Introduction
HCC accounts for 70 to 90% of primary liver cancer
of the world, and is the third major cause of cancer-related
mortality worldwide (1,2). In China, HCC is the second leading
cause of cancer-related mortalities (3,4).
Recently, progress has been made in HCC treatment; however, the
prognosis of HCC patients has not improved significantly. There are
so many challenges remaining. Moreover, it is difficult to diagnose
HCC in the early stage, and many patients develop cancer recurrence
after liver resection (5).
Therefore, the development of novel therapeutic molecular targets
is urgently needed.
MiRNAs play important roles in the pathogenesis of
HCC (6–22). For example, miR-940 levels in HCC
tissues were lower than normal liver tissues, lower miR-940
expression in HCC tissues significantly correlated with the reduced
patient survival rate, low level of miR-940 promoted HCC cell
growth and inhibited cell apoptosis (23). Another study showed that miR-99a was
an independent predictor for the prognosis of HCC patients
(24).
A previous study showed that miR-4782-3p level in
NSCLC tissues is lower than in normal lung tissues. High expression
of miR-4782-3p indicated the favorable prognosis of NSCLC patients.
The targeted genes were USP14, ZEB2 and XIAP (25). Another study showed that USP14
activation promoted tumor progression in HCC (26). Thus we considered that miR-4782-3p
may also play an important role in HCC via USP14.
In this study, we assayed miR-4782-3p expression in
HCC tissues and tested the function of miR-4782-3p in HCC cells.
Our data elucidated the function of miR-4782-3p in HCC.
Materials and methods
Patients
Twenty-seven HCC specimens were collected from the
Department of Hepatobiliary and Gastrointestinal Surgery, Sichuan
Provincial Cancer Hospital (Chengdu, China). Tissue samples were
immediately frozen in liquid nitrogen after isolation. Informed
consent was obtained from each patient. The Ethics Commitment of
Sichuan Provincial Cancer Hospital and the Ethic Commitment of
Sichuan University approved this study. The senior pathologists of
Sichuan Provincial Cancer Hospital evaluated the histological
features of the specimens.
Cell culture
HCC cell lines HepG2 and SMMC-7721 and, L-02 cells
were purchased from the American Type Culture Collection (ATCC;
Rockville, MD, USA). Human HCC HepG2, SMMC-7721 and L-02 cells were
maintained in Dulbecco's modified Eagle's medium (DMEM) with 10%
fetal bovine serum (FBS; Invitrogen Corporation, Grand Island, NY,
USA).
Detection of miR-4782-3p level in HCC
tissues or cells
Total RNAs of HCC tissues and cells were extracted
by TRIzol reagent (Invitrogen Corporation) according the
manufacturer's instruction. The total RNAs was reverse-transcribed
to cDNA by using All-in-One™ miRNA First-Strand cDNA Synthesis kit
(Invitrogen Corporation). The primers were designed and synthesized
by Shengong Company (Shengong, Shanghai, China). Real-time PCR
assay was performed as described previously (24,27).
Oligonucleotides and cell
transfection
miR-4782-3p antisense oligonucleotides (miR-4782-3p
ASO), miR-4782-3p mimics and negative control miRNA were purchased
from RiboBio (Guangdong, China). miR-4782-3p ASO or miR-4782-3p
mimics were transfected into cells by using Lipofectamine
(Invitrogen, Shanghai, China), according to the manufacturer's
instruction (27).
Cell proliferation by MTT analysis
HepG-2 and SMMC-7721 cells (5×103/well)
were seeded into 96-well plates. Then MTT experiments were
performed as previously described (28,29).
Absorbance in each well was measured by using a microplate reader
set at 570 nm.
Apoptosis analysis
The apoptosis of HepG-2 and SMMC-7721 cells
transfected with miR-4782-3p ASO or miR-4782-3p mimics were
analyzed by using PI/Annexin V staining and fluorescence-activated
cell-sorting (FACS) flow cytometer (Biosciences, Beijing, China).
Details in brief, HepG2 and SMMC-7721 cells were seeded into
12-well plates. Twelve hours after transfection, cells were
cultured with serum-depleted medium for 12 h. Then cells were
suspended in binding buffer followed by staining with PI/Annexin
V-FITC for 15 min. The apoptotic rates were calculated on a flow
cytometer (30).
