (10). Astragalus saponins have been
studied extensively on tumor growth inhibition, decrease of cell
invasiveness and angiogenesis, and induction of apoptosis (6,11–14).
In combined therapy, 5-FU in combination with Astragalus
polysaccharides enhances chemosensitivity of hepatoma cells
(15). When in co-treatment with
vinorelbine and cisplatin (VC) for patients with advanced non-small
cell lung cancer, Astragalus polysaccharides have improved patients
quality of life compared with VC alone (16).
Microarray technology provides a high throughput
method to simultaneously analyze the gene expression levels of a
given tissue or cell. It provides diagnostic and prognostic values
in clinical trials because it allows investigators to identify the
differentially expressed genes (DEGs) between samples. The
differential labels are often referred to as biomarkers (17). Many common CRC genes have been
characterized over the last decade. The three proposed classes of
genes are oncogenes, tumor suppressor genes and stability genes
(18). In addition to the expected
frequently mutated genes (APC, TP53, SMAD4,
PIK3CA and KRAS) found in CRC, other genes such as
ARID1A, SOX9 and FAM123B were also identified
(19). Furthermore, the emerging
role of microRNA (miRNA) in cancer chemoprevention was recently
described (20–23). However, the changes in miRNA
expression levels and how they are regulated by AM remains
unclear.
Recently, the database Library of Integrated
Network-based Cellular Signatures (LINCS) was produced (24). It is an expression-based
high-throughput screening application for repurposing biomedicines
and for accelerating drug discovery. It works by comparing recorded
profiles of gene expression induced by bioactive molecular
perturbations to those the users had derived in their studies. The
data was made available by the L1000 technology that generated
approximately one million gene expression profiles from 22,412
unique perturbations applied to 56 different human primary cell
lines and human cancer cell lines. LINCS does not only provide
chemical perturbations, but also gives genetic perturbations such
as knockdowns and overexpression of a single gene, a unique feature
among similar applications.
In the present study, we investigated the antitumor
effect of AM on human colorectal cancer HCT116 mouse xenograft and
observed that AM can effectively reduce the tumor growth in nude
mice. Using microarray analysis, we identified genes and miRNAs the
expression levels and molecular mechanisms, and inferred that these
changes may be associated with AM. Utilizing the LINCS database, we
were able to identify chemopreventive drugs that have similar
expression profile that represents the effect of AM. Our results
suggested that AM, through gene regulation, could be a promising
drug for cancer therapy.
Materials and methods
Ethics statement
All animal work was conducted according to relevant
national and international guidelines. The Animal Use Protocol was
reviewed and approved by the Institutional Animal Care and Use
Committee (IACUC) of Taichung Veterans General Hospital, Taiwan.
The IACUC approval number is La-1021107. Period of protocol was
valid from 04/01/2013 to 03/31/2014.
Cell culture condition
Human colorectal cancer cell line HCT116 was
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). The cancer cells were maintained in McCoy's 5A
medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal
bovine serum, 100 U/ml penicillin and 100 mg/ml streptomycin
(Biowest, Nuaille, France). Cells were cultured at 37°C in a
humidified atmosphere of 5% CO2: 95% air.
Preparation of AM
The crude extract of AM was purchased from Sun Ten
Pharmaceutical Corporation and identified by Brion Research
Institute of Taiwan (batch no. M21051). The dried AM (100 g) was
soaked in 1.5 l water for 24 h. The next day, the stock was
decocted in 90°C distilled water for 6 h. The decoction liquid was
filtered and concentrated using a rotary evaporator, and
lyophilized into powder. The dried powder was stored at −20°C until
use. The voucher specimen (accession no. BP002) is deposited in
Professor Li-Jen Su laboratory, National Central University,
Taiwan.
Tumor xenografts in nude mice
Three-week-old male
BALB/cAnN.Cg-Foxnlnu/CrlNarl mice were purchased from
National Applied Research Laboratories (Taipei, Taiwan). All
animals were housed in appropriate cages at 25°C on a 12-h
light/dark cycle with access to food and water ad libitum.
Animals were allowed a period of acclimatization before any
experimentation. Tumors were established by subcutaneous injection
of 1×106 HCT116 cells into the dorsal skin of mice using
25-G needles. Twelve days later, the animals were fed with 500
mg/kg AM or only distilled water every day for 28 days (n=8 for
each group). Tumor growth was monitored once a week for 4 weeks by
measuring two perpendicular diameters. Tumor volume was calculated
according to the formula: a × b × (b/2), where a and b are the
largest and smallest diameters, respectively. The eight p-values of
tumor volume were calculated based on Benjamini-Hochberg method,
which renders a false discovery rate of 0.1. All animals were
euthanized on day 40 and sacrificed by CO2 inhalation.
The tumors were then collected for further analysis.
Microarray experiment
Total RNA was extracted using PureLink®
RNA Mini kit (Ambion, Carlsbad, CA, USA) from cells treated with
control or high doses of AM, according to the manufacturer's
protocol. The concentration, purity and quality of total RNA for
microarray analysis were determined using Nanodrop 2000 (Thermo
Scientific, Wilmington, DE, USA) and had an
OD260/OD280 ratio ranging from 1.9 to 2.1.
The gene expression data was generated using Affymetrix Human
Transcriptome Array 2.0 (HTA-2.0; Affymetrix, Santa Clara, CA,
USA). The cDNA synthesis and labeling were carried out according to
Affymetrix GeneChip® WT PLUS reagent protocol
(Affymetrix). In brief, the total RNAs were first reverse
transcribed to complementary DNA (cDNA) and then to complimentary
RNAs (cRNAs) by in vitro transcription. The cRNAs were
purified and quantified and the second cycle single strand cDNA
synthesis was performed using cRNAs as template. The RNA was
removed and single strand cDNA was fragmented and hybridized to
arrays. All the arrays were scanned with Affymetrix GeneChip 3000
7G (Affymetrix). The Affymetrix quality control report was produced
to ensure the data quality of the processed arrays. All the arrays
have passed the suggested values provided by Affymetrix and
published on GEO (GEO accession no. GSE70772. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=wnaxqakubzglvip&acc=GSE70772).
Analysis of microarray gene expression
profile
The genes differentially expressed in this study
were selected using GeneSpring software, version GX 7.3 (Agilent,
Santa Clara, CA, USA). The log2-transformed expression
intensities with Robust Multiarray Average (RMA) normalization and
Principle Component Analysis (PCA) from 16 arrays (n=8 in each
group) were performed. According to PCA, a total of only 8 arrays
were used (n=4 in each group) to calculate the correlation
coefficient in each cluster set and heatmap was constructed. The
t-test p-value and fold-change for comparing the differentially
gene expression in the treatment and control group were calculated.
A p-value <0.05 and a fold-change of >1.5 were considered as
differentially expressed. The Fisher's exact test was applied to
identify overrepresented Gene Ontology (GO) terms (25) (http://geneontology.org). A P<0.001 was considered
statistically significant.
