Introduction
Colorectal cancer is the third most common cancer
worldwide and the second leading cause of cancer-related deaths
(1). Elucidating the molecular
mechanisms involved in the proliferation, migration and invasion of
colorectal cancer not only will aid in the further understanding of
the pathogenesis and progression of the disease, but may also
elucidate new targets for effective therapies.
Aberrant regulation of cholesterol homeostasis has
been associated with multiple types of cancer. Numerous studies
have shown increased levels of cholesterol in tumors when compared
to the level in normal tissue (2–7).
Moreover, it has been suggested that cholesterol intake may
increase colorectal cancer risk (8). Multiple pathways to increase
intracellular cholesterol have been observed in various types of
cancer cells. These include upregulation of
3-hydroxy-3-methylglutaryl CoA reductase (HMG-CoAR) activity, the
rate-limiting step of the cholesterol synthesis pathway (9–11),
loss of feedback inhibition of HMGCoAR by cholesterol (12–14),
increased uptake of extracellular cholesterol through the
low-density lipoprotein receptor (15–17),
and decreased expression of the cholesterol exporter termed
ATP-binding cassette transporter A1/cholesterol exporter (ABCA1)
(16,18–20).
The ABCA1 protein mediates the transfer of cellular cholesterol
across the plasma membrane to apolipoprotein A-I (ApoAI), the major
apolipoprotein component of high-density lipoprotein (HDL)
(21). ABCA1 gene, a key player in
cholesterol metabolism, prompted us to investigate its role in
colon cancer.
MicroRNAs (miRNAs) are small, non-coding RNAs that
post-transcriptionally regulate gene expression and play
significant roles in maintaining normal cellular functions,
including regulation of cholesterol homeostasis (22–26).
Deregulation of miRNA expression leads to the onset of diverse
types of diseases, including cancer as exemplified by their
differential expression in carcinomas, sarcomas and hematologic
tumors (27–31). miR-183 can function as an oncogene
by targeting the transcription factor EGR1 and was found to promote
cell migration in colon cancer (32). Yet, the mechanism remains
unknown.
In the present study, we initially confirmed that
ABCA1 is aberrantly expression in colon cancer and colon cancer
cells. Its overexpression inhibited the proliferation of colon
cancer HCT116 cells while the silencing of ABCA1 promoted
proliferation and inhibited apoptosis in colon cancer LDL1 cells.
Upregulation of specific miRNAs can contribute to downregulation of
tumor-suppressive genes (33–35).
Thus, we aimed to ascertain whether ABCA1 is downregulated by
overexpression of a specific miRNA in colon cancer. We screened
microRNAs that may target ABCA1 by miRanda which is a commonly used
prediction algorithm. We found that miR-183 targets the 3′UTR of
ABCA1 mRNA. Subsequent experiments confirmed that miR-183 degraded
ABCA1 mRNA in the colon cancer cells. Finally, we demonstrated that
miR-183 promoted proliferation and inhibited apoptosis in the
cells. Thus, we conclude that miR-183 promotes proliferation and
inhibits apoptosis by regulating ABCA1 in colon cancer.
Materials and methods
Colon cancer tissues and colon cancer
cell lines DLD1, Caco-2, HCT116, DiFi, Lim1215 and HCA7
All colon cancer patients were recruited from the
Jinan Second People's Hospital. The use of human tissue samples and
research conducted on humans followed internationally recognized
guidelines as well as local and national regulations. All
participants provided informed consent. Colon cancer cell lines
DLD1, Caco-2, HCT116, DiFi, Lim1215 and HCA7 were purchased from
the Cell Bank of the Chinese Academy of Sciences (Shanghai, China).
Cells were maintained in RPMI-1640 medium supplemented with 10%
fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA) and
penicillin/streptomycin at 37°C in a humidified atmosphere with 5%
CO2.
ABCA1-expressing plasmids/empty vectors,
shABCA1 plasmids/scramble, pre-miR-183/control miR,
anti-miR-183/scramble and transfection experiments
ABCA1-expressing plasmids/empty vector and shMRTF-A
plasmids/scramble were obtained from Boston University (Boston, MA,
USA). Pre-miR-183/control miR and anti-miR-183/scramble were
purchased from Ambion Inc. (Austin, TX, USA). Before transfection,
the cells were cultured in serum-free medium without antibiotics
for 24 h. On the following day, cells at ~90% confluency were
transfected with transfection reagent (Lipofectamine 2000;
Invitrogen, Carlsbad, CA, USA), according to the manufacturer's
instructions. After incubation for 6 h, the medium was removed and
replaced with normal culture medium for 48 h.
Western blot analysis
Tissue and cell lysates, normalized for cell protein
content, were analyzed by western blotting (36). Mainly, after incubation with the
primary antibody: anti-ABCA1 (1:500), anti-c-myc (1:500), anti-p53
(1:500), anti-CDK2 (1:500), anti-p21 (1:500), anti-cyclin D1
(1:500), anti-BCL2 (1:500), anti-MCL1 (1:500) or anti-β-actin
(1:500) (all from Abcam, Cambridge, MA, USA), the secondary
antibodies were used to bind to the primary antibodies.
MTT assay
Cells were seeded in 96-well plates and divided into
different groups. After treatment, 20 μl MTT solution (5
mg/ml; Sigma, St. Louis, MO, USA) was added to each well and
incubated for 4 h at 37°C. After that, the culture medium was
removed and 150 μl DMSO (Sigma) was added to each well. The
absorbance was measured at 490 nm.
Migration and invasion assays
Cells were transferred into the top chamber with a
non-coated membrane at 5×104 cells/well (24-well plate,
pore size, 8.0 μm; BD Biosciences, San Jose, CA, USA) in the
Transwell migration assay. For the Transwell invasion assay,
5×104 cells/well were plated in the top chamber with a
Matrigel-coated membrane (24-well plate, pore size, 8.0 μm;
BD Biosciences). In both assays, medium with 10% FBS was placed in
the lower chamber, while cells were plated in medium without serum
or growth factors in the upper chamber. After 24 h of incubation,
the non-invading and non-migrating cells were removed by wiping the
upper surface of the filter with a cotton swab. The remaining cells
on the lower surface of the membrane were stained and counted.
Bioinformatic methods
Analysis of potential microRNA target sites was
carried out using commonly used prediction algorithm, miRanda
(http://www.microrna.org/microrna/home.do).