Target prediction
Bioinformatics methods were applied for the
prediction of the targeted genes of miR-4782-3p. Moreover, the
bioinformatic algorithms from TargetScanHuman were used (31–34).
Dual-luciferase assays
To assess and confirm whether miR-4782-3p binds
USP14 directly, a dual-luciferase assay was performed (25,30).
The 3′UTR of USP14 was amplified by using PCR from genomic DNA. The
production was inserted downstream of USP14 3′UTR reporter plasmids
(pRL-USP14; Biotech, Chengdu, China), and mutants of USP14 3′UTR
were generated by Site-Directed Mutagenesis kit (Shanghai, China).
Then the whole plasmid was confirmed by sequencing. Mutation in the
miR-4782-3p binding site of the USP14 3′UTR was constructed by
Shengong Company (Shengong, Chengdu, China). The luciferase
reporter containing mutant was also constructed. For luciferase
assays, HepG2 cells were transfected with luciferase reporter
plasmid along with miR-4782-3p mimics or negative control by using
Lipofectamine 2000 (Invitrogen). Twenty-four hours after
transfection, these cells were analyzed by using a Luciferase assay
kit (Promega, Madison, WI, USA) (35).
Statistical analysis
Two-tailed Student's t-test was used to analyze the
difference between two groups. ANOVA was used to analyze the