Real-time PCR validation
For each sample, 2.5 µg of RNA was converted
into cDNA using SuperScript® VILO™ MasterMix
(Invitrogen, Carlsbad, CA, USA) according to manufacturer's
protocol. All primer sets were designed using NCBI Primer-BLAST and
each primer pair was checked for primer-dimer using Beacon designer
software version 8.13 from Premier Biosoft International (Palo
Alto, CA, USA). The primers were further checked using melt curve
analysis. The reaction was performed on ABI ViiA™ 7 Real-Time PCR
system (Applied Biosystems, Life Technologies, Foster City, CA,
USA) using RealQ Plus 2X Master Mix Green as dye (Ampliqon, Odense,
Denmark) according to manufacturer's manual. The reaction condition
is as followed: 95°C 15 min followed by (98°C: 15 sec, 60°C: 30
sec, 72°C: 30 sec) × 40 cycles. All reactions were carried out with
four biological replicates, and each analysis consists of two
technical replicates. The signal of housekeeping gene GAPDH was
used for normalization and the relative expression levels were
calculated using the 2−ΔΔCt method. The primer sequences
are shown in Table I.
 | Table IPrimer sequences used in this
study. |
Table I
Primer sequences used in this
study.
| Gene | Forward | Reverse | Refseq |
|---|
| BRD2 |
ATTCCAGCTCCTCCTCTTCC |
GCAGAGCCAGCTCTCCTAGA | NM 001113182.2 |
| CREBBP |
GGAAACCTTGAGCCATGTGT |
CACAGGATGCAGACTCCAAA | NM 001079846.1 |
| HNRNPU |
CAGAGCCAAATCTCCTCAGC |
CTTTGCCTTTTGACACACCA | NM 004501 |
| KMT2D |
CTCTGGATGGGATTGATGCT |
CGTGGCTCTTCCTGTTCTTC | NM 003482 |
| MYCBP2 |
TCATCCATCCCTGTGTCTCA |
AAAGAGGCAAGCACAGGAAA | NM 015057.4 |
| TRRAP |
ACCTTGGTGTGTGGTGTCAA |
GCCACACGGATGTATGTCTG | NM 003496 |
Database analysis for differentially
expressed miRNAs
Cytoscape was utilized to construct the miRNA-target
gene network (26). The target
genes of miRNAs were identified from miRTarBase (http://mirtarbase.mbc.nctu.edu.tw) (27) and were subjected to DAVID for GO
biological process analysis. The selected terms were based on
Benjamini corrected P<0.05. miRNA pathway analysis was predicted
using DIANA-miRPath (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mirpath/index)
(28) with FDR corrected
P<0.05.
Identification of compound signature
using LINCS
LINCS or the Library or Integrated Network-based
Cellular Signatures project is the extensive version of CMAP
(29) which is available on the
http://lincscloud.org website. Detail of the
website can be found as published (24). In brief, the up and downregulated
gene lists are inserted to the textbox on the search interface.
Click on the search button will reveal lists of matching
experiments in a table format. The three tables are 'compound
connection', 'consensus knockdown connection' and 'overexpression
connection'. For this study, we only focus on compound connection.
The compound selection criteria were based on the mean connectivity
score across four cell lines with a magnitude between −90 and −99,
which corresponds to significant reverse connections.
Statistical analysis
Statistical analysis was performed using Student's
t-test. Differences were considered statistically significant with
P<0.05 unless otherwise specified.
Results
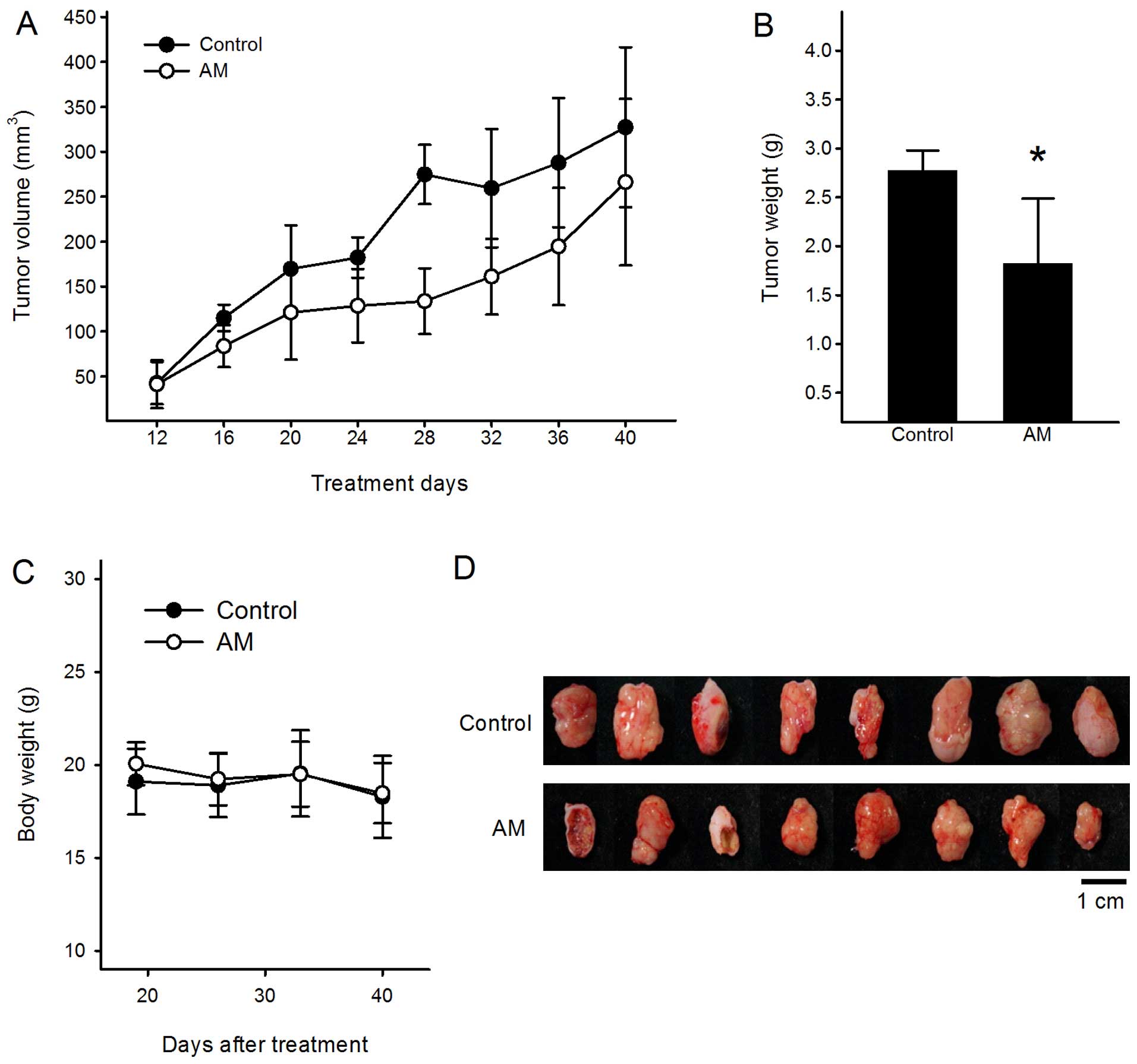
The inhibitive effects of AM in vivo
To test the effect of AM on tumor growth inhibition
in vivo, a human colorectal cancer xenograft nude mouse
model was generated. The mice were orally fed with water or 500
mg/kg AM for 28 days. On the final day, mice were sacrificed and
tumors were inoculated for further studies. As shown in Fig. 1A and B, the tumor volume and final
tumor weight of 500 mg/kg AM groups were reduced by 47.5% (380±109
mm3 in control vs. 199±69 mm3 in AST-treated
mice after 28 days) and 48.4% (2.56±0.49 g in control vs. 1.32±0.74
g in AST-treated mice after 28 days) compared to control,
respectively. However, in no group throughout the experiment, was
significant changes in body weight observed (Fig. 1C), or mortality recorded, unlike the
common side effects of traditional chemotherapy. In addition, the
non-tumor-bearing mouse groups fed with 500 mg/kg AM did not show
any toxicity or change in body weight during the course of
experiment (data not shown). Next, the excised tumors were
evaluated for microarray analysis (Fig.