Immunofluorescence analyses
Cells were plated onto coverslips in 6-well plates
and transfected with 30 nM pre-miR-183 or control miR. After
transfection for 36 h, the coverslips were stained with the
anti-ABCA1 antibody. Anti-rabbit IgG antibody was used as the
secondary antibody (Invitrogen). Coverslips were counterstained
with DAPI (Invitrogen Molecular Probes, Eugene, OR, USA) for
visualization of nuclei. Results were observed using a confocal
laser scanning microscope (Leica Microsystems, Bensheim, Germany)
and analyzed by ImageJ software.
RT-PCR and qRT-PCR for ABCA1
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen) according to the manufacturer's protocol.
qRT-PCR was carried out with a Power SYBR Green PCR Master Mix
(Applied Biosystems, Carlsbad, CA, USA) according to the
manufacturer's instructions. The primers were: ABCA1 forward,
5′-TTAAACGCCCTCACCAAAGAC-3′ and reverse,
5′-AAAAGCCGCCATACCTAAACTCAT-3′.
Real-time PCR for miRNA
Total RNA was isolated from the cultured cells using
the mirVana miRNA isolation kit (Ambion). Detection of the mature
form of miRNAs was performed using the mirVana qRT-PCR miRNA
detection kit (Ambion). The U6 small nuclear RNA was used as an
internal control.
TUNEL staining
TUNEL staining analysis was performed as previously
reported (37).
Statistical analysis
Data are presented as mean ± SEM. Student's t-test
(two-tailed) was used to compare two groups (P<0.05 was
considered significant), unless otherwise indicated (χ2
test).
Results
Aberrant expression of ABCA1 in colon
cancer tissues
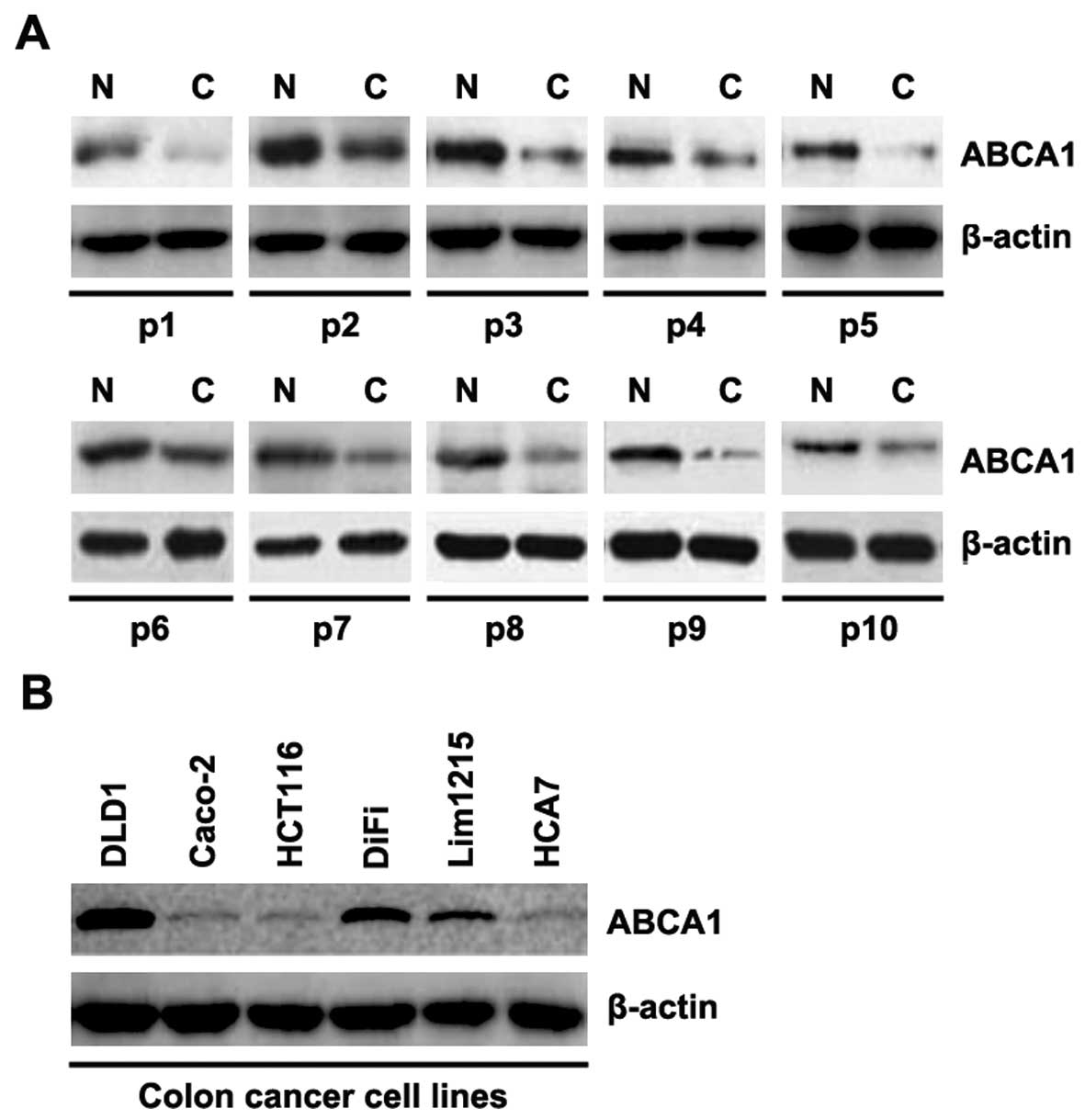
In order to determine ABCA1 protein expression in
colon cancer tissues, we performed western blot assay to detect
ABCA1 protein in colon cancer tissues compared to the level in the
adjacent normal tissues. We found that ABCA1 was decreased in the
cancer tissues of 10 patients, when compared with that in the
adjacent normal tissues (Fig. 1A).
The data implied that ABCA1 may be a tumor-suppressive gene in
colon cancer. In an attempt to identify the ABCA1 protein
expression in the different colon cancer cell lines, we performed
western blotting in DLD1, Caco-2, HCT116, DiFi, Lim1215 and HCA7
cells. ABCA1 protein varied in the different colon cancer cell
lines (Fig. 1B).
