Introduction
Copines are a family of C2 domain-containing
calcium-dependent lipid-binding proteins first identified in
Paramecium tetraurelia and are evolutionally conserved from
Arabidopsis to Homo sapiens (1). To date, nine human Copine family
members have been identified (2,3). The
various functions of this protein family have been shown to be
related to cell growth, cancer development and neuronal
differentiation (4–6). Copine3, a member of this family, is
known to be a membrane binding protein with two tandem C2 domains,
designated C2A and C2B, at the N-terminus followed by an ʻA domainʼ
in the C-terminal region (7).
Previously, Copine3 was shown to bind to the phosphorylated Tyr1248
of receptor tyrosine kinase 2 (ErbB2), indicating that it may play
a regulatory role in ErbB2-dependent breast cancer cell motility
(8). However, the full mechanism of
Copine3 in ErbB2 function is largely unknown.
Jun activation domain-binding protein 1 (Jab1) was
originally cloned as a co-activator of activator protein 1 (AP-1)
(9). Jab1, also named Cops5, is the
fifth component of the COP9 signalosome complex (10), and is evolutionarily conserved in
mammals, plants and yeast (11).
Furthermore, the deletion of Jab1 in mice results in early
embryonic lethality due to impaired cellular proliferation and
accelerated apoptosis (12,13). It has also been shown that Jab1 is
involved in cell cycle progression, apoptosis, signal transduction
and proliferation and is overexpressed in various types of cancer,
including breast cancer (14–16).
In addition, several binding partners of Jab1 have
been reported, further widening its function. For example,
following interaction with Jab1, cyclin-dependent kinase inhibitor
1B (p27Kip1) and tumor-suppressor p53 are
degraded in the proteasome (17,18).
In contrast, Jab1 appears to stabilize hypoxia-inducible factor 1α
(HIF-1α) activation as well as the c-Jun/AP-1 complex (9,19). The
binding of Jab1 to its various partners is likely mediated by one
of its four binding domains, which include a Jun binding domain at
the N-terminus followed by an Mpr1p and PAD1p N-terminal (MPN)
domain. Other proteins with this MPN domain include proteasome
regulatory subunits, eukaryotic initiation factor 3 (eIF3)
subunits, and regulators of transcription factor expression and
function (20). A nuclear export
signal and p27 binding domain are also found in the C-terminal
region of Jab1 (14). The presence
of these various binding motifs likely mediates the binding of Jab1
to a number of its protein and protein complex partners (21).
In the present study, we have shown that Copine3
binds to Jab1 in vitro and in vivo, and have further
investigated the binding region between these two proteins.
Furthermore, we also provide data supporting the ability of Jab1 to
regulate Copine3-mediated ErbB2 signaling activity, whereby
Jab1/Copine3 binding appears to increase ErbB2 protein activity as
well as downstream cellular migration in SKBr3 breast cancer cells.
To our knowledge, this is the first time Copine3/Jab1 binding and
its downstream effects have been investigated.
Materials and methods
Plasmid constructs and mutagenesis
Full-length Copine3 cDNA (GenBank Accession No.
NM_003909) was obtained using rT-PCR with rNA isolated from HEK
293T cells. The full-length clone was generated using this cDNA as
a template via a PCR-based gateway cloning method previously
described (22). C2A, C2B and A
domain mutants were obtained using directed PCR in conjunction with
primers specific for each region, thus deleting the other domains
in the full-length cDNA transcript. Full-length Jab1 cDNA (GenBank
Accession No. NM_006837) was obtained from the Korea research
Institute of Bioscience and Biotechnology (KRIBB; Daejoen, Korea;
hMU009054). N-, MPN and C-terminal domains mutants were obtained
using PCR primers specific for each domain. The resulting PCR
products were cloned into the destination vector pDEST-AD-GFP using
the gateway cloning system (Invitrogen).
Cell culture
The HEK 293T, COS7, SKBr3 and human breast cancer
cell lines used in the present study were all maintained in
Dulbecco's modified Eagle's medium (DMEM) supplemented with 1%
penicillin-streptomycin and 10% fetal bovine serum (FBS) at 37°C
with 5% CO2.
Yeast two-hybrid assay
The Copine3 gene was ligated into pGBKT7, which
encodes a GAL4 DNA binding domain (BD), while the Jab1 gene was
cloned into pGADT7, which encodes an activation domain (AD). To
assess the protein-protein interaction between Copine3 and Jab1,
both BD/Copine3 and AD/Jab1 were co-transformed into the yeast
strain AH109. The transformed yeast was then cultured in SD
synthetic medium lacking leucine, tryptophan and histidine.