1D).
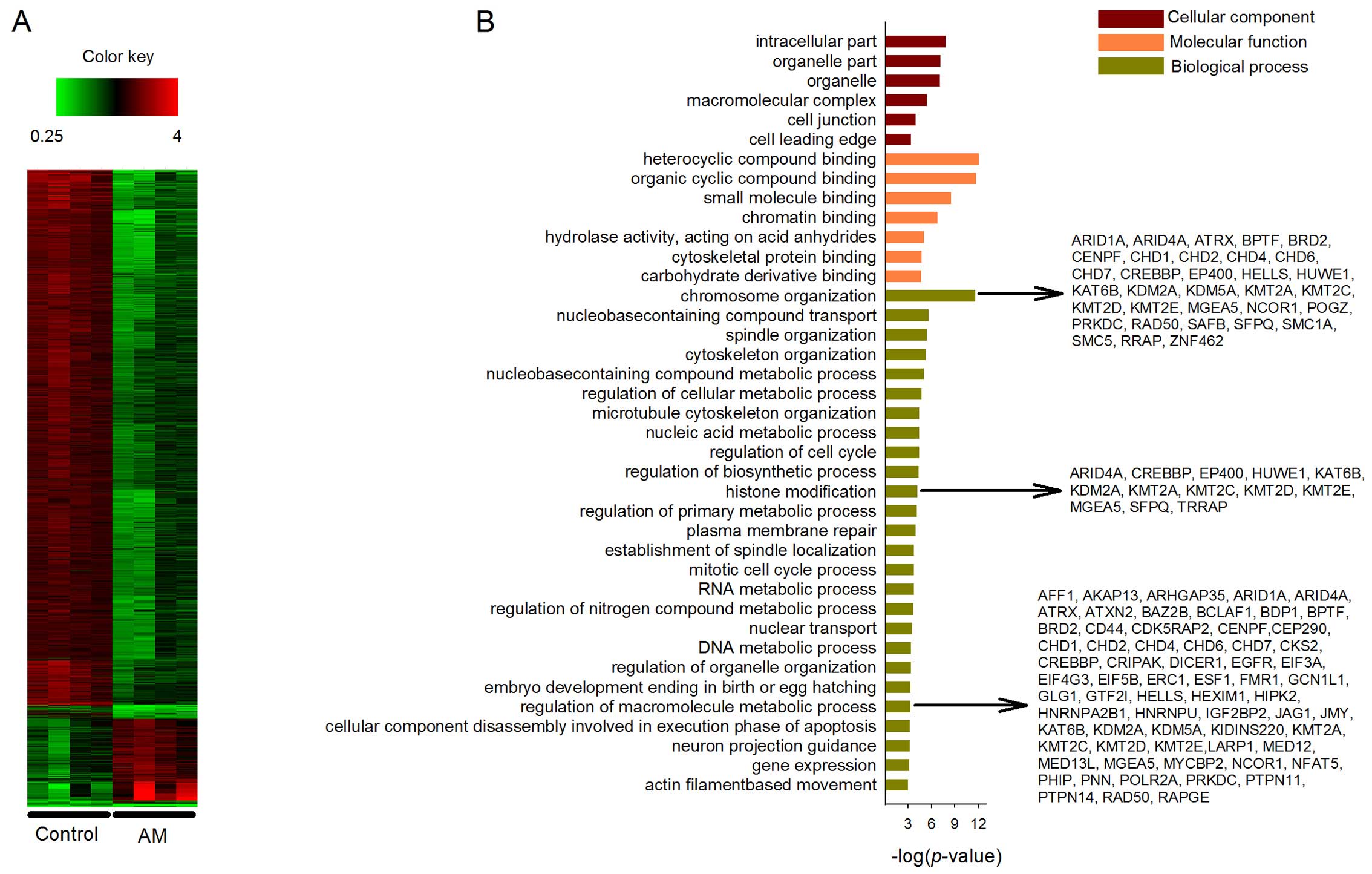
Bioinformatics analysis of molecular
mechanisms
The tumors were processed for microarray
investigation to determine the differential transcriptional
profiles between control and AM-treated groups. After normalization
of the microarray data, a total of 1,454 DEGs were identified,
including 1,257 upregulated and 197 downregulated genes at the
threshold of P<0.05 and fold-change >1.5. The heatmap of gene
expression patterns between control and treatment groups is shown
in Fig. 2A. The significant changes
between control and treatment groups warrant further study.
The DEGs were mapped onto GO for cellular component,
molecular function and biological process analyses. A total of 182
enriched terms covering 224 genes were in the upregulated GO
categories. In Fig. 2B, 6 out of 38
terms were listed under cellular component category. The listed
terms include 'intracellular part', 'organelle part', 'organelle',
'macromolecular complex', 'cell junction' and 'cell leading edge'.
In the molecular function category, 7 out of 45 terms were listed.
The enriched terms include 'heterocyclic compound binding',
'organic cyclic compound binding', 'small molecule binding',
'chromatin binding', 'hydrolase activity, acting on acid
anhydrides', 'cytoskeletal protein binding' and 'carbohydrate
derivative binding'. In the biological process category, a total of
26 out of 99 enriched terms were listed. AM has been widely studied
for its capability of immunopotentiation (30), apoptosis induction (6,31) and
inhibition of inflammation (32)
and invasion (12). The
possibilities that AM may function through suppressing other
emerging cancer hallmarks, such as genome destabilization and
cellular energetics deregulation, remain rarely studied. To
understand such possibilities, we focus the following discussion on
three of the enriched biological processes, which are 'chromosome
organization' (36 genes, P<0.001), 'histone modification' (13
genes, P<0.001) and 'regulation of macromolecule metabolic
process' (86 genes, P<0.001). More specifically at the gene
level, we selected six of the DEGs to quantitatively demonstrate
the effects of AM on their expression levels. The selected genes
are bromodomain containing 2 (BRD2), CREB binding protein
(CREBBP), heterogeneous nuclear ribonucleoprotein U
(HNRNPU), lysine (K)-specific methyltransferase 2D
(KMT2D), MYC binding protein 2, E3 ubiquitin protein ligase
(MYCBP2), and transformation/transcription domain-associated
protein (TRRAP) (Fig. 3A).
