Introduction
Esophageal carcinoma is one of the most common
malignancies and has been ranked as the sixth leading cause of
cancer-related death worldwide (1–4).
Esophageal squamous cell carcinoma (ESCC), accounting for
approximately 90% of all esophageal carcinoma cases, is more
predominant in Asian countries (5).
In the past decades, although tremendous strides have been made in
therapeutic strategies including surgical resection, radiotherapy
and chemotherapy, tumor invasion and metastasis still invariably
heralds a poor prognosis and a low 5-year survival rate in ESCC
patients (6–8). Therefore, a better understanding of
the molecular mechanisms involved in tumor invasion and metastasis
and discovery of a more effective target for anticancer therapy may
help to prolong the survival and improve the life quality of
patients with ESCC.
The a disintegrin and metalloproteases (ADAMs), a
family of zinc-dependent transmembrane proteins, contain a
metalloprotease and a disintegrin-like domain (9–11) and
are closely associated with the degradation of the basement
membrane, cell migration, cell fusion and signal transduction
(12,13), thus playing important roles in the
tumorigenesis, proliferation, differentiation, adhesion, invasion
and metastasis of cancers (14–16).
To date, approximately 40 ADAM members have been identified in this
family, however only 21 are thought to function in humans (10). ADAM10 is an important
proteolytically active member of this family (17). Emerging evidence suggests that
ADAM10 is overexpressed in a variety of cancers and is associated
with the tumor progression of breast cancer (18), non-small cell lung cancer (NSCLC)
(19), nasopharyngeal carcinoma
(20), oral squamous cell carcinoma
(21,22) tongue squamous cell carcinoma
(23), hepatocellular carcinoma
(24,25), gastric cancer (26), colon cancer (27), pancreatic carcinoma (28), bladder cancer (29) and renal cell carcinoma (30). Proteolytically active ADAM10 acts as
an ectodomain-sheddase, which can release the extracellular domain
of membrane-bound proteins, such as adhesion molecules, growth
factors, cytokines, chemokines and receptors; through these actions
they are able to disrupt the tumor microenvironment and modulate
key processes involved in tumor cell proliferation, migration and
angiogenesis (31,32). E-cadherin is a transmembrane
molecule that functions as an adhesion molecule, and is reported as
an ADAM10 substrate (33,34). ADAM10 activity could lead to
elevated shedding of E-cadherin and loss of cell-cell contact
(35), further enhancing migratory
and metastatic behavior (36).
However, to date there has been no related study concerning the
role of ADAM10 in the tumorigenesis, invasion and metastasis of
ESCC. Moreover, whether the potential mechanism by which ADAM10
promotes tumor progression is associated with E-cadherin shedding
is still unclear.
Herein, in the present study, we first detected
expression of ADAM10 protein in ESCC tissues in vivo and
compared it to E-cadherin expression and clinicopathological
parameters associated with tumor invasion and metastasis, including
tumor differentiation, depth of invasion, lymph node metastasis and
TNM stage. In addition, ADAM10 expression in ESCC cell lines was
examined in vitro and compared to cell invasion and
migration ability. Furthermore, we inhibited the expression of
ADAM10 in ESCC cell lines by using ADAM10 siRNA and analyzed the
effect of the downregulation of ADAM10 on cell proliferation,
invasion and migration and the expression levels of E-cadherin
in vitro.
Materials and methods
Statement of ethics
This study was performed according to ethical
protocol approved by the First Affiliated Hospital of Zhengzhou
University, Henan, China. All of the clinical tissue samples used
in this study were collected after obtaining written informed
consents. All efforts were made to minimize suffering.
Tissue specimens
A total of 122 ESCC tissue specimens and 60 adjacent
normal esophageal mucosa were obatined from patients (73 male and
49 female) who underwent surgical resection at The First Affiliated
Hospital of Zhengzhou University, Henan, China. None of the
patients had previously received neoadjuvant radiotherapy,
chemotherapy or immunotherapy. Tissue sections were cut at a
thickness of 4 µm for in situ hybridization (ISH) and
immunohistochemistry (IHC). Clinical features of all the samples
are listed in Table I. The
diagnosis of ESCC was reconfirmed by two board-certified
pathologists.
 | Table IProtein and mRNA expression of ADAM10
and E-cadherin in ESCC tissues and corresponding normal esophageal
mucosa. |
Table I
Protein and mRNA expression of ADAM10
and E-cadherin in ESCC tissues and corresponding normal esophageal
mucosa.
| n | ADAM10
| χ2 | P-value | E-cadherin
| χ2 | P-value |
|---|
| − | + | − | + |
|---|
| Protein |
| Normal mucosa | 60 | 49 | 11 | 37.164 | 0.000 | 7 | 53 | 51.120 | 0.000 |
| ESCC | 122 | 41 | 81 | | | 83 | 39 | | |
| mRNA |
| Normal mucosa | 60 | 51 | 9 | 31.550 | 0.000 | 0 | 60 | 59.946 | 0.000 |
| ESCC | 122 | 50 | 72 | | | 73 | 49 | | |
In situ hybridization (ISH) and
evaluation of ISH staining
Paraffin-embedded slides were deparaffinized,
rehydrated, and immersed in 3% hydrogen peroxide. Then the slides
were digested in pepsin solution, and incubated with
pre-hybridization solution and hybridization solution sequentially.