Adenovirus amplification and
infection
Adenovirus was prepared and propagated in HEK 293A
cells using the ViraPower™ Adenoviral Expression System
(Invitrogen). pDEST-AD-GFP- and pDEST-AD-mcherry-tagged genes
expression vectors were transfected into HEK 293A cells to obtain
adenovirus particles. After 7–10 days, virus particles were
harvested from the cells and media, followed by purification via
centrifugation at 3,000 rpm for 15 min. For adenoviral infection,
COS7 or SKBr3 cells were plated into 6-well plates at a density of
1×105 cells/ml and infected with adenovirus at a
multiplicity of infection (MOI) of 100. The infected cells were
then incubated for 48 h at 37°C.
Confocal microscopy
For our imaging analysis, we used the COS7 cells
infected with GFP-Copine3 and mcherry-Jab1 genes. Further, several
randomly chosen fields from multiple wells of cells were
photographed using a confocal microscope (Olympus FluoView
FV1000).
Western blot analysis and
immunoprecipitation (IP)
HEK 293T and SKBr3 cells were lysed with RIPA buffer
(50 mM Tris-HCl, pH 7.4, 150 mM EDTA, 1 mM PMSF and 1% NP-40)
containing a protease-inhibitor cocktail. Whole-cell lysates were
incubated on ice for 30 min and then cleared at 20,000 × g for 20
min at 4°C. For the IP analysis, the resulting supernatants were
incubated with the indicated antibodies for 3 h, at 4°C with an
additional incubation for 2 h after the addition of protein A/G
plus agarose (Santa Cruz Biotechnology). The immunocomplexes
captured in the agarose gel were then washed three times with RIPA
buffer and eluted by boiling with SDS gel-loading buffer. The
immunoprecipitates were then analyzed by western blotting.
For the western blot analysis, the supernatants were
separated by SDS-PAGE using 10% gels and blotted onto PVDF
membranes. The blots were then probed with anti-Jab1 (1:1,000),
anti-Copine3 (1:1,000) (both from Santa Cruz Biotechnology),
anti-ErbB2 (1:2,000, Abcam), anti-PI3 kinase (1:1,000),
anti-phospho-PI3 kinase (1:1,000) (both from Cell Signaling),
anti-AKT1/2/3 (1:1,000), anti-phospho-AKT1/2/3 (S473; 1:1,000) and
anti-GAPDH (1:3,000) (all from Santa Cruz Biotechnology)
antibodies. Blots were then washed and incubated with horseradish
peroxidase (HRP)-conjugated anti-mouse or anti-rabbit secondary
antibodies, followed by additional washing and the detection with
enhanced chemiluminescence (ECl; AbFrontier).
Wound-healing assay
SKBr3 cells (untreated, GFP-Copine3- and
mcherry-Jab-infected) were inoculated into 6-well plates, and a 100
µl pipette tip was used to create a wound line across the
cell monolayer. The cells that moved into the interspace of the
scratched wound line were counted 48 h later using a phase contrast
microscope. This assay was performed in triplicate.
Statistical analysis
Statistical analyses were performed with Origin8.0
software using an analysis of variance (ANOVA). Statistical
significance was set at P<0.005. Data are reported as the means
± standard deviation (SD) of three independent experiments.