All the six genes were downregulated under AM treatment. The
respective downregulated and upregulated DEGs in the control and
treatment groups did not enrich any GO category.
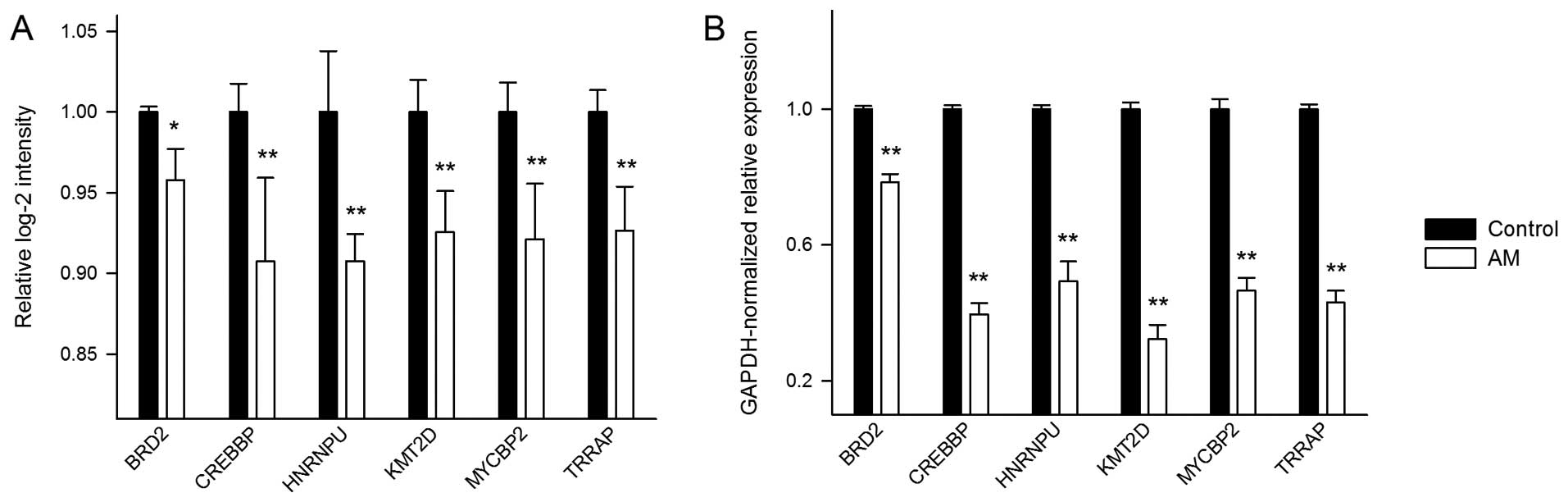
To confirm the above microarray results, we
performed real-time PCR to quantify the expression changes of the
six selected genes (Fig. 3B). The
results show that the genes have wide ranges of Ct values from
20.54 to 33.72. Overall, the real-time PCR expression changes
exhibit a consistent pattern with that from the microarray
analysis.
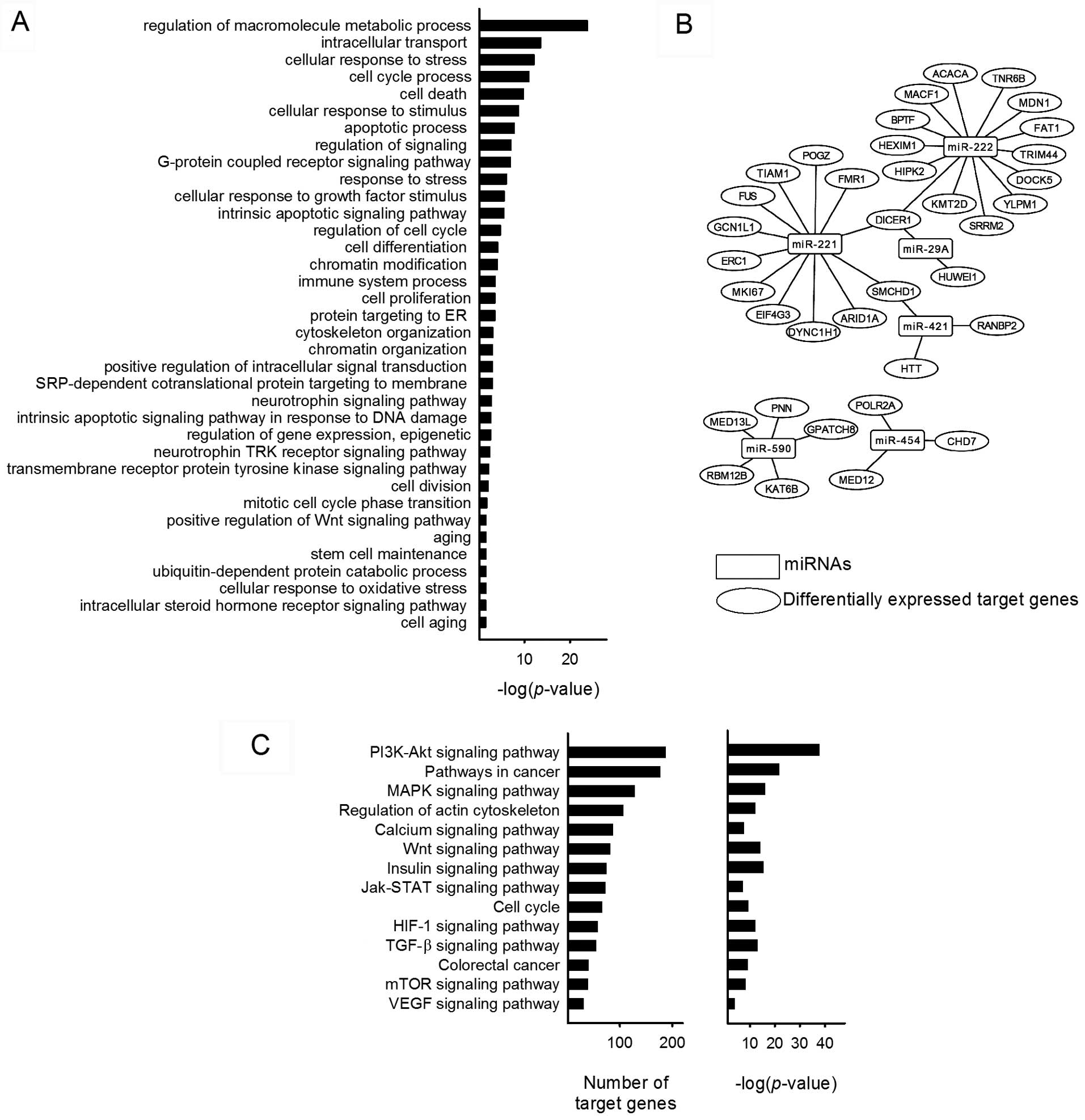
Focal analysis on miRNA
Interestingly, the Affymetrix HTA 2.0 array covers
>40,000 non-coding transcripts on the chip and a total of 29
miRNAs were expressed among the 1,425 DEGs presented on the
heatmap. To eludicate the functions of the miRNAs, miRTarBase was
utilized to identify the target genes. A total of 885 target genes
were identified including 37 DEGs. GO enrichment biological process
analysis of the target genes was associated with terms such as
'regulation of macromolecule metabolic process', 'intracellular
transport', 'cellular response to stress', 'cell cycle process',
'cell death', 'regulation of signaling', 'chromatin modification',
'immune system process', 'cell proliferation', 'transmembrane
receptor protein tyrosine kinase signaling pathway' and 'positive
regulation of Wnt signaling pathway' (Fig. 4A). The network of miRNAs and
differentially expressed target genes are shown in Fig. 4B. These genes were tumor
suppressor-related (ARID1A, FAT1, HIPK2,
MDN1, MED13L, RANBP2, PNN, and
SMCHD1), invasion and migration-related (BPTF,
DOCK5, MED12, TIAM1 and TRIM44), and
cell proliferation-related (BPTF, DICER1, FUS,
HEXIM1, KAT6B, MKI67, and POLR2A). The
pathway analysis of miRNA predicted target genes were analyzed
using DIANA-miRPath. A total of 100 biological pathways were
identified, including many well-known cancer-related pathways such
as PI3k/Akt signaling pathway, MAPK signaling pathway, HIF-1
signaling pathway, JAK/STAT signaling pathway and VEGF signaling
pathway. PI3K/Akt pathway has the most identified target genes
among the predicted biological pathways and miR-590 was present in
all of the selected 14 pathways (Fig.