Overexpression of ABCA1 inhibits
proliferation, but does not affect the migration and invasion of
colon cancer HCT116 cells
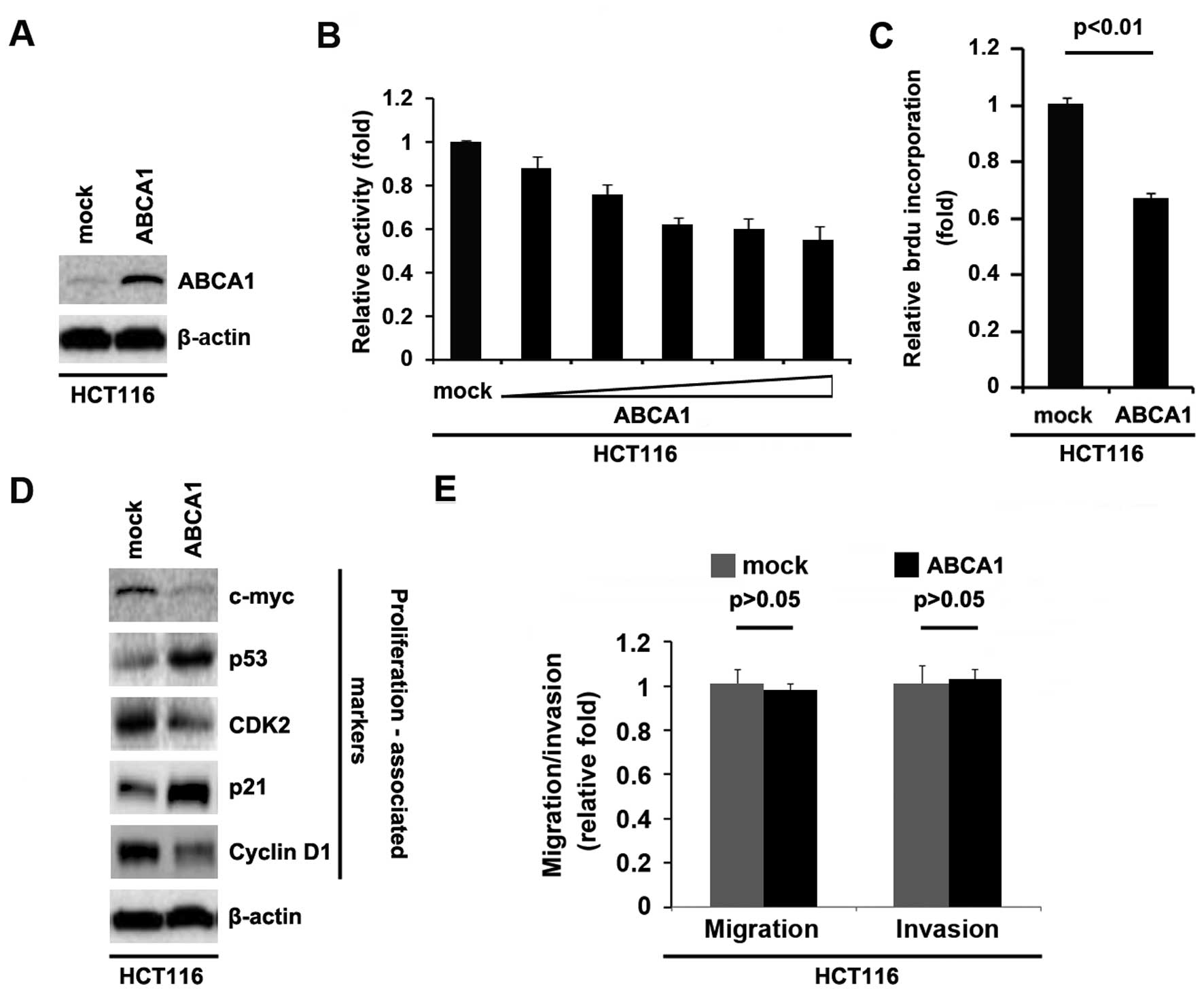
In an attempt to determine whether ABCA1 regulates
the proliferation of HCT116 cells, the cells were transfected with
ABCA1-expressing plasmids. After stable transfection, ABCA1
expression was detected by western blotting. The results showed
that the ABCA1-expressing plasmids evidently upregulated ABCA1
protein expression in the HCT116 cells (Fig. 2A). We next performed an MTT assay to
detect the proliferation of the HCT116 cells transfected with the
ABCA1-expressing plasmid. The results showed that ABCA1 inhibited
the proliferation of HCT116 cells after 48 h of transfection and
the inhibition was dose-dependent (Fig.
2B). To further show the effects of ABCA1 on proliferation, we
performed BrdU incorporation assay to detect DNA synthesis in the
cells. The results confirmed that ABCA1 significantly inhibited DNA
synthesis in the cells (Fig. 2C).
In order to further identify the effect of ABCA1 on proliferation,
we performed western blotting to confirm that ABCA1 could affect
proliferation markers. The results of western blotting demonstrated
that c-myc, CDK2 and cyclin D1 protein were downregulated and p53
and p21 were upregulated by ABCA1 in the HCT116 cells (Fig. 2D).
Given that ABCA1 obviously promotes HCT116 cell
proliferation, we next sought to determine whether ABCA1 would have
an impact on migration and invasion in the HCT116 cells. The
migration and invasion assays showed that overexpression of ABCA1
did not affect the migration and invasion of the HCT116 cells
(Fig. 2E).