Results
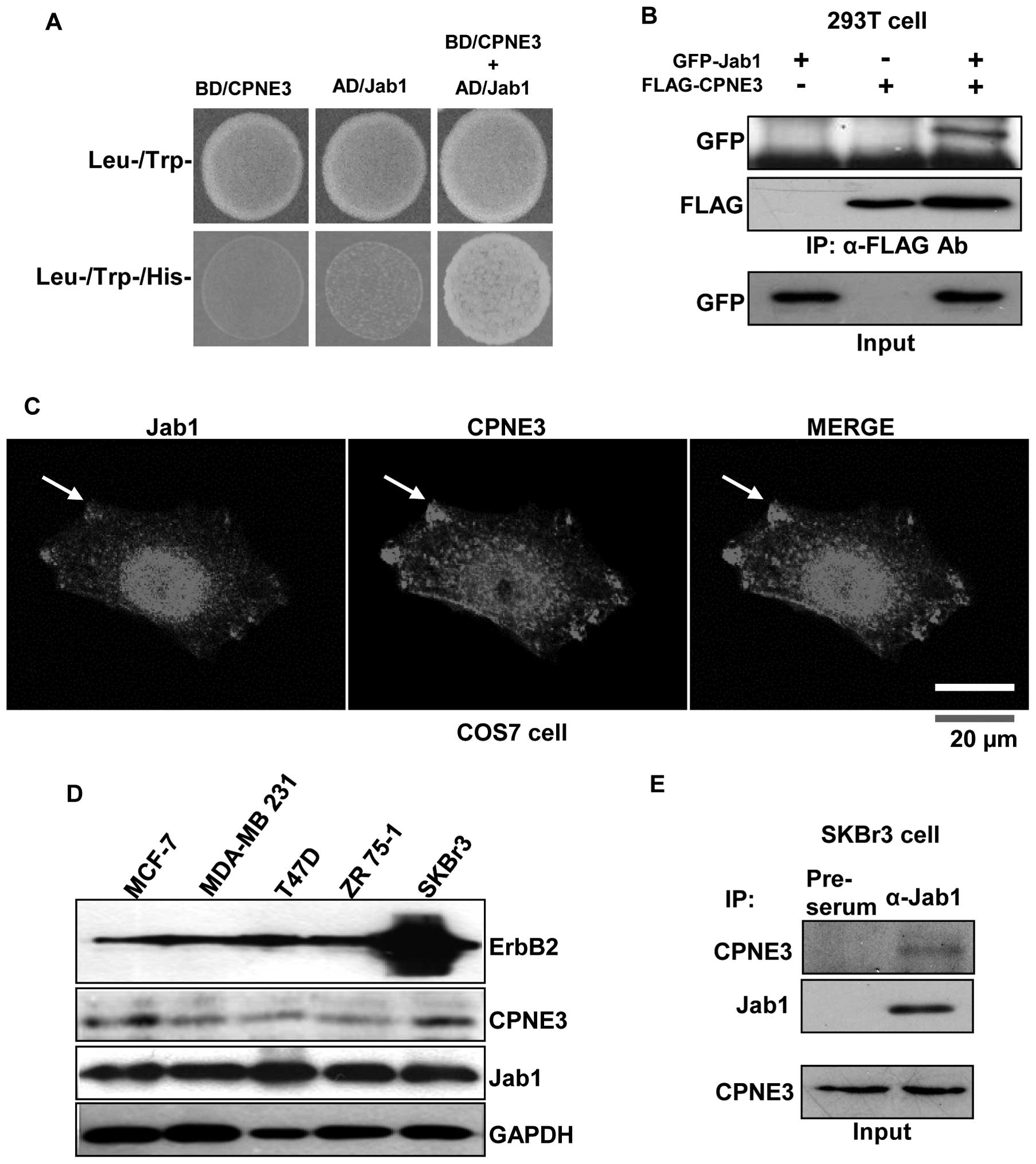
Protein-protein interaction between
Copine3 and Jab1 in vitro and in vivo
In order to identify novel Copine3 binding proteins,
we performed a yeast two-hybrid screening using Copine3 as bait.
One of the positive clones was determined to be Jab1 (data not
shown). This protein-protein interaction between Copine3 and Jab1
was also confirmed using Jab1 as bait in a yeast two-hybrid assay
(Fig. 1A) as well as an in
vitro binding assay performed in 293T cells (Fig. 1B). These data clearly show that
Copine3 binds directly to Jab1. Next, the interaction between these
two proteins was further confirmed with confocal microscopy of
Ad-mcherry-Jab1 and Ad-GFP-Copine3 infected COS7 cells. It appears
that Jab1 localizes to the nucleus and cytosol region, particularly
around the membrane, while Copine3 localizes to the around nucleus
and cytosol (Fig. 1C). The areas
where these two proteins are co-localized in the membrane are
marked with an arrow (Fig. 1C).
We then evaluated the endogenous protein expression
of ErbB2, Copine3 and Jab1 in several breast cancer cell types
(Fig. 1D). To investigate the
interaction between Jab1 and Copine3 and the effects of this
interaction on ErbB2/Copine3-related pathways in breast cancer,
endogenous co-IP experiments were carried out in SKBr3 cells.
Notably, IP with anti-Jab1 and pre-serum antibodies, followed by
western blotting with an anti-Copine3 antibody, showed that Jab1 is
in fact associated with Copine3 in this cell type (Fig. 1E).