After visualization of ISH using BCIP/NBT, the slides were
counter-stained with Nuclear Fast Red. The digoxin-labeled probe
sequences used in this study were as follows: ADAM10 forward
primer, 5-GAGGAGTGTACGTGTGCCAGTT-3 and reverse primer,
5-GACCACTGAAGTGCCTACTCCA-3; E-cadherin forward primer,
5-AAAGGCCCATTTCCTAAAAACCT-3 and reverse primer,
5-TGCGTTCTCTATCCAGAGGCT-3. ADAM10- and E-cadherin-positive staining
was viewed as kyano-purple granule-like material that was located
in the cytoplasm. The staining index was calculated by adding the
scores for the percentage of positively stained cells (1, 0–10%; 2,
>10–30%; 3, >30–70%; and 4, >70%) and the staining
intensity (1, weak; 2, moderate; and 3, strong). A score of 0–2 was
designated as a negative score (−), and ≥3 as a positive score (+).
All of the staining analyses were independently performed by two
pathologists.
Immunohistochemistry (IHC) and evaluation
of IHC staining
Paraffin-embedded slides were deparaffinized,
rehydrated, and subjected to antigen retrieval by using a microwave
oven. Then the slides were incubated with primary antibodies
anti-ADAM10 (1:200, Abcam, USA) or anti-E-cadherin (1:100, Santa
Cruz, CA, USA) respectively, and subsequently incubated with
biotinylated anti-rabbit IgG secondary antibody. After
visualization of the immunoreactivity using DAB, the slides were
counterstained with hematoxylin. ADAM10-and E-cadherin-positive
staining was viewed as brown-yellow granule-like material that was
located in the membrane and/or cytoplasm. The staining index was
calculated by adding the scores for the percentage of positively
stained cells (1, 0–10%; 2, >10–50%; and 3, >50%) and the
staining intensity (1, weak; 2, moderate; and 3, strong). A score
of 0–2 was designated as a negative score (−), and ≥3 as a positive
score (+). All of the staining analyses were independently
performed by two pathologists.
Cell lines and culture
Human ESCC cell line EC-1 was supplied by Professor
Shihua Cao (Hong Kong University). Eca109 and TE-1 cells were
maintained in the Key Laboratory of Tumor Pathology of Zhengzhou
University. Normal esophageal epithelial cells (NEECs) of primary
culture were obtained from fresh biopsies of esophageal mucosal
tissues, during our previous experiment. All cells were cultured in
RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS),
100 µg/ml streptomycin and 100 U/ml penicillin, in a
humidified incubator containing 5% CO2 at 37°C.
Additionally, 1.0 ng/ml of epidermal growth factor (EGF) was added
to the medium for the NEECs.
ADAM10 siRNA transfection of EC-1
cells
The sequences of ADAM10 siRNA and the control
nonspecific siRNA were as follows: sense strand,
AAAGGAUUCCCAUACUGAC and antisense strand, GUCAGUAUGGGAAUCCUUU for
ADAM10; sense strand, AUCUUGAUCUUCAUUGUGC and antisense strand,
GCACAAUGAAGAUCAAGAU for the nonspecific control. EC-1 cells grown
until 80–90% confluency were transfected with 100 nM ADAM10 siRNA
using Lipofectamine 2000 (Invitrogen, Carlsbad, USA) according to
the manufacturer's instructions. EC-1 cells untreated or
transfected with the nonspecific siRNA were used as control. The
cells were cultured for an additional 48 h in a humidified
incubator containing 5% CO2 at 37°C before being
harvested for further assays.
RNA extraction, reverse transcription
(RT) and real-time PCR
Total RNA was extracted from the cultured cells
using the TRIzol reagent (Invitrogen, CA, USA) according to the
manufacturer's protocol. Two micrograms of total RNA was used for
cDNA synthesis using the HiScript II RT kit (Vazyme, Nanjing,
China). To evaluate ADAM10 and E-cadherin mRNA expression,
real-time PCR was performed using a PRISM 7300 Sequence Detection
system (Applied Biosystems, Foster City, CA, USA). β-actin was
selected as an internal control. The primer sequences were as
follows: forward, 5-CACGAGAAGCTGTGATTGCC-3 and reverse,
5-ATTCCGGAGAAGTCTGTGGTC-3 for ADAM10; forward,
5-TGATTCTGCTGCTCTTGCTG-3 and reverse, 5-CAAAGTCCTGGTCCTCTTCTCC-3
for E-cadherin; forward, 5-TTCGAGCAAGAGTGGCCA-3 and reverse,
5-AGGTAGTTTCGTGGATGCCA-3 for β-actin. ADAM10 and E-cadherin
expression levels were normalized to β-actin and analyzed by using
the 2−0394ΔCt method (37). All experiments were carried out in
triplicate.