4C). Around 40% of the pathways are implicated in
carcinogenesis, suggesting the novel role of AM on miRNA
regulation.
Drug predictions using LINCS
LINCS derives perturbagents of both aggravate or
reverse directions, thus allowing users to identify lists of
harmful or beneficial drugs for possible therapeutic treatments. To
search for small molecules that have similar effect as AM, that it,
decrease in gene expression after administration of drug, we have
identified a total of 71 significant perturbagens. The top 14
perturbagens are listed in Table
II. The drugs include antitumor effect such as SN-38,
teniposide, amsarcrine, topotecan, camptothecin, irinotecan and
etoposide. Other therapeutic drugs such as mitomycin-c, celastrol
and mycophenolic acid were also listed. The results suggested that
the effect of AM is similar to the perturbagens listed in Table II. This provides support for the
reliability of our microarray study.
 | Table IIThe top perturbagen hits from
LINCS. |
Table II
The top perturbagen hits from
LINCS.
| Perturbagen ID | Perturbagen
name | Scorea | Therapeutic
use |
|---|
| BRD-A02481876 | Importazole | −100.0 | None |
| BRD-A36630025 | SN-38 | −99.9 |
Anti-neoplastic |
| BRD-A48237631 | Mitomycin-c | −99.7 | Antibiotic |
| BRD-A80960055 | Celastrol | −99.6 |
Anti-inflammation |
| BRD-A35588707 | Teniposide | −99.4 |
Anti-neoplastic |
| BRD-K98490050 | Amsacrine | −99.4 |
Anti-neoplastic |
| BRD-A59985574 | Topotecan | −99.3 |
Anti-neoplastic |
| BRD-A30437061 | Camptothecin | −99.2 |
Anti-neoplastic |
| BRD-K08547377 | Irinotecan | −98.5 |
Anti-neoplastic |
| BRD-K31542390 |
Mycophenolic-acid | −98.4 |
Immunosuppressant |
| BRD-K80622725 | STK-397047 | −97.8 | None |
| BRD-K31342827 |
Bisindolylmaleimide | −97.6 | None |
| BRD-A18419789 | Etoposide | −97.4 |
Anti-neoplastic |
| BRD-K53792571 | Inhibitor-BEC | −97.4 | None |
Discussion
In the present study, we have demonstrated that AM
can effectively reduce the tumor growth in nude mice without
significantly altering the mouse body weight. DEGs extracted from
microarray gene expression profile indicate statistically
significant functional changes including catalytic activity,
binding, cellular component, biological process, biological
regulation and cellular component organization. These changes
correlate well with morphological changes in mouse tumors. For
example, terms such as chromosome organization, histone
modification and regulation of macromolecule metabolic process were
upregulated and downregulated in control and treatment groups,
respectively. These terms are rarely described in TCM, suggesting a
new therapeutic use of AM.
Our microarray data revealed that many
epigenetic-related genes including KMT2D, BRD2,
CREBBP, ARID1A, were altered in AM-treated groups.
Hsieh et al tested 3,294 TCMs and found that ~29.8% have the
potential to affect the epigenome of the human cells (33). Epigenetics is the change in gene
expression independent of alteration in DNA sequences (34). Its modification involves changes in
DNA methylation, histone modification and miRNAs expression
(23). In cancer cells, two forms
of epigenetics are commonly observed. One is hypomethylation of the
proto-oncogenes or increase in expression of genes implicated in
tumor progression. On the other hand, tumor suppressor genes were
silenced due to hypermethylation on their promoter region. These
reactions are catalyzed by a family of enzymes called DNA
methyltransferases (DNMTs), histone acetyltransferase (HATs) and
histone deacetylase (HDACs) (35,36).
KMT2D is an important component of the multi-protein complex
that directs epigenetics regulated genes and embryonic development
(37,38). KMT2D are frequently mutated
in malignant cells. For instance, KMT2D contains recurrent
mutation in 90% of the cases in follicular lymphoma (39). Furthermore, high levels of
KMT2D are associated with breast and colon cancer
malignancies (40). Another
property of KMT2D is its ability to promote cell migration
(41); thus it is possible that the
reduction of KMT2D in the treatment group was affected by AM
since the roots of Astragalus membranaceus and/or
Astragalus saponins have been shown to attenuate invasion
and migration of cancer cell lines (12,42).
Interestingly, KMT2D also plays a part in regulating calcium
signaling pathways (41,43) and deregulation of this pathway may
mediate tumor growth (44–46). Astragaloside IV, one of the main
constituents found in AM, was able to reduce the excess
intracellular calcium by improving the activity of calcium pump in
myocardial cells (8,47), suggesting astragaloside IV is
valuable in enhancing tumor inhibition by implicating the calcium
signaling pathway.
Small molecules targeting bromodomain, especially
the BET family (BRD1, BRD2, BRD3, BRD4,
and BRDT), can be a promising therapeutic strategy in cancer
treatment. The bromodomain is an evolutionarily conserved protein
that act as 'reader' of histone acetylation and regulates
protein-protein interaction via lysine residue (34,48).