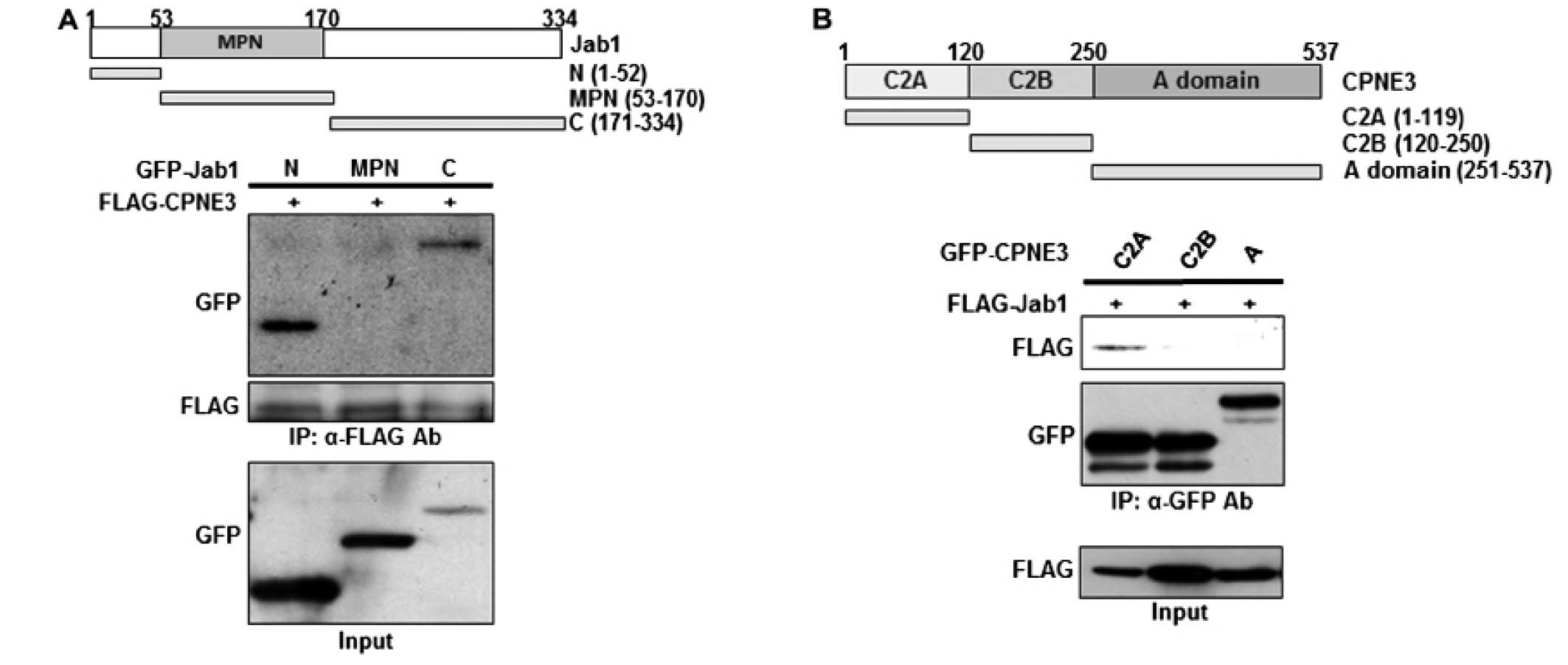
Identification of the binding regions
involved in the Jab1/Copine3 interaction
To identify the specific binding regions involved
for both proteins involved in the Jab1/Copine3 interaction, the
N-terminus, MPN domain and C-terminus of Jab1 as well as the C2A,
C2B, and A domains of Copine3 were isolated and N-terminally tagged
with GFP (Fig. 2A and B for Jab1
and Copine3, respectively). These truncated mutants were then
co-transfected into 293T cells with either FLAG-Copine3 (for the
Jab1 mutants) or FLAG-Jab1 (for the Copine3 mutants). Our data
indicate that, while Copine3 does not bind with the MPN domain of
Jab1, it does interact with both the N- and C-terminal domains
(Fig. 2A), indicating that the
tertiary structure of Jab1 may be important during this binding
interaction. Notably, Jab1 appears to bind only with the C2A domain
of Copine3 (Fig. 2B), demonstrating
that this region contains the binding site of Jab1. Generally, the
C2 domains in Copine3 are known for their Ca++ and lipid
binding capabilities; however, our data indicate that at least the
C2A domain also functions as a putative binding domain.