Protein extraction and western blot
analysis
Protein was extracted from the cultured cells using
RIPA lysis buffer (Well, Shanghai, China) according to the
manufacturer's protocol. The protein concentration was determined
using the BCA kit (Bioss, Beijing, China). For western blot
analysis, 50 µg of protein extract from each cell line was
separated by SDS-PAGE and transferred onto a PVDF membrane. After
blocking with 5% non-fat milk in PBST for 1 h at room temperature,
the membrane was incubated with the primary antibody (ADAM10,
1:500, 84 kDa; E-cadherin, 1:200, 120 kDa) at 4°C overnight.
β-actin (1:5,000, 43 kDa) was selected as a loading control.
Protein bands were detected with the ECL system (Well, Shanghai,
China) and exposed to X-ray film. Quantification was performed by
using ImageJ analysis tool. All experiments were carried out in
triplicate.
Matrigel invasion assay
Tumor cell invasion ability was analyzed using an
invasion chamber (Corning®, Corning, NY, USA). Briefly,
a polycarbonate membrane with 8-µm pores was coated with
Matrigel and dried at room temperature for 1 h in a 24-well plate.
Cells in serum-free medium were seeded into the upper chamber of
the wells and RPMI-1640 with 10% FBS was added into the lower
chamber as a chemoattractant. Following incubation at 37°C in 5%
CO2 for 24 h, cells on the upper surface of the membrane
were scrubbed away by a cotton swab. Cells invading through the
membrane and adhering to the lower surface of the membrane were
fixed with 95% alcohol, stained with 0.05% crystal violet for 1 h,
and quantitated by measuring the number of stained cells in five
individual fields at a ×400 magnification under a light
micro-scope. All experiments were carries out in triplicate.
Scratch wound healing assay
Cell migration ability was measured by a scratch
wounding healing assay. Cells were seeded into 12-well plates and
incubated in RPMI-1640 with 10% FBS medium. Following incubation
for 24 h, a scratch would was made across the center of the
monolayer cells by using a sterile 200-µl pipette tip.
Subsequently, the cells were washed with PBS to remove the detached
cells and incubated with serum-free medium for an additional 48 h.
Images of the cells that had migrated into the cell-free scratch
wound area were acquired at a ×200 magnification under a reversed
light microscope. Cell migration ability was quantitated by
measuring the width of the wounds in at least five representative
fields and is expressed as 1 minus the average percent of the wound
closure compared with the initial wound area measured. All
experiments were carried out in triplicate.
Cell proliferation assay
Cell proliferation ability was measured by Cell
Counting Kit-8 (CCK-8) assay. Briefly, the cells were seeded into
96-well plates at a density of 5×103 cells/well and
incubated for 0, 24, 48 and 72 h, respectively. At different time
intervals, 10 µl of CCK-8 solution (Sangon, Shanghai, China)
was added to the corresponding wells and incubated for an
additional 4 h. The staining intensity in the medium was measured
by determining the absorbance (OD values) at 450 nm to obtain cell
growth curves. All experiments were carried out in triplicate.
Statistical analysis
All the data were analyzed using SPSS version 17.0
statistical package. The difference in counted data was examined by
the χ2 test. Meanwhile, quantitative data are expressed
as mean ± SD, and differences in quantitative data were examined by
the t-test. A P-value <0.05 was defined as indicating a
statistically significant result.
Results
ADAM10 expression is elevated and
E-cadherin expression is reduced in ESCC tissues
Emerging evidence suggests that ADAM10 is
overexpressed in a variety of cancers and participates in tumor
progression (18–30). Thus, IHC was initially performed to
determine the protein expression level of ADAM10 in ESCC tissues
and corresponding normal esophageal mucosal tissues.
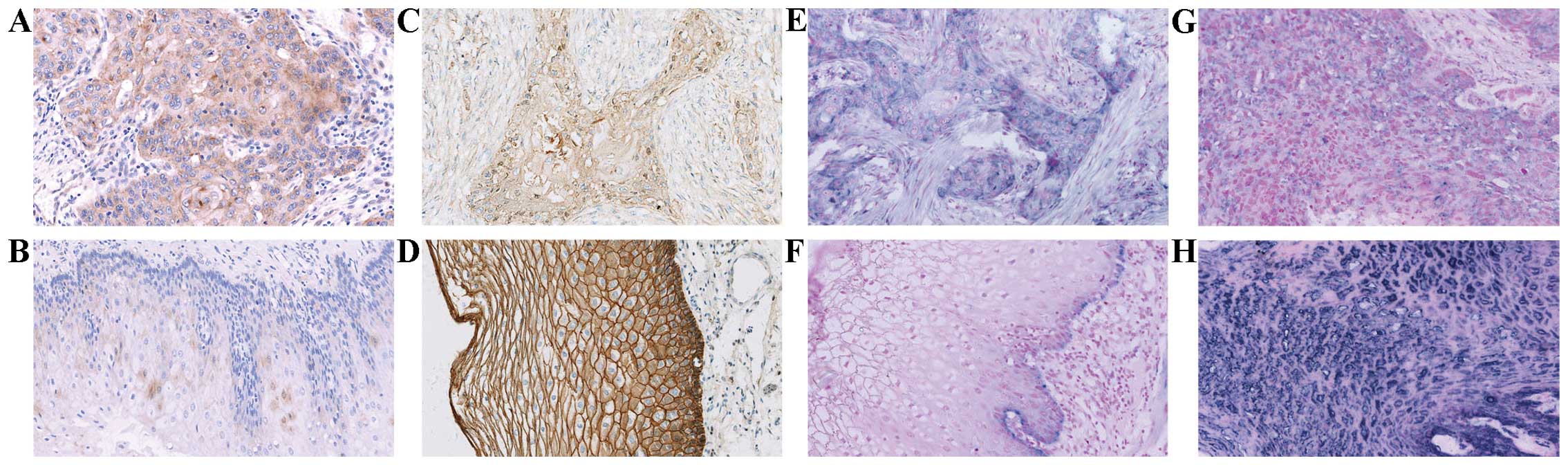
ADAM10-positive signals showed brown-yellow granules in the
membrane and/or cytoplasm (Fig. 1A and
B). Cytoplasmic expression of ADAM10 protein was strong in the
ESCC tissues (Fig. 1A), but weak in
the normal esophageal mucosa (Fig.