BRD2, the potential oncogene in humans, was downregulated in
our mouse xenograft treatment group. A few BET protein inhibitors
were designed to target the acetylated histones and the
bromodomains (49) and inhibition
of BET proteins can ultimately reduce the transcription of
proto-oncogenes such as MYC, BCL2 and CDK6
(50). Furthermore, BRD2
expression can result in B cell maglinancy, which is activated by
nuclear factor-κB (NF-κB)-regulated genes (51). Consistent with this finding, another
study has shown that BRD2 can directly regulate NF-κB
activity in a macrophage cell line and downregulation of
BRD2 ultimately decreases NF-κB activity (52). NF-κB is pivatol in the control of
inflammation, cell proliferation and apoptosis. AM has long been
known for its anticancer effect by activating anti-inflammatory and
immune boost properties. For instance, saponins found in AM were
able to induce tumor growth inhibition and promote apoptosis
through ERK-independent NF-κB signaling pathway (11). We suspected that AM regulates NF-κB
activity by altering BRD2 expression.
Energy metabolism reprogramming is one of the
emerging hallmarks of many cancers (53,54).
One of the factors facilitating uncontrollable proliferation of
cancer cells is their high rates of aerobic glycolysis, a
phenomenon known as the 'Warburg effect' (55,56).
The factor affords tumor cells even more abundant ATPs than can be
produced through oxidative phosphorylation. The degraded glucose
through glycolysis recursively fuels proliferation by providing
intermediates needed for synthesizing amino acids and nucleosides
(53,57), the respective building blocks of
protein and DNA. MYC is one of the main oncogenes in cancer
cells by promoting glucose transporter 1 (GLUT1) and
contributes to the Warburg effect (54). In our microarray profile, we have
identified MYC-associated genes such as MYCBP2
(58), HNRNPU (59) and TRRAP (60), all of which play a role in
regulating MYC expression, as having been downregulated by
AM. Studies have shown that MYCBP2 positively regulates with
mTOR signaling (61), which
controls the metabolic reprogramming of cancer cells (62), and during metabolic stress, mTOR is
inactivated by AMP-activated protein kinase (AMPK) (54). Astragalus polysacchride is known to
activate AMPK to alleviate glucose toxicity in diabetes (63). Furthermore, drugs such as metaformin
and phenformin that were developed to target type 2 diabetes may
have use in treating cancers by triggering AMPK in cells (64,65),
suggesting AM, like meta-formin or phenformin, is an excellent
adjuvant drug possibly by activating AMPK and ultimately decreasing
mTOR and MYCBP2 expression.
miRNAs are a family of small, evolutionary
conserved, endogenous and non-coding RNAs that regulate gene
expression in broad range of animals, plants and viruses. They play
critical regulatory roles in a large number of biological processes
such as cellular differentiation, proliferation, migration and
apoptosis (66,67). Several studies had focused on TCMs
and their effect on miRNA expression. For example, curcumin and its
derivatives were able to alter miRNA signature profiles in various
cancers including colon (23,68).
Bufalin, one of the active ingredients of Venenum bufonis,
can inhibit colon cancer metastasis by acting synergistically with
miR-497 (69). However, studies on
AM in modulating miRNA expression in cancers are limited. Here, we
identified 29 differentially expressed miRNAs and found that
miR-590 has the most identified target genes and is involved in all
of the selected miRNA predictive pathways. miR-590, a possible
oncomiR, was overexpressed in many tumors (70–72).
Database mining identified TGFBR2 is one of the target genes
of miR-590-5p. Studies have found that mutation of TGFBR2
occurs in 25% of CRC and in >90% of microsatellite
instability-high CRC (73). These
findings shed light on the additional roles of AM in miRNA
regulation.
The results from LINCS indicate that many
chempreventive drugs have profiles similar to that of AM.
Interestingly, many of the identified antitumor agents were of
natural origin. For example, etoposide and teniposide are the
derivative of podophyllotoxin which was found in the plant
Podophyllum peltatum, SN-38, topotecan and irinotecan are
the derivatives of camptothecin which was isolated from
Camptotheca acuminata (74).
Celastrol is another TCM-isolated chemical compound from the root
of Trypterigium wilfordii. More recently, studies have shown
that celastrol, in addition to its anti-inflammatory activities,
can inhibit proliferation of various human cancer cells (75). We have also identified curcumin
(score: −92.5) as another natural product that have similar gene
expression effect as AM. Curcumin is by far the most studied
TCM-based anticancer drug. It can modulate the growth of tumor
cells by regulating several pathways such as NF-κB, PI3K/Akt,
JAK/STAT, AMPK and mTOR, all of which were also observed in AM.
Although more work is needed to further clarify the
effects of AM at the molecular level, our preliminary results show
that crude extract of AM inhibits growth of CRC in vivo
without apparent toxicity and side effect. Our bioinformatics
analysis provides preliminary insight into the molecular mechanisms
of AM, while the identified genes and miRNAs offer clues to
explaining the reduced growth of mouse xenografted tumors. Our
derived gene expression profile provides valuable information for
selecting biomarkers and could contribute to future discovery of
novel drugs for CRC.
Acknowledgments
The present study was supported by grants from the
Ministry of Science and Technology (NSC 101-2320-B-008-001-MY3 and
MOST-104-2320-B-182A-010). AM is kindly provided by Sun Ten
Pharmaceutical Corporation. The authors thank Professor Sun-Chong
Wang from National Central University for valuable comments and
technical support from the Core Facility of High Throughput
Experimental Analysis from Institute of Systems Biology and
Bioinformatics, National Central University, Taiwan. The Core
Facility of High Throughput Experimental Analysis is supported by
Aim for Top University Project from the Ministry of Education.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
2
|
Su SY, Huang JY, Jian ZH, Ho CC, Lung CC
and Liaw YP: Mortality of colorectal cancer in Taiwan, 1971–2010:
Temporal changes and age-period-cohort analysis. Int J Colorectal
Dis. 27:1665–1672. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dietvorst MH and Eskens FA: Current and
novel treatment options for metastatic colorectal cancer: Emphasis
on Aflibercept. Biol Ther. 3:25–33. 2013. View Article : Google Scholar
|
|
4
|
Wong CS, Wong VW, Chan CM, Ma BB, Hui EP,
Wong MC, Lam MY, Au TC, Chan WH, Cheuk W, et al: Identification of
5-fluorouracil response proteins in colorectal carcinoma cell line
SW480 by two-dimensional electrophoresis and MALDI-TOF mass
spectrometry. Oncol Rep. 20:89–98. 2008.PubMed/NCBI
|
|
5
|
Qi F, Li A, Inagaki Y, Gao J, Li J, Kokudo
N, Li XK and Tang W: Chinese herbal medicines as adjuvant treatment
during chemo- or radio-therapy for cancer. Biosci Trends.
4:297–307. 2010.
|
|
6
|
Tin MM, Cho CH, Chan K, James AE and Ko
JK: Astragalus saponins induce growth inhibition and apoptosis in
human colon cancer cells and tumor xenograft. Carcinogenesis.