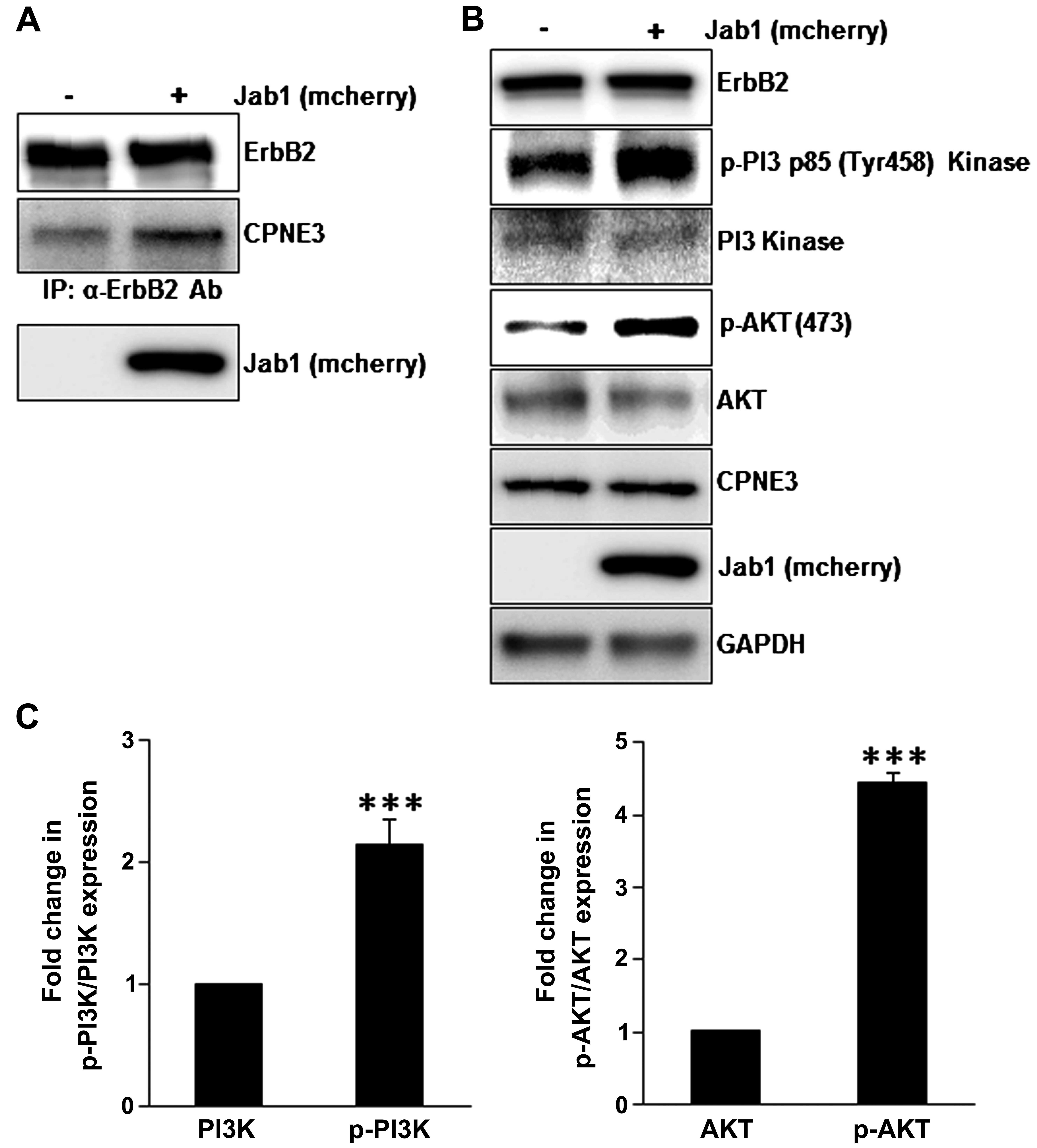
Jab1/Copine3 binding induces interaction
between Copine3 and ErbB2, activating downstream ErbB2 signaling
pathways in SKBr3 cells
It has been reported that Copine3 interacts with
ErbB2 in various breast cancer cell types (8). To investigate the effect of Jab1
binding on the interaction between Copine3 and ErbB2, co-IP
experiments were carried out in SKBr3 cells overexpressing Jab1
(Fig. 3A). It appears that the
interaction between Copine3 and ErbB2 was markedly increased when
Jab1 was overexpression in this cell type. In addition, to further
confirm ErbB2 pathway activation, various downstream signaling
proteins were investigated. Notably, the levels of phosphorylated
PI3 p85 kinase and AKT appear to increase when Jab1 is
overexpressed in SKBr3 cells (Fig. 3B
and C), indicating that high levels of Jab1, and thus an
increase in Jab1/Copine3 binding, activate ErbB2-related signaling
in this cell type. Based on these data, we conclude that Jab1
regulates ErbB2 downstream signaling pathway via binding to Copine3
and inducing the interaction between Copine3 and ErbB2.