1B). The positive rate of ADAM10 was 66.4% (81/122) in the ESCC
tissues, significantly higher than that in the corresponding normal
esophageal mucosa [18.3% (11/60); P=0.000) (Table I).
Since ADAM10 mediates E-cadherin shedding and
regulates epithelial cell-cell adhesion (33), we next detected the expression level
of E-cadherin in the ESCC tissues and the corresponding normal
esophageal mucosa tissues. E-cadherin-positive signals were
presented as brown-yellow granules in the membrane and/or cytoplasm
(Fig. 1C and D). Membranal and/or
cytoplasmic expression of E-cadherin protein was negative or
moderate (Fig. 1C) in the ESCC
tissues, but strong in the normal esophageal mucosa (Fig. 1D). The positive rate of E-cadherin
was 32.0% (39/122) in the ESCC tissues, significantly lower than
that in the corresponding normal esophageal mucosa [88.3% (53/60);
P=0.000) (Table I).
Furthermore, in situ hybridization (ISH) was
used to detect the mRNA expression levels of ADAM10 and E-cadherin
in the ESCC tissues and corresponding normal esophageal mucosa
tissues. ADAM10-and E-cadherin-positive signals both showed
kyano-purple granule in cytoplasm (Fig.
1E–H). Expression of ADAM10 mRNA was strong in the ESCC tissues
(Fig. 1E), but was present only in
the basement of the normal esophageal mucosa (Fig. 1F). Expression of E-cadherin mRNA was
negative or weak (Fig. 1G) in the
ESCC tissues, but strong in the normal esophageal mucosa (Fig. 1H). The positive rates of ADAM10 and
E-cadherin were 59.0 (72/122) and 40.2% (49/122) in the ESCC
tissues, and 15.0 (9/60) and 100.0% (60/60) in the corresponding
normal mucosa, respectively. Moreover, the expression of ADAM10
mRNA in the ESCC tissues was significantly higher than that in the
corresponding normal esophageal mucosa (P=0.000), whereas contrary
results were observed in the expression of E-cadherin mRNA
(P=0.000) (Table I).
Taken together, the above data demonstrated that
overexpression of ADAM10 contributes to carcinogenesis of ESCC and
this action may be attributable to ADAM10-mediated E-cadherin
shedding.
Elevated ADAM10 and reduced E-cadherin
protein levels are associated with tumor progression in ESCC
tissues
To investigate the clinical significance of ADAM10
and E-cadherin in ESCC tissues, we analyzed the correlation of
their protein expression levels with clinicopathological features
including histological grade, depth of invasion, lymph node
metastasis and TNM stage. As shown in Table II, ADAM10 expression was positively
correlated with depth of invasion, lymph node metastasis and TNM
stage (P<0.05, respectively), but not correlated with
histological grade (P>0.05). On the contrary, E-cadherin
expression was negatively correlated with histological grade, depth
of invasion, lymph node metastasis and TNM stage (P<0.05,
respectively). These data demonstrated that in the ESCC tissues
elevated ADAM10 and reduced E-cadherin levels play a pivotal role
in tumor progression, such as invasion and metastasis.
 | Table IICorrelation of ADAM10 and E-cadherin
proteins with clinicopathological parameters in the ESCC
tissues. |
Table II
Correlation of ADAM10 and E-cadherin
proteins with clinicopathological parameters in the ESCC
tissues.