28:1347–1355. 2007. View Article : Google Scholar
|
|
7
|
Ma XQ, Shi Q, Duan JA, Dong TT and Tsim
KW: Chemical analysis of Radix Astragali (Huangqi) in China: A
comparison with its adulterants and seasonal variations. J Agric
Food Chem. 50:4861–4866. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ren S, Zhang H, Mu Y, Sun M and Liu P:
Pharmacological effects of Astragaloside IV: A literature review. J
Tradit Chin Med. 33:413–416. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cui R, He J, Wang B, Zhang F, Chen G, Yin
S and Shen H: Suppressive effect of Astragalus membranaceus Bunge
on chemical hepatocarcinogenesis in rats. Cancer Chemother
Pharmacol. 51:75–80. 2003. View Article : Google Scholar
|
|
10
|
Auyeung KK, Law PC and Ko JK: Novel
anti-angiogenic effects of formononetin in human colon cancer cells
and tumor xenograft. Oncol Rep. 28:2188–2194. 2012.PubMed/NCBI
|
|
11
|
Auyeung KK, Law PC and Ko JK: Astragalus
saponins induce apoptosis via an ERK-independent NF-κB signaling
pathway in the human hepatocellular HepG2 cell line. Int J Mol Med.
23:189–196. 2009.PubMed/NCBI
|
|
12
|
Auyeung KK, Woo PK, Law PC and Ko JK:
Astragalus saponins modulate cell invasiveness and angiogenesis in
human gastric adenocarcinoma cells. J Ethnopharmacol. 141:635–641.
2012. View Article : Google Scholar
|
|
13
|
Law PC, Auyeung KK, Chan LY and Ko JK:
Astragalus saponins downregulate vascular endothelial growth factor
under cobalt chloride-stimulated hypoxia in colon cancer cells. BMC
Complement Altern Med. 12:1602012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang T, Xuan X, Li M, Gao P, Zheng Y, Zang
W and Zhao G: Astragalus saponins affect proliferation, invasion
and apoptosis of gastric cancer BGC-823 cells. Diagn Pathol.
8:1792013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tian QE, De Li H, Yan M, Cai HL, Tan QY
and Zhang WY: Effects of Astragalus polysaccharides on
P-glycoprotein efflux pump function and protein expression in H22
hepatoma cells in vitro. BMC Complement Altern Med. 12:942012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo L, Bai SP, Zhao L and Wang XH:
Astragalus polysaccharide injection integrated with vinorelbine and
cisplatin for patients with advanced non-small cell lung cancer:
Effects on quality of life and survival. Med Oncol. 29:1656–1662.
2012. View Article : Google Scholar
|
|
17
|
Chu W, Ghahramani Z, Falciani F and Wild
DL: Biomarker discovery in microarray gene expression data with
Gaussian processes. Bioinformatics. 21:3385–3393. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nat Med. 10:789–799. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cancer Genome Atlas Network: Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karius T, Schnekenburger M, Dicato M and
Diederich M: MicroRNAs in cancer management and their modulation by
dietary agents. Biochem Pharmacol. 83:1591–1601. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y, Kong D, Wang Z and Sarkar FH:
Regulation of microRNAs by natural agents: An emerging field in
chemoprevention and chemotherapy research. Pharm Res. 27:1027–1041.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Neelakandan K, Babu P and Nair S: Emerging
roles for modulation of microRNA signatures in cancer
chemoprevention. Curr Cancer Drug Targets. 12:716–740. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Teiten MH, Dicato M and Diederich M:
Curcumin as a regulator of epigenetic events. Mol Nutr Food Res.
57:1619–1629. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Duan Q, Flynn C, Niepel M, Hafner M,
Muhlich JL, Fernandez NF, Rouillard AD, Tan CM, Chen EY, Golub TR,
et al: LINCS Canvas Browser: Interactive web app to query, browse
and interrogate LINCS L1000 gene expression signatures. Nucleic
Acids Res. 42:W449–W460. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar :
|
|
27
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res. 42:D78–D85.
2014. View Article : Google Scholar :
|
|
28
|
Vlachos IS, Kostoulas N, Vergoulis T,
Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD,
Prionidis K, Dalamagas T and Hatzigeorgiou AG: DIANA miRPath v.2.0:
Investigating the combinatorial effect of microRNAs in pathways.
Nucleic Acids Res. 40:W498–W504. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lamb J: The Connectivity Map: A new tool
for biomedical research. Nat Rev Cancer. 7:54–60. 2007. View Article : Google Scholar
|
|
30
|
Shao BM, Xu W, Dai H, Tu P, Li Z and Gao
XM: A study on the immune receptors for polysaccharides from the
roots of Astragalus membranaceus, a Chinese medicinal herb. Biochem
Biophys Res Commun. 320:1103–1111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Auyeung KK, Mok NL, Wong CM, Cho CH and Ko
JK: Astragalus saponins modulate mTOR and ERK signaling to promote
apoptosis through the extrinsic pathway in HT-29 colon cancer
cells. Int J Mol Med. 26:341–349. 2010.PubMed/NCBI
|
|
32
|
Qin Q, Niu J, Wang Z, Xu W, Qiao Z and Gu
Y: Astragalus membranaceus inhibits inflammation via phospho-P38
mitogen-activated protein kinase (MAPK) and nuclear factor (NF)-κB
pathways in advanced glycation end product-stimulated macrophages.
Int J Mol Sci. 13:8379–8387. 2012. View Article : Google Scholar
|
|
33
|
Hsieh HY, Chiu PH and Wang SC: Epigenetics
in traditional chinese pharmacy: A bioinformatic study at
pharmacopoeia scale. Evid Based Complement Alternat Med.
2011:8167142011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Virani S, Colacino JA, Kim JH and Rozek
LS: Cancer epigenetics: A brief review. ILAR J. 53:359–369. 2012.
View Article : Google Scholar
|
|
36
|
Feinberg AP and Tycko B: The history of
cancer epigenetics. Nat Rev Cancer. 4:143–153. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Demers C, Chaturvedi CP, Ranish JA, Juban
G, Lai P, Morle F, Aebersold R, Dilworth FJ, Groudine M and Brand
M: Activator-mediated recruitment of the MLL2 methyltransferase
complex to the beta-globin locus. Mol Cell. 27:573–584. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Glaser S, Lubitz S, Loveland KL, Ohbo K,
Robb L, Schwenk F, Seibler J, Roellig D, Kranz A, Anastassiadis K,
et al: The histone 3 lysine 4 methyltransferase, Mll2, is only
required briefly in development and spermatogenesis. Epigenetics
Chromatin. 2:52009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Morin RD, Mendez-Lago M, Mungall AJ, Goya
R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field
M, et al: Frequent mutation of histone-modifying genes in
non-Hodgkin lymphoma. Nature. 476:298–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Natarajan TG, Kallakury BV, Sheehan CE,
Bartlett MB, Ganesan N, Preet A, Ross JS and Fitzgerald KT:
Epigenetic regulator MLL2 shows altered expression in cancer cell
lines and tumors from human breast and colon. Cancer Cell Int.
10:132010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guo C, Chen LH, Huang Y, Chang CC, Wang P,
Pirozzi CJ, Qin X, Bao X, Greer PK, McLendon RE, et al: KMT2D
maintains neoplastic cell proliferation and global histone H3
lysine 4 monomethylation. Oncotarget. 4:2144–2153. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu JJ, Sun WY, Hu SS, Zhang S and Wei W: A
standardized extract from Paeonia lactiflora and Astragalus
membranaceus induces apoptosis and inhibits the proliferation,
migration and invasion of human hepatoma cell lines. Int J Oncol.