difference of three groups. Kaplan-Meier analysis was employed to
evaluate the overall survival of HCC patients. The correlation
between miR-4782-3p and USP14 was performed by Spearman's
correlation analysis. All statistical analysis was performed by
SPSS version 19. Values are expressed as the mean ± SD from three
tests. All values of P<0.05 are marked with an asterisk in the
figures, and was considered statistically significantly
different.
Results
The low expression of miR-4782-3p in HCC
tissues and its role in the survival of HCC
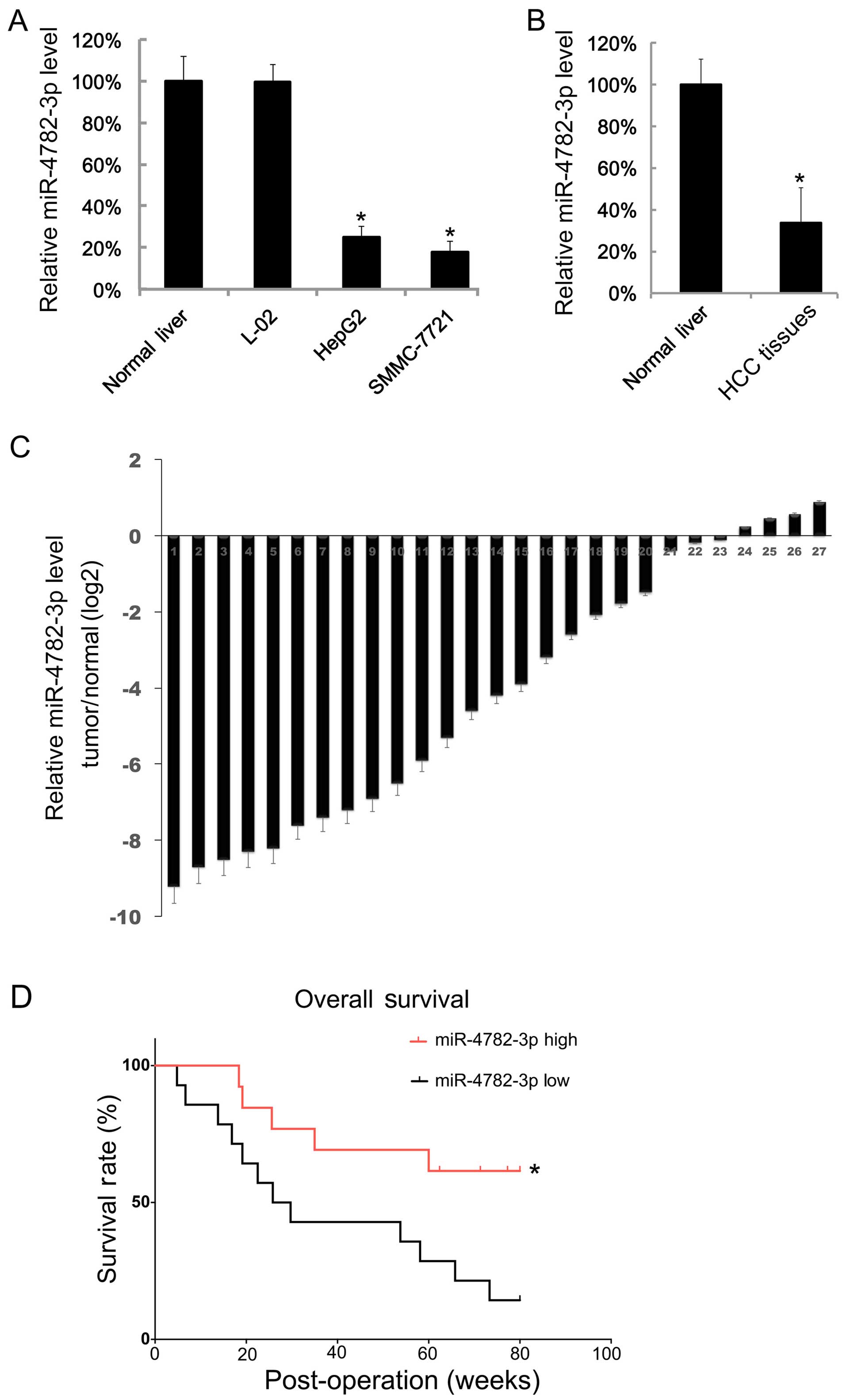
We first examined the miR-4782-3p level in HCC cell
lines; the miR-4782-3p level in normal liver tissue was treated as
a negative control. We found that the miR-4782-3p levels in HepG2
and SMMC-7721 cells were lower than in normal liver tissue and L-02
cells (Fig. 1A). Then we assayed
the miR-4782-3p levels in 27 HCC tissues and adjacent normal liver
tissues. We found that the mean value of miR-4782-3p levels in
tumor tissues is lower than the mean value of adjacent normal
tissues (Fig. 1B). The relative
expression of miR-4782-3p in each HCC tissue and the matched
adjacent normal liver tissue are shown in Fig. 1C. There were only 4 pairs, in which