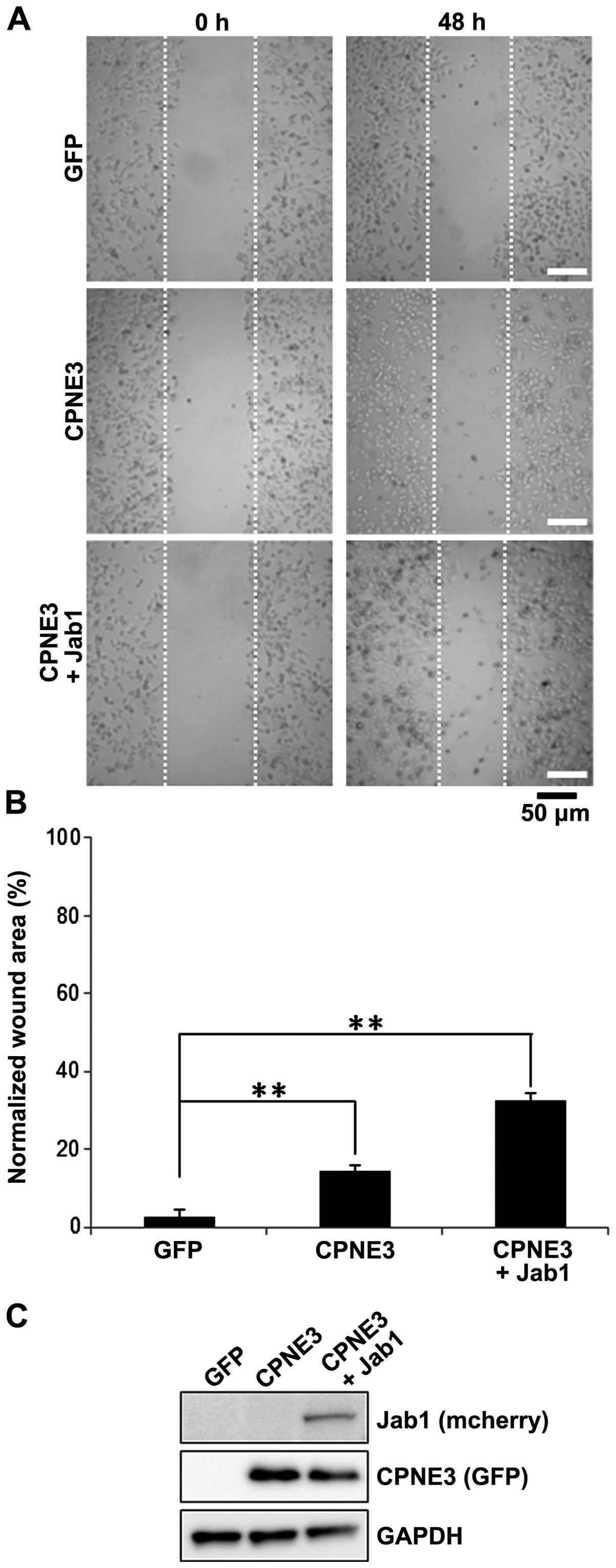
Jab1/Copine3 binding activates tumor cell
migration in SKBr3 cells
In addition to investigating the activation of
downstream proteins involved in the ErbB2-mediated signaling
cascade, we also examined the effect of Jab1/Copine3 binding on
ErbB2-dependent breast cancer cell motility. To do so, we utilized
a wound-healing assay using SKBr3 cells. One day following
infection with Ad-GFP, Ad-Copine3 or Ad-Copine3 + Ad-Jab1, we
created a ʻwoundʼ in the monolayered SKBr3 cells. Cell migration
into the wound was evaluated after 48 h (Fig. 4A). Basal migration of the GFP
infected control cells was low. Not surprisingly, the number of
migrating cells into the wound area increased ~15% in the cells
overexpressing Copine3 alone (Fig. 4A
and B). However, the number of cells that migrated into the
wound area was ~35% higher in the Copine3 and Jab1 co-infected
cells compared to the controls. The expression levels of Copine3
and Jab1 were also confirmed using western blot analysis (Fig. 4C). Notably, in this analysis, GAPDH
protein was similarly expressed in each group. Taken together, our
data show that overexpression of Jab1 in conjunction with Copine3
overexpression, and their subsequent binding, markedly increases
SKBr3 cell migration.
Discussion
In the present study, we revealed a novel binding
relationship between Copine3 and Jab1. This interaction was first
demonstrated with a yeast two-hybrid screen using Copine3 as bait.
Notably, we then confirmed this interaction in a reverse yeast
two-hybrid screen with Jab1 as bait as well as using in
vitro and in vivo assays. These assays further confirmed
the co-localization of Copine3 and Jab1 and direct binding between
the two proteins in breast cancer cells. The decision to focus on
the role of these proteins specifically in breast cancer cells
stemmed from a previous study that Copine3 expression correlates
with ErbB2 amplification in breast cancer and preferentially binds
to the pTyr1248 residue of this oncogene (8). This previous study also indicated that
this Copine3/ErbB2 interaction occurs at the plasma membrane.