43:1643–1651. 2013.PubMed/NCBI
|
|
43
|
Guo C, Chang CC, Wortham M, Chen LH,
Kernagis DN, Qin X, Cho YW, Chi JT, Grant GA, McLendon RE, et al:
Global identification of MLL2-targeted loci reveals MLL2's role in
diverse signaling pathways. Proc Natl Acad Sci USA.
109:17603–17608. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kim SY, Yang D, Myeong J, Ha K, Kim SH,
Park EJ, Kim IG, Cho NH, Lee KP, Jeon JH, et al: Regulation of
calcium influx and signaling pathway in cancer cells via
TRPV6-Numb1 interaction. Cell Calcium. 53:102–111. 2013. View Article : Google Scholar
|
|
45
|
Monteith GR, Davis FM and Roberts-Thomson
SJ: Calcium channels and pumps in cancer: Changes and consequences.
J Biol Chem. 287:31666–31673. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang H, Zhang Q, He J and Lu W: Regulation
of calcium signaling in lung cancer. J Thorac Dis. 2:52–56.
2010.PubMed/NCBI
|
|
47
|
Li ZP and Cao Q: Effects of astragaloside
IV on myocardial calcium transport and cardiac function in ischemic
rats. Acta Pharmacol Sin. 23:898–904. 2002.PubMed/NCBI
|
|
48
|
Zeng L and Zhou MM: Bromodomain: An
acetyl-lysine binding domain. FEBS Lett. 513:124–128. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Belkina AC and Denis GV: BET domain
co-regulators in obesity, inflammation and cancer. Nat Rev Cancer.
12:465–477. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dawson MA, Prinjha RK, Dittmann A,
Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C,
Savitski MM, et al: Inhibition of BET recruitment to chromatin as
an effective treatment for MLL-fusion leukaemia. Nature.
478:529–533. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Greenwald RJ, Tumang JR, Sinha A, Currier
N, Cardiff RD, Rothstein TL, Faller DV and Denis GV: E mu-BRD2
transgenic mice develop B-cell lymphoma and leukemia. Blood.
103:1475–1484. 2004. View Article : Google Scholar
|
|
52
|
Belkina AC, Nikolajczyk BS and Denis GV:
BET protein function is required for inflammation: Brd2 genetic
disruption and BET inhibitor JQ1 impair mouse macrophage
inflammatory responses. J Immunol. 190:3670–3678. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Muñoz-Pinedo C, El Mjiyad N and Ricci JE:
Cancer metabolism: Current perspectives and future directions. Cell
Death Dis. 3:e2482012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Warburg O, Posener K and Negelein E: Ueber
den stoffwechsel der tumoren. Biochem Z. 152:319–344. 1924.In
German.
|
|
56
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
DeBerardinis RJ, Lum JJ, Hatzivassiliou G
and Thompson CB: The biology of cancer: Metabolic reprogramming
fuels cell growth and proliferation. Cell Metab. 7:11–20. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Han S, Kim S, Bahl S, Li L, Burande CF,
Smith N, James M, Beauchamp RL, Bhide P, DiAntonio A, et al: The E3
ubiquitin ligase protein associated with Myc (Pam) regulates
mammalian/mechanistic target of rapamycin complex 1 (mTORC1)
signaling in vivo through N- and C-terminal domains. J Biol Chem.
287:30063–30072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Weidensdorfer D, Stöhr N, Baude A, Lederer
M, Köhn M, Schierhorn A, Buchmeier S, Wahle E and Hüttelmaier S:
Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic
RNPs. RNA. 15:104–115. 2009. View Article : Google Scholar :
|
|
60
|
Nikiforov MA, Chandriani S, Park J,
Kotenko I, Matheos D, Johnsson A, McMahon SB and Cole MD:
TRRAP-dependent and TRRAP-independent transcriptional activation by
Myc family oncoproteins. Mol Cell Biol. 22:5054–5063. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Han S, Witt RM, Santos TM, Polizzano C,
Sabatini BL and Ramesh V: Pam (protein associated with Myc)
functions as an E3 ubiquitin ligase and regulates TSC/mTOR
signaling. Cell Signal. 20:1084–1091. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yecies JL and Manning BD: Transcriptional
control of cellular metabolism by mTOR signaling. Cancer Res.
71:2815–2820. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zou F, Mao XQ, Wang N, Liu J and Ou-Yang
JP: Astragalus polysaccharides alleviates glucose toxicity and
restores glucose homeostasis in diabetic states via activation of
AMPK. Acta Pharmacol Sin. 30:1607–1615. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhou G, Myers R, Li Y, Chen Y, Shen X,
Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, et al: Role of
AMP-activated protein kinase in mechanism of metformin action. J
Clin Invest. 108:1167–1174. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
de Krijger I, Mekenkamp LJ, Punt CJ and
Nagtegaal ID: MicroRNAs in colorectal cancer metastasis. J Pathol.
224:438–447. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Dong H, Lei J, Ding L, Wen Y, Ju H and
Zhang X: MicroRNA: Function, detection, and bioanalysis. Chem Rev.
113:6207–6233. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Roy S, Levi E, Majumdar AP and Sarkar FH:
Expression of miR-34 is lost in colon cancer which can be
re-expressed by a novel agent CDF. J Hematol Oncol. 5:582012.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Qiu YY, Hu Q, Tang QF, Feng W, Hu SJ,
Liang B, Peng W and Yin PH: MicroRNA-497 and bufalin act
synergistically to inhibit colorectal cancer metastasis. Tumour
Biol. 35:2599–2606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chu Y, Ouyang Y, Wang F, Zheng A, Bai L,
Han L, Chen Y and Wang H: MicroRNA-590 promotes cervical cancer
cell growth and invasion by targeting CHL1. J Cell Biochem.
115:847–853. 2014. View Article : Google Scholar
|
|
71
|
Jiang X, Xiang G, Wang Y, Zhang L, Yang X,
Cao L, Peng H, Xue P and Chen D: MicroRNA-590-5p regulates
proliferation and invasion in human hepatocellular carcinoma cells
by targeting TGF-β RII. Mol Cells. 33:545–551. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yang H, Zheng W, Zhao W, Guan C and An J:
Roles of miR-590-5p and miR-590-3p in the development of
hepatocellular carcinoma. Nan Fang Yi Ke Da Xue Xue Bao.
33:804–811. 2013.In Chinese. PubMed/NCBI
|
|
73
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar
|
|
74
|
Efferth T, Li PC, Konkimalla VS and Kaina
B: From traditional Chinese medicine to rational cancer therapy.
Trends Mol Med. 13:353–361. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kannaiyan R, Manu KA, Chen L, Li F,
Rajendran P, Subramaniam A, Lam P, Kumar AP and Sethi G: Celastrol
inhibits tumor cell proliferation and promotes apoptosis through
the activation of c-Jun N-terminal kinase and suppression of PI3
K/Akt signaling pathways. Apoptosis. 16:1028–1041. 2011. View Article : Google Scholar : PubMed/NCBI
|