HCC tissues showed higher levels of miR-4782-3p. Next, the 27 HCC
patients were separated into two groups by the median of the
miR-4782-3p expression, and their progress was followed for about
100 weeks. We found that HCC patients with higher miR-4782-3p level
had a longer survival period (Fig.
1D).
The effect of miR-4782-3p mimics on cells
growth and apoptosis
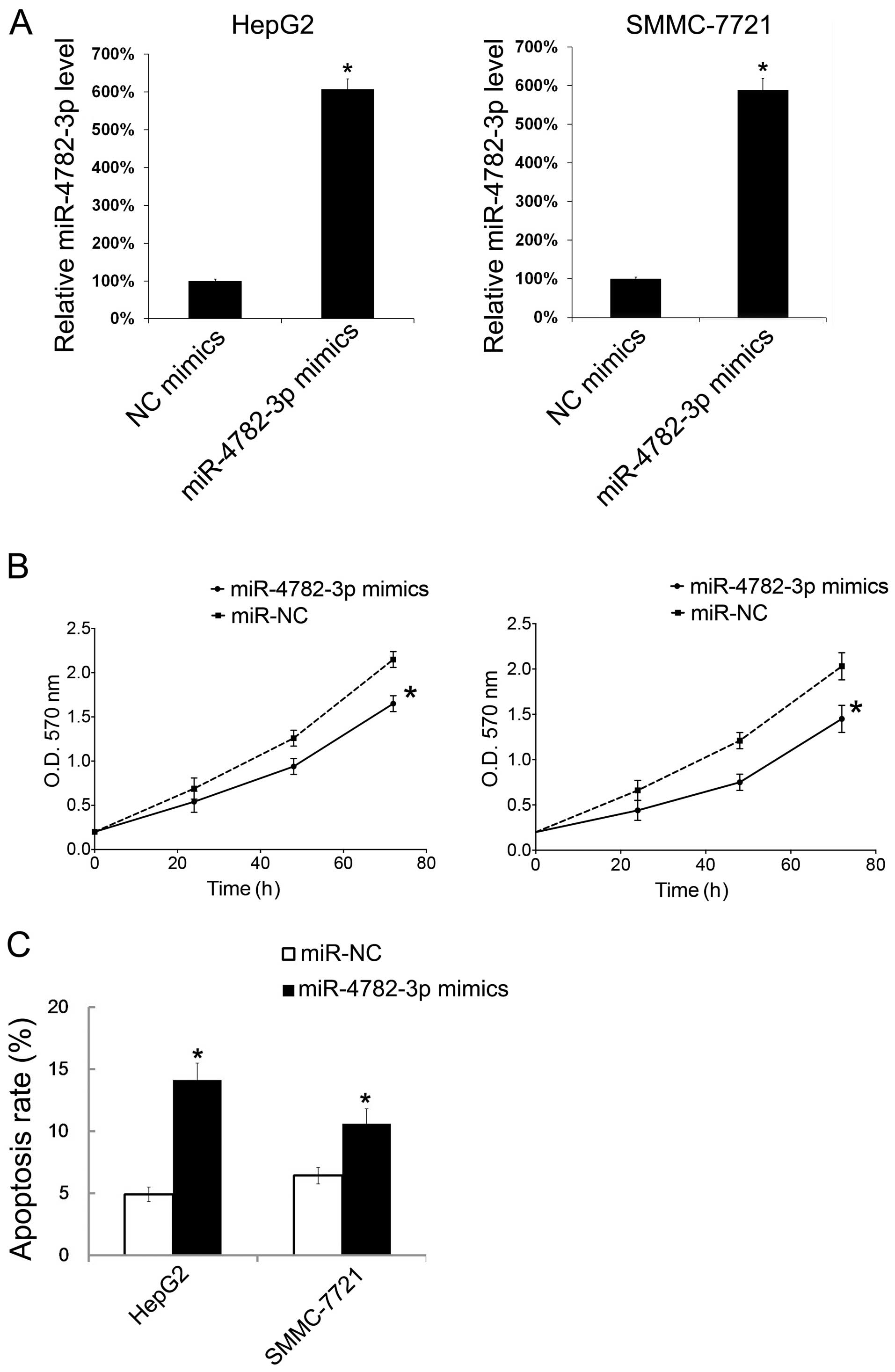
To investigate the effect of miR-4782-3p in cell
growth and apoptosis, HepG2 and SMMC-7721 cells were transiently
transfected with miR-4782-3p mimics, and then the cell growth and
apoptosis were evaluated. Forty-eight hours after transfection of
the miR-4782-3p mimic, the miR-4782-3p level in HepG2 and SMMC-7721
cells was tested by qRT-PCR. It showed that the miR-4782-3p levels
in both cell types were increased (Fig.
2A). We then detected the effect of miR-4782-3p on cell growth
by MTT. With transfection of miR-4782-3p mimic, HepG2 and SMMC-7721
cells showed significant decrease in cellular growth (Fig. 2B). To verify whether miR-4782-3p
could influence HCC cells apoptosis, PI/Annexin V double staining
were performed to evaluate apoptosis. The results showed that
miR-4782-3p mimic transfection increased apoptosis about 3–5%
(Fig. 2C).