Furthermore, Jab1 protein is also largely expressed in invasive
breast cancer (23). Thus, the
proximity of these proteins in breast cancer cells, confirmed in
the present study, spurred us to investigate their possible
influence on each other's expression and function. Notably, in a
preliminary analysis, we did not find any evidence supporting the
direct binding of Jab1 with ErbB2 (data not shown), further
focusing our investigation on the relationship between Jab1 and
Copine3.
It is generally known that the A domain found in
members of the Copine family is the primary protein binding domain
involved in regulation, while the C2 domains are typically related
to Ca++ and membrane binding (2). However, Benes et al revealed
that the C2 domain of protein kinase Cδ (PKCδ) directly binds to
phosphotyrosine peptides in a sequence-specific manner (24). In the present study, we have shown
that the C2A domain of Copine3 is the only region involved in Jab1
binding. As this is one of the few studies identifying
non-Ca++/membrane-dependent C2 domain-mediated binding,
additional study is necessary to determine the full function of
this binding motif.
Furthermore, Jab1 has been shown to bind to and
degrade p27Kip1 and p53, while in other contexts
it can stabilize HIF-1α activation and the c-Jun/AP-1 complex
(9,17–19).
The binding domains involved in these Jab1 functions are found
throughout the structure and include a Jun binding domain at the
N-terminus, an MPN domain, as well as a p27 binding domain at the
C-terminus (17–19). Generally, the function of the MPN
domain has been related to ubiquitin isopeptidase/deubiquitinase in
the ubiquitin-based signaling and protein turnover pathways in
eukaryotes (20). Our data indicate
that Copine3 preferentially binds with the N- and C-terminal
regions of Jab1, thereby excluding the MPN domain. Notably, this
distinct exclusion of MPN domain binding possibly indicates Jab1
binding-mediated activation of Copine3. In fact, our results show
that binding of Jab1 and Copine3 activates Copine3/ErbB2 binding as
well as the downstream ErbB2 signaling cascade and ErbB2-mediated
cell migration. We believe that this Jab1/Copine3-mediated
activation may involve Jab1 binding-induced conformation changes in
the tertiary structure of Copine3, making this protein more
accessible and/or more susceptible to ErbB2 binding. Additional
studies identifying the specific binding sites and structural
organization of the Jab1/Copine3 complex are warranted.
In conclusion, we have investigated the binding
complex formed by Jab1 and Copine3 in breast cancer cells. Our data
indicate that this binding complex not only forms in breast cancer
cells, but also influences the ErbB2 binding capabilities of
Copine3. It is likely that Copine3 is stabilized through the
binding of Jab1, potentially allowing it to bind more strongly to
ErbB2. Thus, Copine3 appears to be the link between Jab1 expression
and ErbB2-mediated downstream signal pathways and cellular
motility. The present study is the first to describe the
Copine3/Jab1 interaction and further enhances our understanding of
the signaling cascades involved in breast cancer.
Acknowledgments
The present study was supported by the Basic Science
Research Program (NRF-2012R1A1A2005035) and the Bio-Synergy
research Project (NrF-2014M3A9C4066463) through the National
research Foundation of Korea.
References
|
1
|
Creutz CE, Tomsig JL, Snyder SL, Gautier
MC, Skouri F, Beisson J and Cohen J: The copines, a novel class of
C2 domain-containing, calcium-dependent, phospholipid-binding
proteins conserved from Paramecium to humans. J Biol Chem.
273:1393–1402. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tomsig JL and Creutz CE: Copines: A
ubiquitous family of Ca2+-dependent phospholipid-binding
proteins. Cell Mol Life Sci. 59:1467–1477. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maitra R, Grigoryev DN, Bera TK, Pastan IH
and Lee B: Cloning, molecular characterization, and expression
analysis of Copine 8. Biochem Biophys Res Commun. 303:842–847.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang S, Yang H, Grisafi P, Sanchatjate S,
Fink GR, Sun Q and Hua J: The BON/CPN gene family represses cell
death and promotes cell growth in Arabidopsis. Plant J. 45:166–179.