Silencing of ABCA1 promotes
proliferation, but does not affect migration and invasion in colon
cancer DLD1 cells
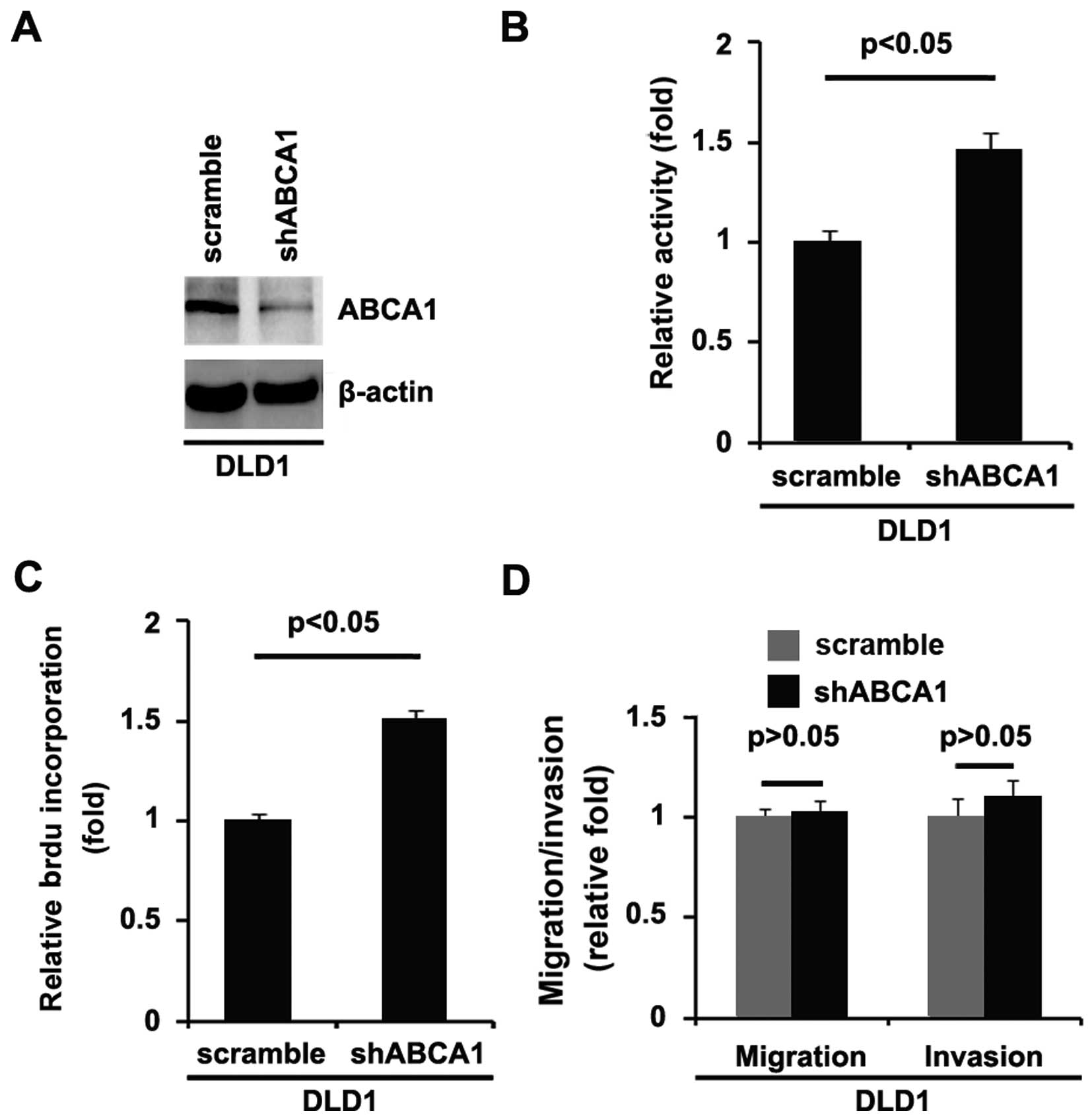
In order to further identify the role of ABCA1 in
the regulation of the proliferation of colon cancer cells, DLD1
cells were transfected with shABCA1 plasmids. We found that ABCA1
protein was significantly decreased by the shABCA1 plasmids
(Fig. 3A). After stable
transfection, the proliferation rates of DLD1 cells were assessed
by MTT assay. The results showed that silencing of ABCA1
significantly increased the proliferation rate of the DLD1 cells
(Fig. 3B). This was further
revealed by BrdU incorporation analysis showing that transfection
with shABCA1 resulted in increased DNA synthesis activity per
viable cell in the DLD1 cells (Fig.
3C).
Given that silencing of ABCA1 promotes DLD1 cell
proliferation, we next sought to determine whether silencing of
ABCA1 would have any impact on the migration and invasion in DLD1
cells. The migration and invasion assays of DLD1 cells showed that
silencing of ABCA1 did not affect migration and invasion (Fig. 3D).
Silencing of ABCA1 inhibits apoptosis in
colon cancer LDL1 cells
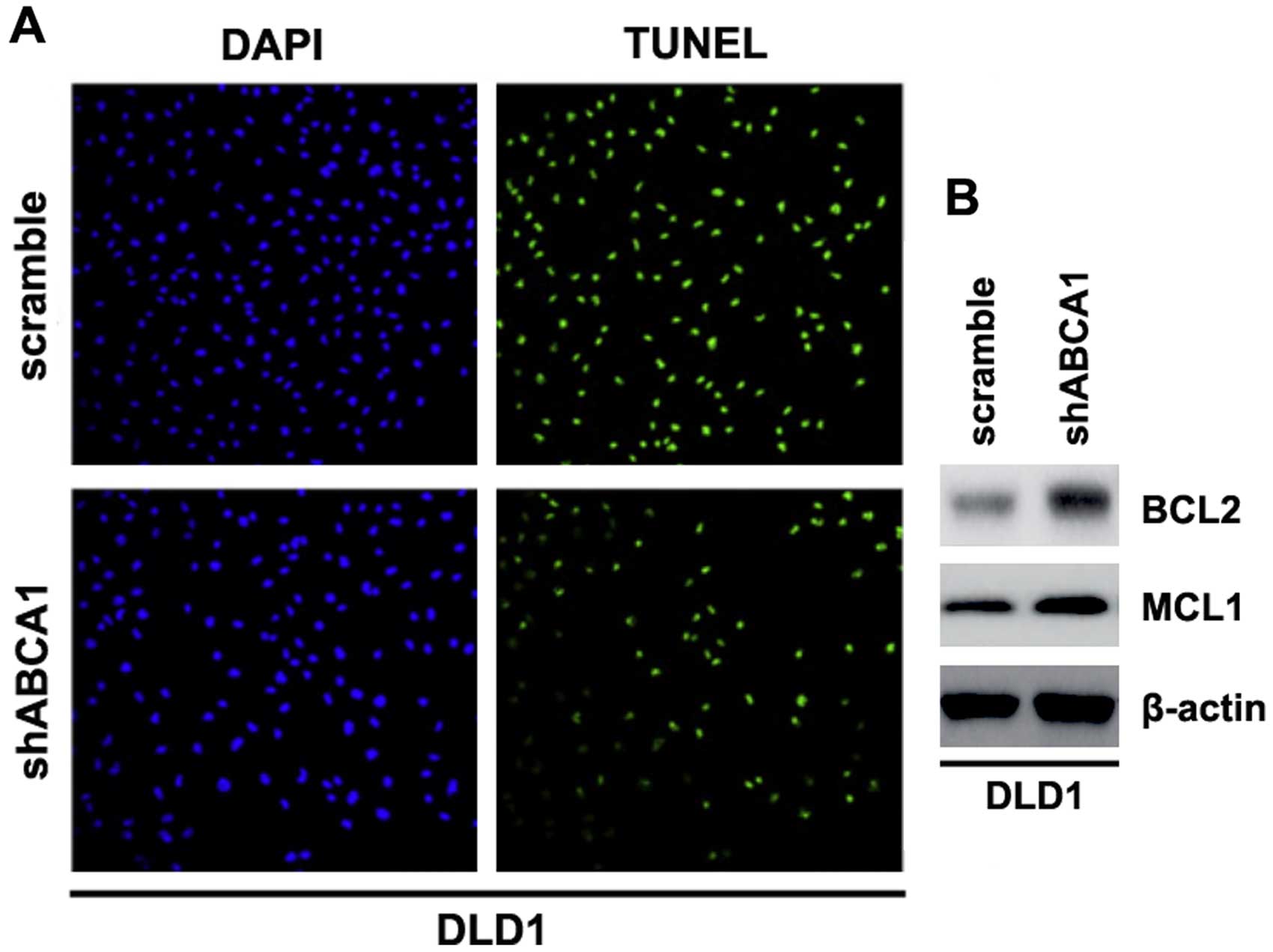
Having demonstrated that silencing of ABCA1 promotes
the proliferation of LDL1 cells, to provide evidence that ABCA1 is
involved in the regulation of apoptosis of LDL1 cells, we performed
TUNEL assay to analyze whether silencing of ABCA1 affects the
apoptosis in LDL1 cells. Through TUNEL assay, we observed a change
in the apoptotic rate in the LDL1 cells transfected with shABCA1.