The effect of miR-4782-3p ASO on cell
growth and apoptosis
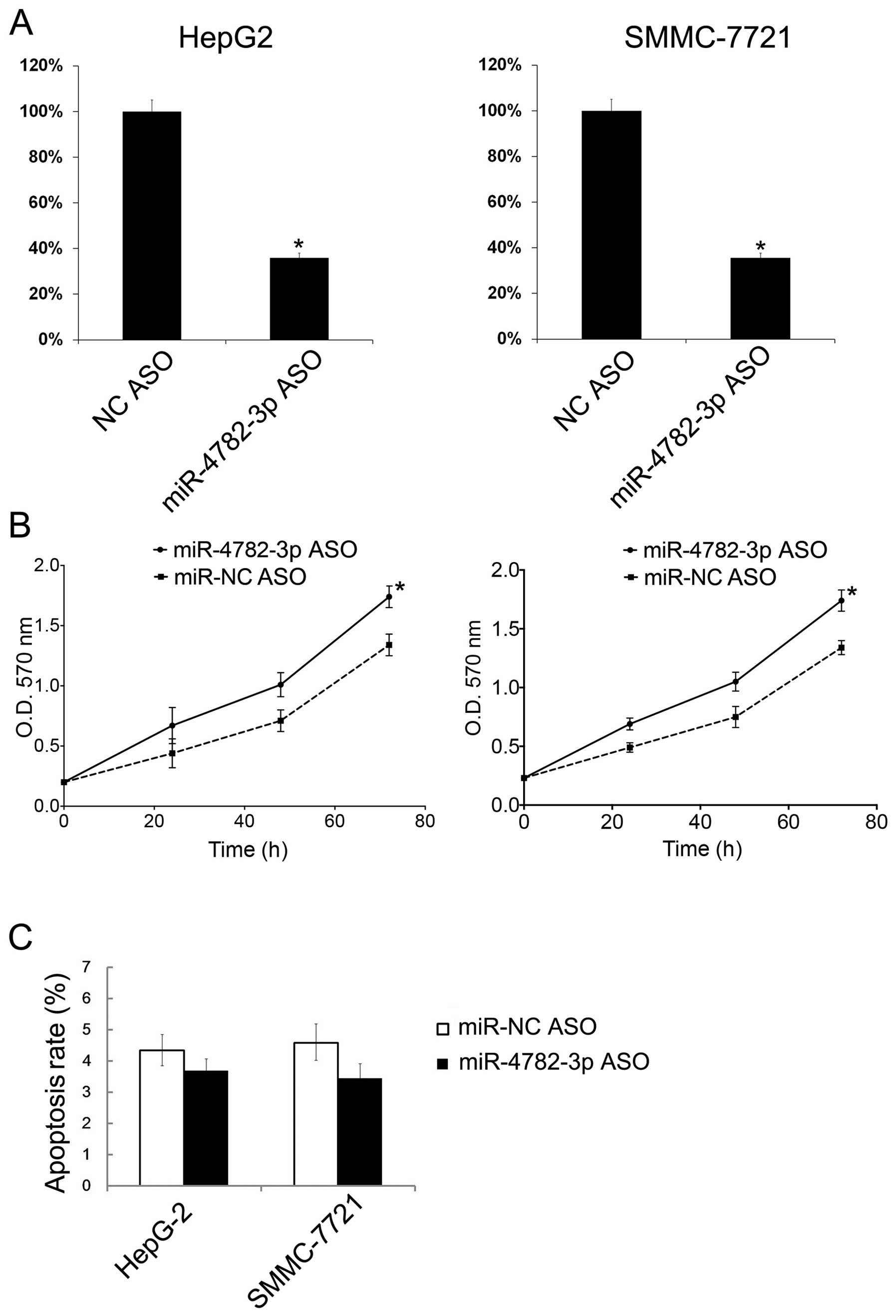
Additionally, HepG2 and SMMC-7721 cells were
transfected with miR-4782-3p ASO to downregulate the miR-4782-3p
levels. Then the miR-4782-3p ASO transfection was tested by
qRT-PCR. The miR-4782-3p levels in both cell lines decreased
following the transfection (Fig.
3A). With miR-4782-3p ASO transfection, HepG2 and SMMC-7721
cells showed significant increase in cellular growth (Fig. 3B). However, the miR-4782-3p ASO
showed no significant effect on cell apoptosis (Fig. 3C).