2006. View Article : Google Scholar
|
|
5
|
Ramsey CS, Yeung F, Stoddard PB, Li D,
Creutz CE and Mayo MW: Copine-I represses NF-kappaB transcription
by endoproteolysis of p65. Oncogene. 27:3516–3526. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Park N, Yoo JC, Ryu J, Hong SG, Hwang EM
and Park JY: Copine1 enhances neuronal differentiation of the
hippocampal progenitor HiB5 cells. Mol Cells. 34:549–554. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Whittaker CA and Hynes RO: Distribution
and evolution of von Willebrand/integrin A domains: Widely
dispersed domains with roles in cell adhesion and elsewhere. Mol
Biol Cell. 13:3369–3387. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Heinrich C, Keller C, Boulay A, Vecchi M,
Bianchi M, Sack R, Lienhard S, Duss S, Hofsteenge J and Hynes NE:
Copine-III interacts with ErbB2 and promotes tumor cell migration.
Oncogene. 29:1598–1610. 2010. View Article : Google Scholar
|
|
9
|
Claret FX, Hibi M, Dhut S, Toda T and
Karin M: A new group of conserved coactivators that increase the
specificity of AP-1 transcription factors. Nature. 383:453–457.
1996. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chamovitz DA and Segal D: JAB1/CSN5 and
the COP9 signalosome. A complex situation. EMBO Rep. 2:96–101.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei N, Tsuge T, Serino G, Dohmae N, Takio
K, Matsui M and Deng XW: The COP9 complex is conserved between
plants and mammals and is related to the 26S proteasome regulatory
complex. Curr Biol. 8:919–922. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tomoda K, Yoneda-Kato N, Fukumoto A,
Yamanaka S and Kato JY: Multiple functions of Jab1 are required for
early embryonic development and growth potential in mice. J Biol
Chem. 279:43013–43018. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tian L, Peng G, Parant JM, Leventaki V,
Drakos E, Zhang Q, Parker-Thornburg J, Shackleford TJ, Dai H, Lin
SY, et al: Essential roles of Jab1 in cell survival, spontaneous
DNA damage and DNA repair. Oncogene. 29:6125–6137. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shackleford TJ and Claret FX: JAB1/CSN5: A
new player in cell cycle control and cancer. Cell Div. 5:262010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wei N, Serino G and Deng XW: The COP9
signalosome: More than a protease. Trends Biochem Sci. 33:592–600.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Barnes RO, West NR, Olson M, Chu
JE and Watson PH: Jab1 is a target of EGFR signaling in
ERalpha-negative breast cancer. Breast Cancer Res. 10:R512008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tomoda K, Kubota Y, Arata Y, Mori S, Maeda
M, Tanaka T, Yoshida M, Yoneda-Kato N and Kato JY: The cytoplasmic
shuttling and subsequent degradation of p27Kip1 mediated
by Jab1/CSN5 and the COP9 signalosome complex. J Biol Chem.
277:2302–2310. 2002. View Article : Google Scholar
|
|
18
|
Lee EW, OH W and Song J: Jab1 as a
mediator of nuclear export and cytoplasmic degradation of p53. Mol
Cells. 22:133–140. 2006.PubMed/NCBI
|
|
19
|
Bae MK, Ahn MY, Jeong JW, Bae MH, Lee YM,
Bae SK, Park JW, Kim KR and Kim KW: Jab1 interacts directly with
HIF-1alpha and regulates its stability. J Biol Chem. 277:9–12.
2002. View Article : Google Scholar
|
|
20
|
Birol M and Echalier A: Structure and
function of MPN (Mpr1/Pad1 N-terminal) domain-containing proteins.
Curr Protein Pept Sci. 15:504–517. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Burger-Kentischer A, Finkelmeier D, Thiele
M, Schmucker J, Geiger G, Tovar GE and Bernhagen J: Binding of
JAB1/CSN5 to MIF is mediated by the MPN domain but is independent
of the JAMM motif. FEBS Lett. 579:1693–1701. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Park JY, Hwang EM, Park N, Kim E, Kim DG,
Kang D, Han J, Choi WS, Ryu PD and Hong SG: Gateway RFP-fusion
vectors for high throughput functional analysis of genes. Mol
Cells. 23:357–362. 2007.PubMed/NCBI
|
|
23
|
Kouvaraki MA, Rassidakis GZ, Tian L, Kumar
R, Kittas C and Claret FX: Jun activation domain-binding protein 1
expression in breast cancer inversely correlates with the cell
cycle inhibitor p27Kip1. Cancer Res. 63:2977–2981.
2003.PubMed/NCBI
|
|
24
|
Benes CH, Wu N, Elia AE, Dharia T, Cantley
LC and Soltoff SP: The C2 domain of PKCdelta is a phosphotyrosine
binding domain. Cell. 121:271–280. 2005. View Article : Google Scholar : PubMed/NCBI
|