| n | ADAM10
| χ2 | P-value | E-cadherin
| χ2 | P-value |
|---|
| − | + | − | + |
|---|
| Gender |
| Male | 73 | 44 | 29 | 0.015 | 0.904 | 48 | 25 | 0.434 | 0.510 |
| Female | 49 | 29 | 20 | | | 35 | 14 | | |
| Age (years) |
| ≥60 | 77 | 46 | 31 | 0.001 | 0.977 | 52 | 25 | 0.024 | 0.877 |
| <60 | 45 | 27 | 18 | | | 31 | 14 | | |
| Histological
grade |
| I | 33 | 16 | 17 | 4.510 | 0.105 | 16 | 17 | 7.981 | 0.018 |
| II | 51 | 14 | 37 | | | 38 | 13 | | |
| III | 38 | 11 | 27 | | | 29 | 9 | | |
| Depth of
invasion |
| Superficial
muscularis | 19 | 11 | 8 | 6.014 | 0.049 | 8 | 11 | 7.360 | 0.025 |
| Deep
muscularis | 46 | 14 | 32 | | | 32 | 14 | | |
| Fibrous
membrane | 57 | 16 | 41 | | | 43 | 14 | | |
| Lymph node
metastasis |
| No | 74 | 34 | 40 | 12.835 | 0.000 | 44 | 30 | 6.357 | 0.012 |
| Yes | 48 | 7 | 41 | | | 39 | 9 | | |
| TNM stage |
| I | 17 | 11 | 6 | 8.920 | 0.012 | 7 | 10 | 6.790 | 0.034 |
| II | 44 | 14 | 30 | | | 33 | 11 | | |
| III | 61 | 16 | 45 | | | 43 | 18 | | |
Negative correlation of ADAM10 and
E-cadherin protein levels in ESCC tissues
To explore the role of active ADAM10 involving
E-cadherin in ESCC progression, we further studied the correlation
of ADAM10 protein expression with E-cadherin in the ESCC tissues.
As shown in Table III, ADAM10
protein expression was significantly negatively correlated with
E-cadherin (r=−0.554, P=0.000), implying ADAM10 may control the
invasion and metastasis of ESCC through E-cadherin shedding.
 | Table IIICorrelation of ADAM10 with E-cadherin
protein in the ESCC tissues. |
Table III
Correlation of ADAM10 with E-cadherin
protein in the ESCC tissues.
| E-cadherin | ADAM10
| r | P-value |
|---|
| + | − |
|---|
| + | 11 | 28 | −0.554 | 0.000 |
| − | 70 | 13 | | |
ADAM10 is overexpressed in ESCC cell
lines and high expression of ADAM10 is correlated with increased
cell invasion and migration in vitro
To further investigate the potential biological
roles of ADAM10 in ESCC, cell experiments were carried out. We
first used quantitative real-time PCR and western blot analysis to
detect the expression levels of ADAM mRNA and protein in ESCC
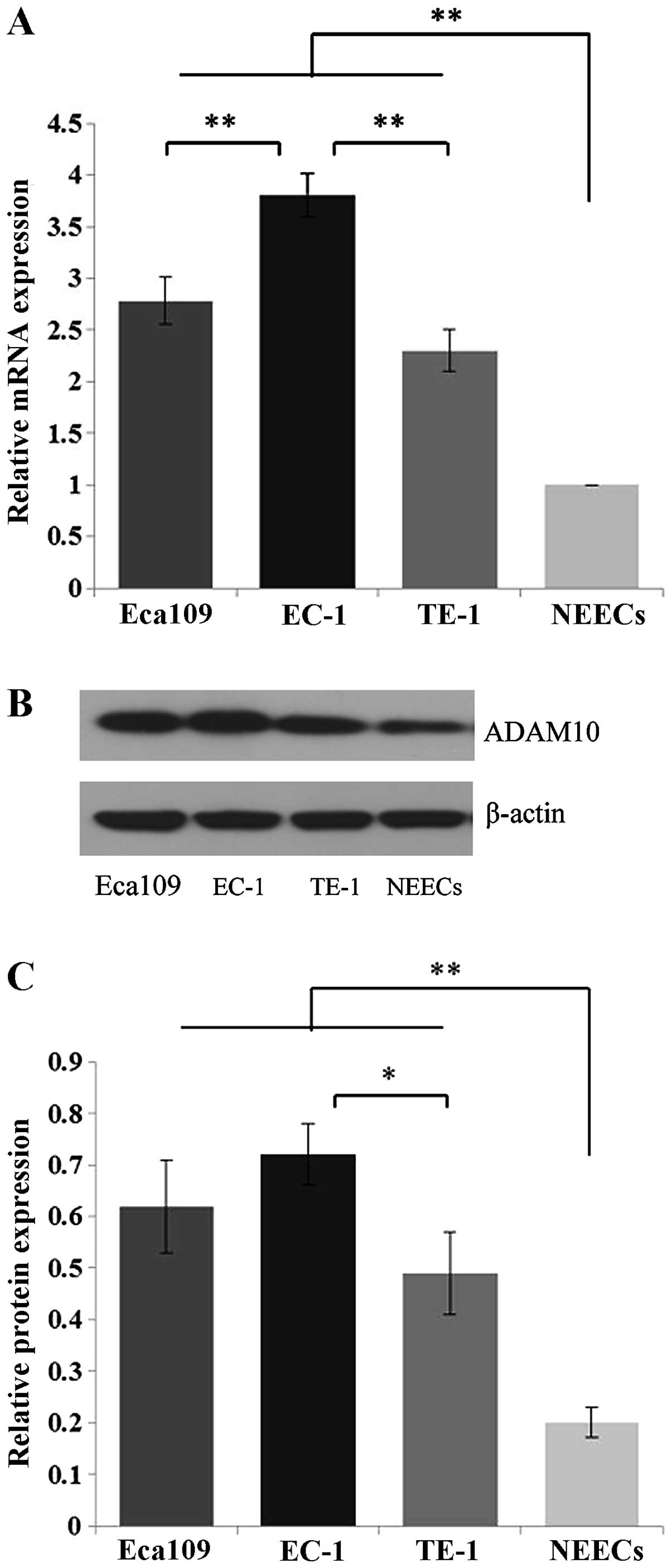
Eca109, EC-1 and TE-1 cells. Compared to the NEECs, Eca109, EC-1
and TE-1 cells showed significant overexpression of ADAM10 at both
the mRNA and protein levels (P<0.01) (Fig. 2). Moreover, this increased
expression was more apparent in the EC-1 cells than that in the
Eca109 and TE-1 cells at the mRNA level (P<0.01), and than that
in the TE-1 cells at the protein level (P<0.05) (Fig. 2).