miR-4782-3p targets USP14
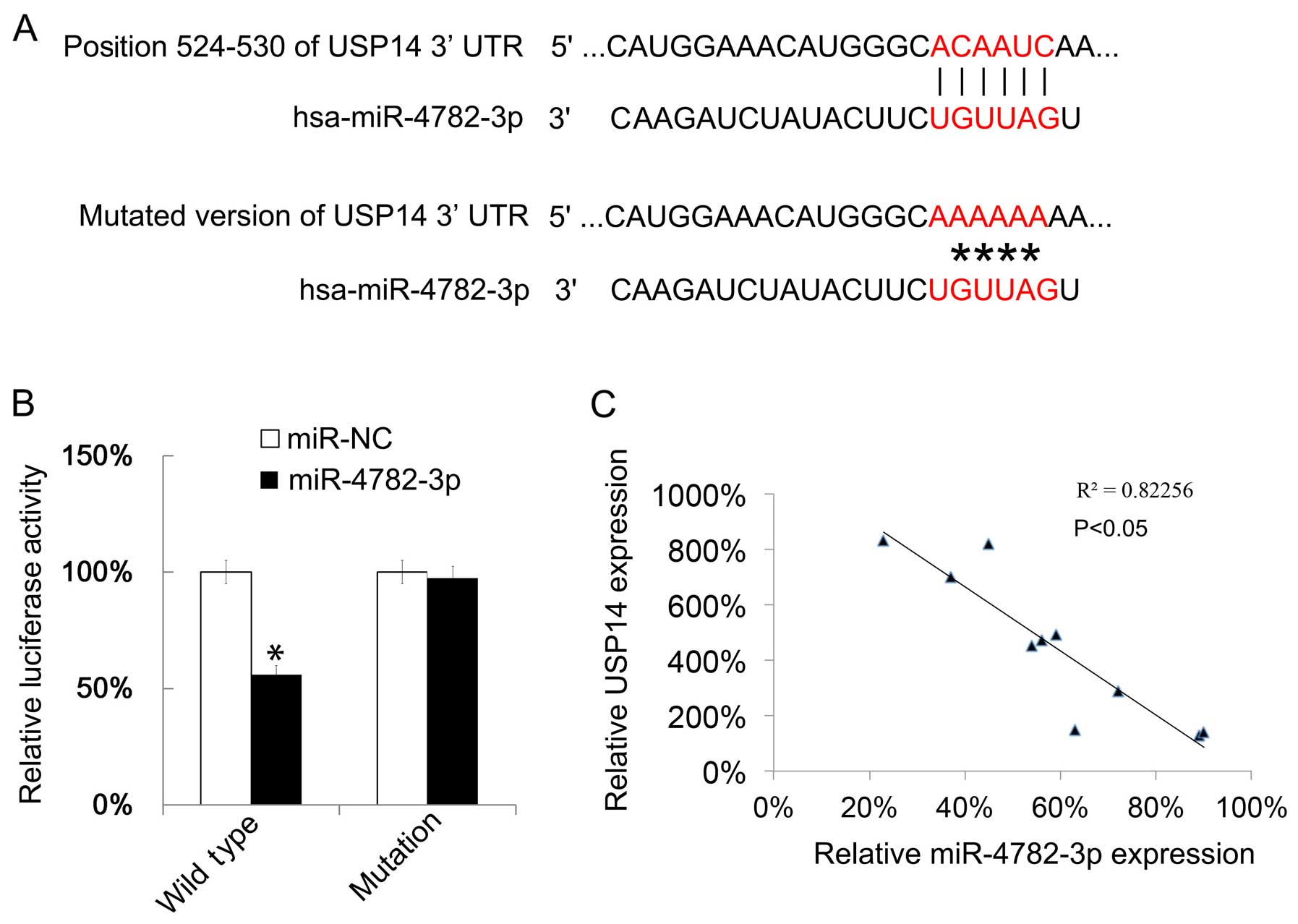
The gene targets of miR-4782-3p were predicted by
TargetScanHuman, this prediction showed that USP14 could be
targeted by miR-4782-3p. Then we mutated the binding site of
miR-4782-3p in USP14 (Fig. 4A). The
effect of miR-4782-3p on USP14 translation was tested by a
luciferase reporter assay. The luciferase reporter plasmid with
wild-type 3′UTR of USP14 or the mutated version as indicated in
Fig. 4A showed that upregulation of
miR-4782-3p reduced the luciferase activity of the reporter gene
with wild-type, but not with the mutant (Fig. 4B). Next the USP14 mRNA expressions
in 10 HCC tissues were examined by qRT-PCR. We found that the
miR-4782-3p level and the USP14 mRNA level were negatively
correlated (Fig. 4C).