Namely, silencing of ABCA1 inhibited the apoptosis of the LDL1
cells (Fig. 4A).
We also performed western blotting to identify
whether protein levels of apoptosis-associated markers were also
affected by shABCA1 in the cells. BCL2 and MCL1 are important
anti-apoptotic molecules (38,39).
We showed that BCL2 and MCL1 expression levels were upregulated by
silencing of ABCA1 in the cells (Fig.
4B).
miR-183 suppresses ABCA1 protein
expression in the colon cancer cells
Having demonstrated that ABCA1 expression is
specifically downregulated in colon cancer and it inhibits the
proliferation of colon cancer cells, we aimed to ascertain the
mechanisms involved in the inhibition of ABCA1 expression in colon
cancer. MicroRNAs (miRNAs) are a new class of small (~22
nucleotide) non-coding RNAs that negatively regulate protein-coding
gene expression by targeting mRNA degradation or translation
inhibition (22,40–44).
Upregulation of specific miRNAs can contribute to tumor-suppressive
gene downregulation (45). Thus, we
hypothesized that ABCA1 is downregulated by overexpression of a
specific miRNA in colon cancer.
To further confirm our hypothesis, we used a
commonly used prediction algorithm - miRanda (http://www.microrna.org/microrna/home.do) to analyze
the 3′UTR of ABCA1. The algorithm predicted that miR-183 may target
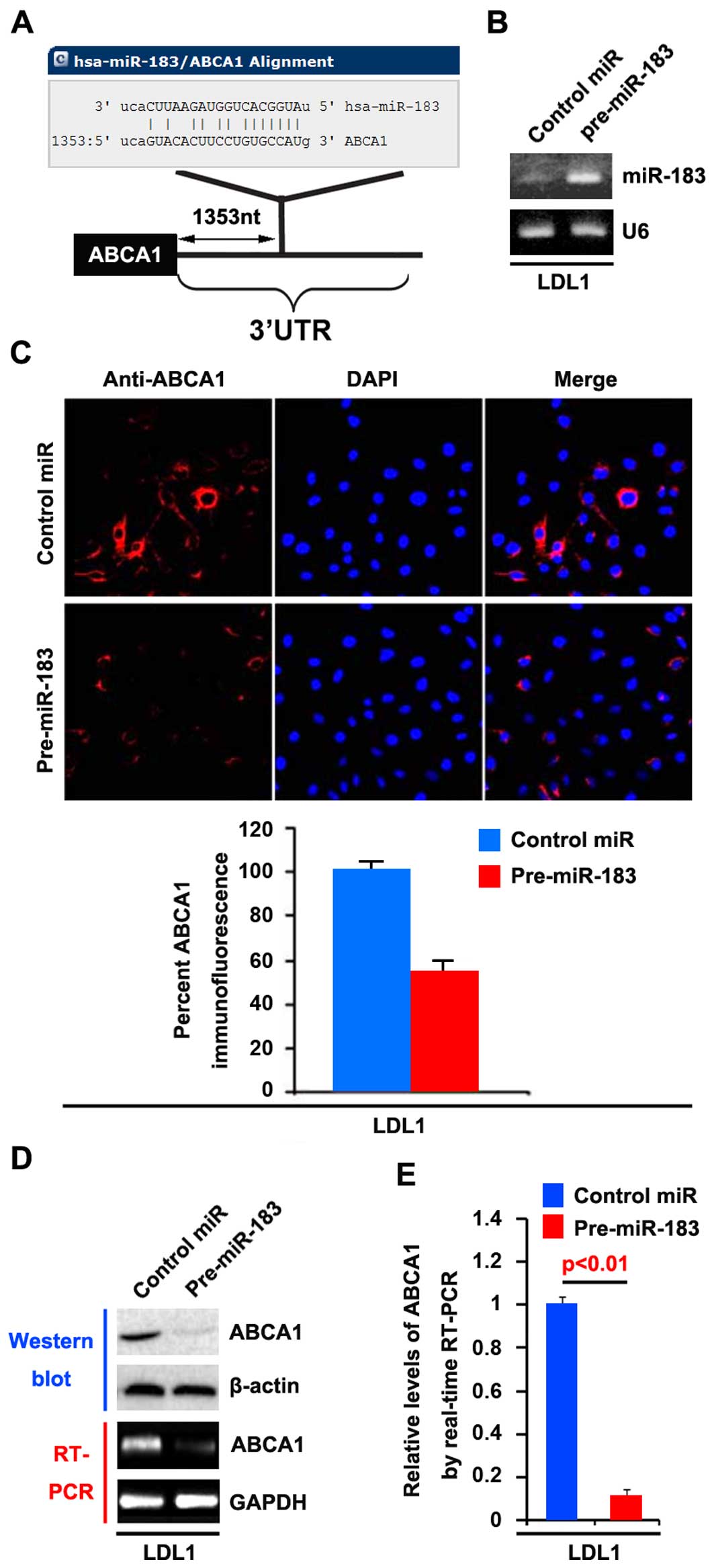
the 3′UTR of ABCA1 (Fig. 5A). Thus,
we reasoned that miR-183 could downregulate ABCA1 expression by
targeting its 3′UTR in colon cancer and that ABCA1 was suppressed
in colon cancer, due to the upregulation of miR-183 (32).
In an attempt to identify the role of miR-183 in the
regulation of ABCA1 expression in LDL1 cells, the cells were
transfected with pre-miR-183 and control miR. After transfection,
miR-183 expression was detected by real-time PCR. The results
showed that miR-183 was increased by pre-miR-183 in the cells
(Fig. 5B).