Based on the above clinical correlation analysis in
the ESCC tissues, it is reasonable to hypothesize that ADAM10 plays
an important role in promoting the invasion and migration of ESCC
cells. Therefore, we next detected the invasion and migration
ability of the ESCC cells using Matrigel invasion and scratch wound
healing assays, respectively. The invasion assay demonstrated that
the EC-1 cells exhibited a significant increase in the number of
cells that traversed the membrane toward a chemoattractant as
compared with the TE-1 cells (P<0.05) (Fig. 3A and B). Additionally, the scratch
assay demonstrated that EC-1 cells showed a significant increase in
the tumor cell migration distance as compared with the Eca109 and
TE-1 cells (P<0.01) (Fig. 3C and
D).
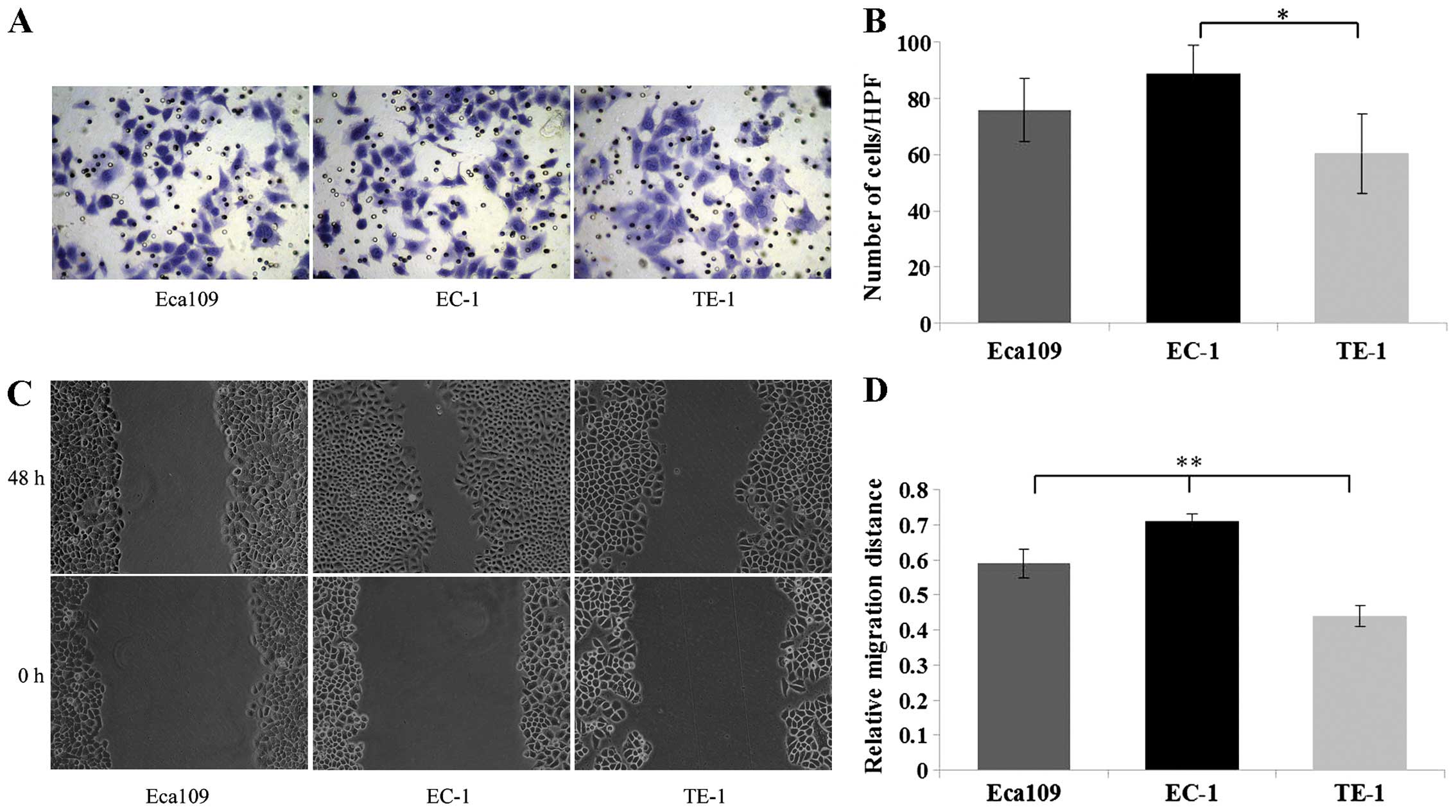 | Figure 3Matrigel invasion and scratch wound
healing assays were used to detect the invasion and migration
ability of ESCC cells. (A) For the Matrigel invasion assay, 3 ESCC
cell lines (Eca109, EC-1, TE-1) in serum-free medium were seeded
into the upper chamber and incubated for 24 h, respectively;
subsequently cells invading through the Matrigel were fixed,
stained and photographed under a light microscope at a ×400
magnification. (B) EC-1 cells exhibited a significant increase in
the number of traversed cells as compared with the TE-1 cells. (C)
For the scratch wound healing assay, 3 ESCC cell lines (Eca109,
EC-1, TE-1) were seeded into 12-well plates and incubated in medium
containing 10% FBS for 24 h, respectively. Subsequently the wounds
were made, and the cells were incubated in serum-free medium for an
additional 48 h and photographed under a reversed light microscope
at a ×200 magnification. (D) The bar graph shows that the EC-1 cell
migration was more rapid than the rate noted in the TE-1 cells.