Discussion
miR-4782-3p has shown its important role in the
pathogenesis of NSCLC. miR-4782-3p in NSCLC tissues was relatively
low. High expression of miR-4782-3p indicated favorable prognosis
of NSCLC patients. The targeted genes were USP14, ZEB2 and XIAP
(25). In this study, the role of
miR-4782-3p in HCC was studied. We demonstrated miR-4782-3p also
showed a low level in HCC tissues. Importantly, HCC patients with
higher miR-4782-3p level had longer survival period. The underlying
mechanism may be that the level of miR-4782-3p in HCC promoted cell
growth and inhibited apoptosis. To the best of our knowledge, this
is the first report on the role of miR-4782-3p in HCC.
USP14 has been proven to be the target gene of
miR-4782-3p, and another study showed that USP14 activation
promotes tumor progression in HCC (26). Thus our study has established a
connection between miR-4782-3p and USP14 in HCC.
USP14 plays important roles in different types of
cancers. For example, overexpression of USP14 in NSCLC was
associated with shorter overall survival of patients (36). High expression of USP14 was related
to poor prognosis of epithelial ovarian cancer patients (37). Thus an inhibitor of USP14, like
miR-4782-3p, may have a potential therapeutic effect.
Our data showed that overexpression of miR-4782-3p
could increase the apoptosis rate of HepG-2 and SMMC-7721 cells,
however, apoptosis of these cell lines did not decrease
significantly after downregulation of miR-4782-3p. We considered
the reason to be that the apoptosis rate of HepG-2 and SMMC-7721 is
very low, thus it is hard to reduce it to even lower apoptosis
rate.
In conclusion, this study revealed the role of
miR-4782-3p in HCC. Our study contributes to finding a new target
for HCC therapy.
References
|
1
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273.e1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gao J, Xie L, Yang WS, Zhang W, Gao S,
Wang J and Xiang YB: Risk factors of hepatocellular carcinoma -
current status and perspectives. Asian Pac J Cancer Prev.
13:743–752. 2012. View Article : Google Scholar
|
|
5
|
Portolani N, Coniglio A, Ghidoni S,
Giovanelli M, Benetti A, Tiberio GA and Giulini SM: Early and late
recurrence after liver resection for hepatocellular carcinoma:
Prognostic and therapeutic implications. Ann Surg. 243:229–235.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down-regulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Budhu A, Jia HL, Forgues M, Liu CG,
Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP and
Zhuang SM: MicroRNA-195 suppresses tumorigenicity and regulates
G1/S transition of human hepatocellular carcinoma cells.
Hepatology. 50:113–121. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y
and Zhuang SM: MicroRNA-101, down-regulated in hepatocellular
carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer
Res. 69:1135–1142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar
|
|
11
|
Braconi C and Patel T: MicroRNA expression
profiling: A molecular tool for defining the phenotype of
hepatocellular tumors. Hepatology. 47:1807–1809. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Murakami Y, Tamori A, Itami S, Tanahashi
T, Toyoda H, Tanaka M, Wu W, Brojigin N, Kaneoka Y, Maeda A, et al:
The expression level of miR-18b in hepatocellular carcinoma is
associated with the grade of malignancy and prognosis. BMC Cancer.
13:992013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gramantieri L, Fornari F, Callegari E,
Sabbioni S, Lanza G, Croce CM, Bolondi L and Negrini M: MicroRNA
involvement in hepatocellular carcinoma. J Cell Mol Med.
12:2189–2204. 2008. View Article : Google Scholar
|
|
14
|
Wang B, Majumder S, Nuovo G, Kutay H,
Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K and Jacob ST:
Role of microRNA-155 at early stages of hepatocarcinogenesis
induced by choline-deficient and amino acid-defined diet in C57BL/6
mice. Hepatology. 50:1152–1161. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Coulouarn C, Factor VM, Andersen JB,
Durkin ME and Thorgeirsson SS: Loss of miR-122 expression in liver
cancer correlates with suppression of the hepatic phenotype and
gain of metastatic properties. Oncogene. 28:3526–3536. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ji J, Shi J, Budhu A, Yu Z, Forgues M,
Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ji J, Yamashita T, Budhu A, Forgues M, Jia
HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH, et al: Identification
of microRNA-181 by genome-wide screening as a critical player in
EpCAM-positive hepatic cancer stem cells. Hepatology. 50:472–480.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song G, Sharma AD, Roll GR, Ng R, Lee AY,
Blelloch RH, Frandsen NM and Willenbring H: MicroRNAs control
hepatocyte proliferation during liver regeneration. Hepatology.