We then performed immunofluorescence analyses in
LDL1 cells transfected with pre-miR-183 or control miR. The results
showed that ABCA1 protein was evidently suppressed in the cells
transfected with pre-miR-183 (Fig.
5C). We next performed RT-PCR and western blotting to detect
ABCA1 expression in LDL1 cells transfected with pre-miR-183 or
control miR. The results showed that ABCA1 protein (Fig. 5D) and mRNA expression (Fig. 5D) were significantly down-regulated
in the cells transfected with pre-miR-183. Consistent with the
results of RT-PCR, real-time PCR demonstrated that ABCA1 mRNA was
reduced in the LDL1 cells transfected with pre-miR-183, compared
with the control miR-transfected group (Fig. 5E). All the data demonstrated that
miR-183 suppresses ABCA1 mRNA and protein expression in colon
cancer cells.
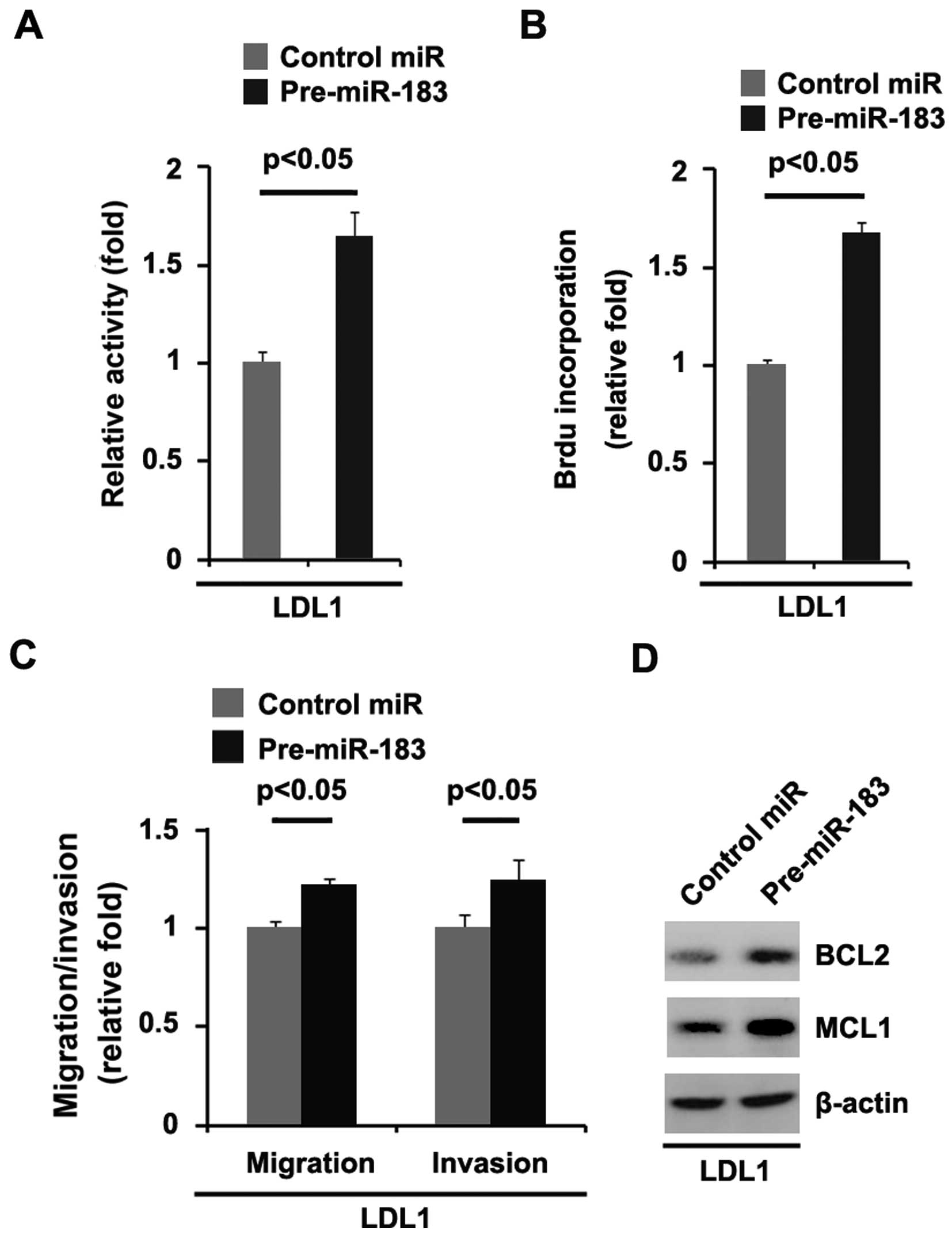
miR-183 promotes the proliferation,
migration and invasion of colon cancer DLD1 cells
In order to further identify the role of miR-183 in
regulating the proliferation of DLD1 cells, the cells were
transfected with pre-miR-183 and control miR. After stable
transfection, the proliferation rates of the DLD1 cells were
assessed by MTT assay. The results showed that miR-183
significantly promoted the proliferation rate of DLD1 cells
(Fig. 6A). This was further
revealed by BrdU incorporation analysis showing that transfection
with pre-miR-183 resulted in increased DNA synthesis activity per
viable cell in the DLD1 cells (Fig.
6B).
Given that miR-183 obviously promotes DLD1 cell
proliferation, we next sought to determine whether miR-183 has any
impact on the migration and invasion of DLD1 cells. The migration
and invasion assays showed that miR-183 promoted the migration and
invasion of the DLD1 cells (Fig.
6C).
We also performed western blot analysis to identify
whether protein levels of apoptosis-associated markers were also
affected by pre-miR-183 in the cells. Our studies showed that BCL2
and MCL1 expression levels were upregulated by pre-miR-183 in the
cells (Fig. 6D).
Discussion
Colorectal cancer is the second leading cause of
death by cancer worldwide (46).
Western populations have a 1 in 20 lifetime risk of developing the
disease and in many countries the rates are increasing (47). Despite major advances in our
understanding of colon cancer, successful treatment remains
dependent on early diagnosis and surgical intervention. Current
oncological treatments such as radiotherapy and chemotherapy have
relatively little impact on long-term survival and currently hope
is pinned on screening to diagnose cancers and remove them even
earlier. Yet, current population-based screening methods can only
reduce colorectal cancer deaths by 20–30% (48) indicating that new approaches based
on better understanding of the disease are needed before colorectal
cancer can be added to the list of treatable or preventable
malignancies.
Cholesterol is necessary for many functions in
mammalian cells. Consequently, under normal conditions the
intracellular cholesterol content is carefully regulated through
processes of synthesis, uptake and efflux, with efflux carried out
mainly by ABCA1 and ABCG1 (ATP-binding cassette transporters)
(49,50). Deregulation of cholesterol
homeostasis in human cancer is associated with the loss of ABCA1
(51). Yet, there is no report in
colon cancer. For the first time, we demonstrated that ABCA1
protein was downregulated in colon cancer. Its overexpression
inhibited proliferation while the silencing of ABCA1 promoted
proliferation and inhibited apoptosis in colon cancer cells,
implying that it is a tumor-suppressive gene in this disease.
Upregulation of specific miRNAs can contribute to
downregulation of tumor-suppressive genes (33–35).
Thus, we hypothesized that ABCA1 is downregulated by overexpression
of a specific miRNA in colon cancer. Recently, it was reported that
miR-183 is an oncogene in cancer (32). For example, the upregulation of
miR-183 is associated with the onset and progression of
hepatocellular carcinoma (HCC) (52). miR-183 was found to be an oncogene
targeting Dkk-3 and SMAD4 in prostate cancer (53), and miR-183 functions as an oncogene
by targeting the transcription factor EGR1 and promoting tumor cell
migration in synovial sarcoma and colon cancer cell lines (32). In addition, miR-183 was found to
inhibit TGF-β1-induced apoptosis by downregulation of PDCD4
expression in human HCC cells (54); The increased expression of miR-183
is closely related to advanced clinical stage, lymph node and
distant metastases, and poor prognosis of colorectal cancer,
indicating that miR-183 may serve as a predictive biomarker for the
prognosis or the aggressiveness of colorectal cancer (55). Yet, the mechanism of miR-183 as an
oncogene keeps emerging. We found that miR-183 promoted the
proliferation and inhibited the apoptosis in colon cancer by
targeting ABCA1. We observed that miR-183 promoted migration and
invasion, while ABCA1 did not affect migration and invasion in
colon cancer cells, implying that miR-183 may regulate migration
and invasion through other target genes.
Recognition of the function of ABCA1 and its
regulation in tumors will ultimately provide a better understanding
of the signaling pathways that can be therapeutically modulated.
Further investigation of the role of ABCA1 in colon cancer is
warranted.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
2
|
Dessí S, Batetta B, Anchisi C, Pani P,
Costelli P, Tessitore L and Baccino FM: Cholesterol metabolism
during the growth of a rat ascites hepatoma (Yoshida AH-130). Br J
Cancer. 66:787–793. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dessì S, Batetta B, Pulisci D, Spano O,
Anchisi C, Tessitore L, Costelli P, Baccino FM, Aroasio E and Pani
P: Cholesterol content in tumor tissues is inversely associated
with high-density lipoprotein cholesterol in serum in patients with
gastrointestinal cancer. Cancer. 73:253–258. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kolanjiappan K, Ramachandran CR and
Manoharan S: Biochemical changes in tumor tissues of oral cancer
patients. Clin Biochem. 36:61–65. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rudling M and Collins VP: Low density
lipoprotein receptor and 3-hydroxy-3-methylglutaryl coenzyme A
reductase mRNA levels are coordinately reduced in human renal cell
carcinoma. Biochim Biophys Acta. 1299:75–79. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schaffner CP: Prostatic cholesterol
metabolism: Regulation and alteration. Prog Clin Biol Res.
75A:279–324. 1981.PubMed/NCBI
|
|
7
|
Yoshioka Y, Sasaki J, Yamamoto M, Saitoh
K, Nakaya S and Kubokawa M: Quantitation by (1)H-NMR of dolichol,
cholesterol and choline-containing lipids in extracts of normal and
phathological thyroid tissue. NMR Biomed. 13:377–383. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Järvinen R, Knekt P, Hakulinen T, Rissanen
H and Heliövaara M: Dietary fat, cholesterol and colorectal cancer
in a prospective study. Br J Cancer. 85:357–361. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Caruso MG, Notarnicola M, Santillo M,
Cavallini A and Di Leo A: Enhanced 3-hydroxy-3-methyl-glutaryl
coenzyme A reductase activity in human colorectal cancer not
expressing low density lipoprotein receptor. Anticancer Res.
19:451–454. 1999.PubMed/NCBI
|
|
10
|
Caruso MG, Notarnicola M, Cavallini A and
Di Leo A: 3-Hydroxy-3-methylglutaryl coenzyme A reductase activity
and low-density lipoprotein receptor expression in diffuse-type and
intestinal-type human gastric cancer. J Gastroenterol. 37:504–508.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Notarnicola M, Messa C, Pricci M, Guerra
V, Altomare DF, Montemurro S and Caruso MG: Up-regulation of
3-hydroxy-3-methylglutaryl coenzyme A reductase activity in
left-sided human colon cancer. Anticancer Res. 24:3837–3842.
2004.
|
|
12
|
Gregg RG, Davidson M and Wilce PA:
Cholesterol synthesis and HMG CoA reductase activity during
hepatocarcinogenesis in rats. Int J Biochem. 18:389–393. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hentosh P, Yuh SH, Elson CE and Peffley
DM: Sterol-independent regulation of 3-hydroxy-3-methylglutaryl
coenzyme A reductase in tumor cells. Mol Carcinog. 32:154–166.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Siperstein MD: Cholesterol,
cholesterogenesis and cancer. Adv Exp Med Biol. 369:155–166. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Graziani SR, Igreja FA, Hegg R, Meneghetti
C, Brandizzi LI, Barboza R, Amâncio RF, Pinotti JA and Maranhão RC:
Uptake of a cholesterol-rich emulsion by breast cancer. Gynecol
Oncol. 85:493–497. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schimanski S, Wild PJ, Treeck O, Horn F,
Sigruener A, Rudolph C, Blaszyk H, Klinkhammer-Schalke M, Ortmann
O, Hartmann A, et al: Expression of the lipid transporters ABCA3
and ABCA1 is diminished in human breast cancer tissue. Horm Metab
Res. 42:102–109. 2010. View Article : Google Scholar
|
|
17
|
Tatidis L, Masquelier M and Vitols S:
Elevated uptake of low density lipoprotein by drug resistant human
leukemic cell lines. Biochem Pharmacol. 63:2169–2180. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Basso K, Margolin AA, Stolovitzky G, Klein
U, Dalla-Favera R and Califano A: Reverse engineering of regulatory
networks in human B cells. Nat Genet. 37:382–390. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ki DH, Jeung HC, Park CH, Kang SH, Lee GY,
Lee WS, Kim NK, Chung HC and Rha SY: Whole genome analysis for
liver metastasis gene signatures in colorectal cancer. Int J
Cancer. 121:2005–2012. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Moustafa MA, Ogino D, Nishimura M, Ueda N,
Naito S, Furukawa M, Uchida T, Ikai I, Sawada H and Fukumoto M:
Comparative analysis of ATP-binding cassette (ABC) transporter gene
expression levels in peripheral blood leukocytes and in liver with
hepatocellular carcinoma. Cancer Sci. 95:530–536. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Attie AD: ABCA1: At the nexus of
cholesterol, HDL and atherosclerosis. Trends Biochem Sci.