51:1735–1743. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ura S, Honda M, Yamashita T, Ueda T,
Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K and Kaneko
S: Differential microRNA expression between hepatitis B and
hepatitis C leading disease progression to hepatocellular
carcinoma. Hepatology. 49:1098–1112. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang
Y, Tantoso E, Li KB, Ooi LL, Tan P, et al: Profiling microRNA
expression in hepatocellular carcinoma reveals microRNA-224
up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific
target. J Biol Chem. 283:13205–13215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yuan B, Liang Y, Wang D and Luo F: miR-940
inhibits hepatocellular carcinoma growth and correlates with
prognosis of hepatocellular carcinoma patients. Cancer Sci.
106:819–824. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li D, Liu X, Lin L, Hou J, Li N, Wang C,
Wang P, Zhang Q, Zhang P, Zhou W, et al: MicroRNA-99a inhibits
hepatocellular carcinoma growth and correlates with prognosis of
patients with hepatocellular carcinoma. J Biol Chem.
286:36677–36685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu N, Zhang C, Bai C, Han YP and Li Q:
miR-4782-3p inhibited non-small cell lung cancer growth via USP14.
Cell Physiol Biochem. 33:457–467. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang G, Li L and Zhou W: USP14 activation
promotes tumor progression in hepatocellular carcinoma. Oncol Rep.
34:2917–2924. 2015.PubMed/NCBI
|
|
27
|
Song B, Zhang C, Li G, Jin G and Liu C:
miR-940 inhibited pancreatic ductal adenocarcinoma growth by
targeting MyD88. Cell Physiol Biochem. 35:1167–1177. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu C, Gao F, Li B, Mitchel RE, Liu X, Lin
J, Zhao L and Cai J: TLR4 knockout protects mice from
radiation-induced thymic lymphoma by downregulation of IL6 and
miR-21. Leukemia. 25:1516–1519. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu C, Zhou C, Gao F, Cai S, Zhang C, Zhao
L, Zhao F, Cao F, Lin J, Yang Y, et al: miR-34a in age and tissue
related radio-sensitivity and serum miR-34a as a novel indicator of
radiation injury. Int J Biol Sci. 7:221–233. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin L, Liang H, Wang Y, Yin X, Hu Y, Huang
J, Ren T, Xu H, Zheng L and Chen X: microRNA-141 inhibits cell
proliferation and invasion and promotes apoptosis by targeting
hepatocyte nuclear factor-3β in hepatocellular carcinoma cells. BMC
Cancer. 14:8792014. View Article : Google Scholar
|
|
31
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar :
|
|
33
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Garcia DM, Baek D, Shin C, Bell GW,
Grimson A and Bartel DP: Weak seed-pairing stability and high
target-site abundance decrease the proficiency of lsy-6 and other
microRNAs. Nat Struct Mol Biol. 18:1139–1146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Grentzmann G, Ingram JA, Kelly PJ,
Gesteland RF and Atkins JF: A dual-luciferase reporter system for
studying recoding signals. RNA. 4:479–486. 1998.PubMed/NCBI
|
|
36
|
Wu N, Liu C, Bai C, Han YP, Cho WC and Li
Q: Over-expression of deubiquitinating enzyme USP14 in lung
adenocarcinoma promotes proliferation through the accumulation of
β-catenin. Int J Mol Sci. 14:10749–10760. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Y, Wang J, Zhong J, Deng Y, Xi Q, He
S, Yang S, Jiang L, Huang M, Tang C, et al: Ubiquitin-specific
protease 14 (USP14) regulates cellular proliferation and apoptosis
in epithelial ovarian cancer. Med Oncol. 32:3792015. View Article : Google Scholar
|