32:172–179. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim VN: Small RNAs: Classification,
biogenesis, and function. Mol Cells. 19:1–15. 2005.PubMed/NCBI
|
|
24
|
Najafi-Shoushtari SH, Kristo F, Li Y,
Shioda T, Cohen DE, Gerszten RE and Näär AM: MicroRNA-33 and the
SREBP host genes cooperate to control cholesterol homeostasis.
Science. 328:1566–1569. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ramirez CM, Dávalos A, Goedeke L, Salerno
AG, Warrier N, Cirera-Salinas D, Suárez Y and Fernández-Hernando C:
MicroRNA-758 regulates cholesterol efflux through
posttranscriptional repression of ATP-binding cassette transporter
A1. Arterioscler Thromb Vasc Biol. 31:2707–2714. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ramírez CM, Rotllan N, Vlassov AV, Dávalos
A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno
A, et al: Control of cholesterol metabolism and plasma high-density
lipoprotein levels by microRNA-144. Circ Res. 112:1592–1601. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mendell JT: miRiad roles for the miR-17-92
cluster in development and disease. Cell. 133:217–222. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Subramanian S, Lui WO, Lee CH, Espinosa I,
Nielsen TO, Heinrich MC, Corless CL, Fire AZ and van de Rijn M:
MicroRNA expression signature of human sarcomas. Oncogene.
27:2015–2026. 2008. View Article : Google Scholar
|
|
30
|
Sarver AL, Phalak R, Thayanithy V and
Subramanian S: S-MED: Sarcoma microRNA expression database. Lab
Invest. 90:753–761. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Calin GA, Cimmino A, Fabbri M, Ferracin M,
Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI,
et al: miR-15a and miR-16-1 cluster functions in human leukemia.
Proc Natl Acad Sci USA. 105:5166–5171. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sarver AL, Li L and Subramanian S:
MicroRNA miR-183 functions as an oncogene by targeting the
transcription factor EGR1 and promoting tumor cell migration.
Cancer Res. 70:9570–9580. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang XW, Xi XQ, Wu J, Wan YY, Hui HX and
Cao XF: Micro-RNA-206 attenuates tumor proliferation and migration
involving the downregulation of NOTCH3 in colorectal cancer. Oncol
Rep. 33:1402–1410. 2015.PubMed/NCBI
|
|
37
|
Bekker-Méndez C, Guzmán-Aguilar RM,
Hernández-Cueto MA, Huerta-Yepez S, Jarillo-Luna RA,
González-Veyrand E and González-Bonilla CR: TUNEL-positive cells in
the surgical border of an amputation due to infected diabetic foot.
Mol Med Rep. 5:363–372. 2012.
|
|
38
|
Ji F, Zhang H, Wang Y, Li M, Xu W, Kang Y,
Wang Z, Wang Z, Cheng P, Tong D, et al: MicroRNA-133a,
downregulated in osteosarcoma, suppresses proliferation and
promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone. 56:220–226.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang H, Cai X, Wang Y, Tang H, Tong D and
Ji F: microRNA-143, down-regulated in osteosarcoma, promotes
apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol
Rep. 24:1363–1369. 2010.PubMed/NCBI
|
|
40
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pasquinelli AE, Reinhart BJ, Slack F,
Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B,
Müller P, et al: Conservation of the sequence and temporal
expression of let-7 heterochronic regulatory RNA. Nature.
408:86–89. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
42
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Farh KK, Grimson A, Jan C, Lewis BP,
Johnston WK, Lim LP, Burge CB and Bartel DP: The widespread impact
of mammalian microRNAs on mRNA repression and evolution. Science.
310:1817–1821. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ma L, Young J, Prabhala H, Pan E, Mestdagh
P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S,
et al: miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin
and cancer metastasis. Nat Cell Biol. 12:247–256. 2010.PubMed/NCBI
|
|
46
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Center MM, Jemal A, Smith RA and Ward E:
Worldwide variations in colorectal cancer. CA Cancer J Clin.
59:366–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Heresbach D, Manfredi S, D'halluin PN,
Bretagne JF and Branger B: Review in depth and meta-analysis of
controlled trials on colorectal cancer screening by faecal occult
blood test. Eur J Gastroenterol Hepatol. 18:427–433. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Oram JF and Lawn RM: ABCA1. The gatekeeper
for eliminating excess tissue cholesterol. J Lipid Res.
42:1173–1179. 2001.PubMed/NCBI
|
|
50
|
Kennedy MA, Barrera GC, Nakamura K, Baldán
A, Tarr P, Fishbein MC, Frank J, Francone OL and Edwards PA: ABCG1
has a critical role in mediating cholesterol efflux to HDL and
preventing cellular lipid accumulation. Cell Metab. 1:121–131.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Solomon KR, Allott EH, Freeman MR and
Freedland SJ: Words of wisdom. Re: Dysregulation of cholesterol
homeostasis in human prostate cancer through loss of ABCA1. Eur
Urol. 63:1128–1129. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liang Z, Gao Y, Shi W, Zhai D, Li S, Jing
L, Guo H, Liu T, Wang Y and Du Z: Expression and significance of
microRNA-183 in hepatocellular carcinoma. Scientific World Journal.
2013:3818742013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ueno K, Hirata H, Shahryari V, Deng G,
Tanaka Y, Tabatabai ZL, Hinoda Y and Dahiya R: microRNA-183 is an
oncogene targeting Dkk-3 and SMAD4 in prostate cancer. Br J Cancer.
108:1659–1667. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li J, Fu H, Xu C, Tie Y, Xing R, Zhu J,
Qin Y, Sun Z and Zheng X: miR-183 inhibits TGF-beta1-induced
apoptosis by downregulation of PDCD4 expression in human
hepatocellular carcinoma cells. BMC Cancer. 10:3542010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhou T, Zhang GJ, Zhou H, Xiao HX and Li
Y: Overexpression of microRNA-183 in human colorectal cancer and
its clinical significance. Eur J Gastroenterol Hepatol. 26:229–233.
2014. View Article : Google Scholar
|