*P<0.05, **P<0.01. |
In line with the aberrant ADAM10 in these 3 types of
ESCC cell lines, EC-1 cells with high ADAM10 expression
demonstrated higher invasion and migration ability, whereas TE-1
cells with low ADAM10 expression demonstrated poor invasion and
migration ability, suggesting that high ADAM10 expression is
consistent with increased cell invasion and migration ability.
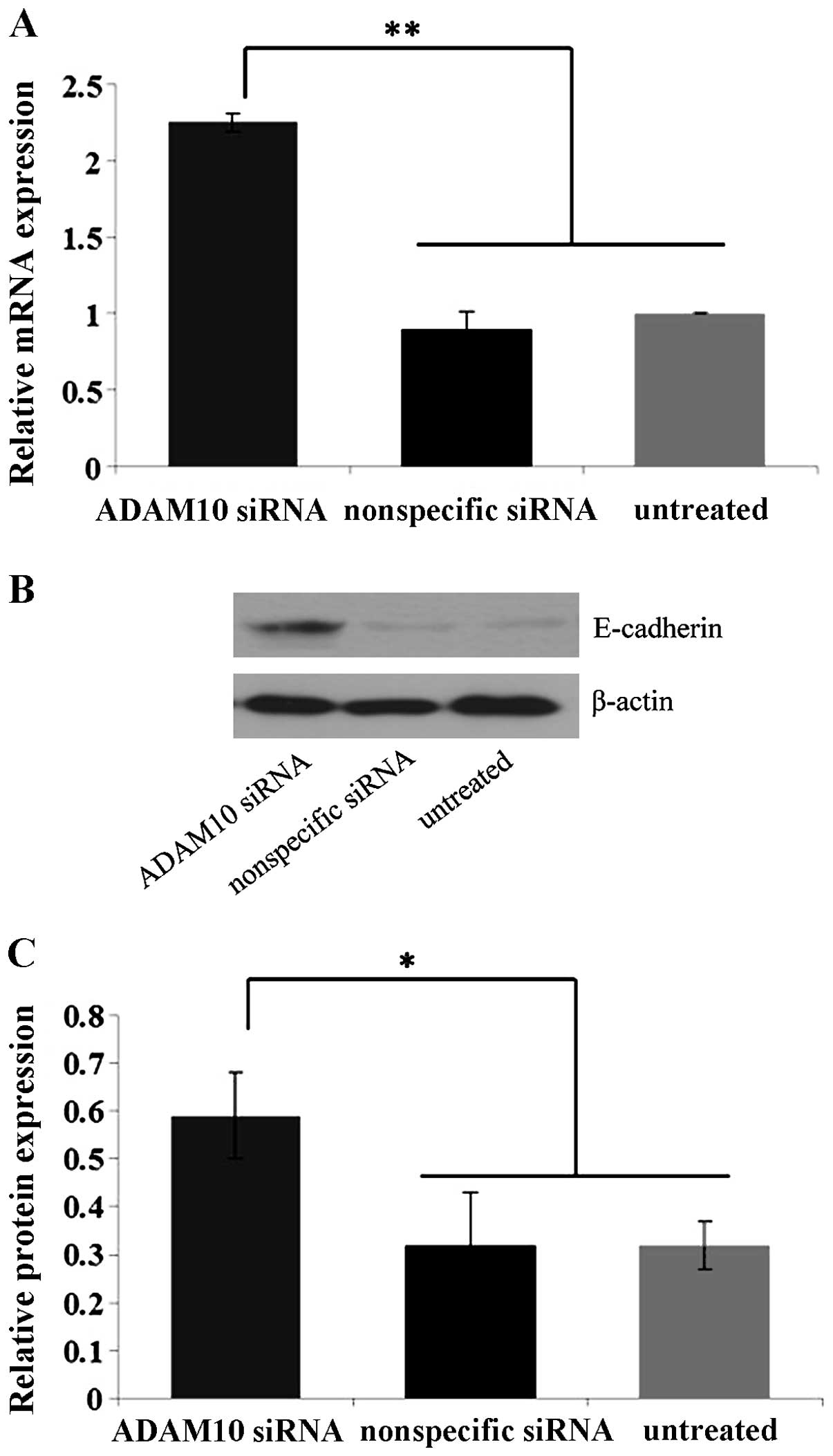
Specific blockage of ADAM10 by ADAM10
siRNA in EC-1 cells in vitro
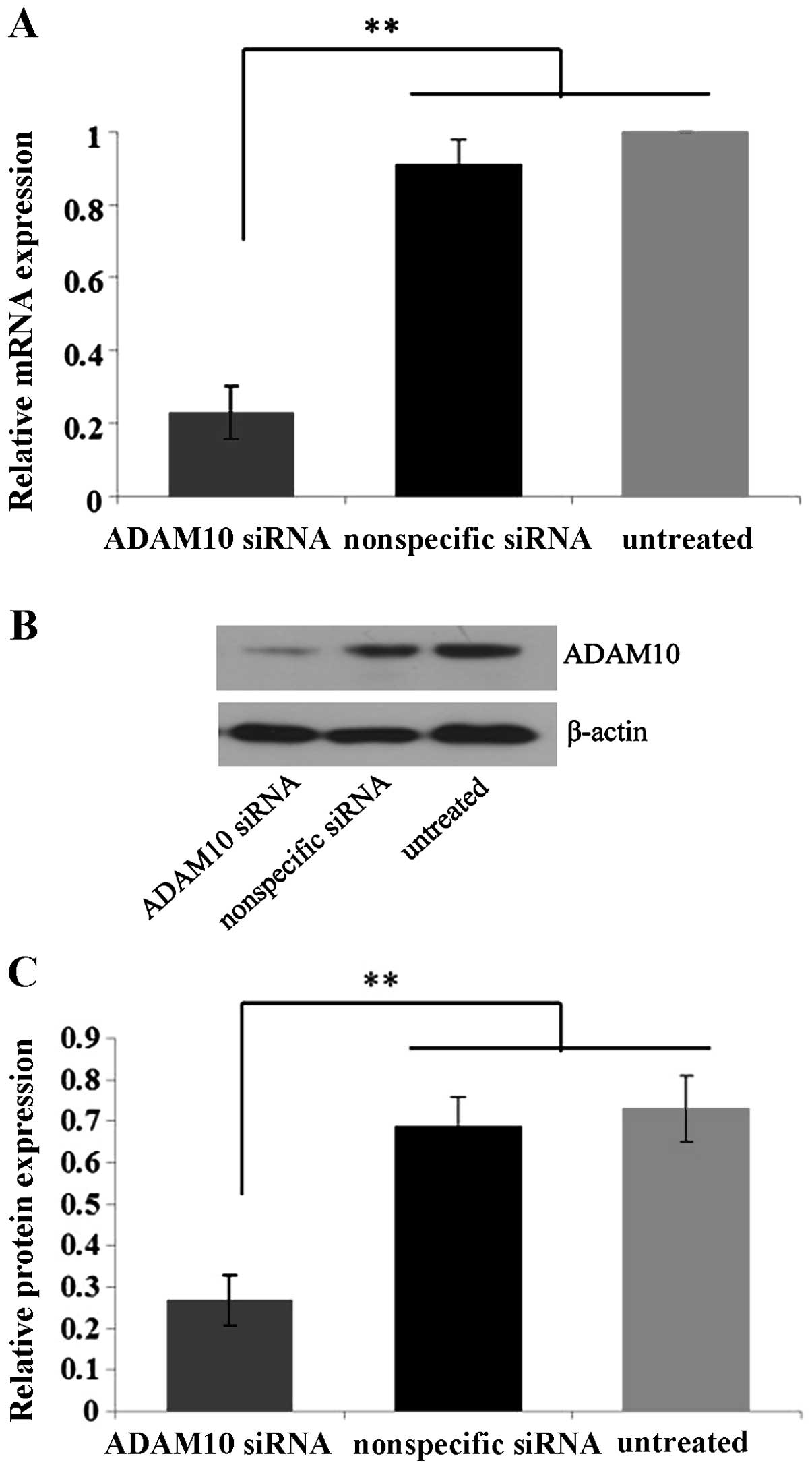
In order to detect the knockdown efficiency of
ADAM10 by ADAM10 siRNA, EC-1 cells were selected due to their
elevated ADAM10 expression and enhanced invasion and migration
ability compared to the Eca109 and TE-1 cell lines. Quantitative
real-time PCR and western blot analysis showed that the expression
levels of ADAM10 mRNA and protein in the ADAM10 siRNA-transfected
cells were suppressed by ~75 and 60%, significantly lower than
those in the control nonspecific siRNA-transfected and untreated
cells (P<0.01) (Fig. 4),
confirming the specific and efficient blockage of ADAM10 by ADAM10
siRNA in the EC-1 cells in vitro.
Knockdown of ADAM10 inhibits the invasion
and migration of EC-1 cells in vitro
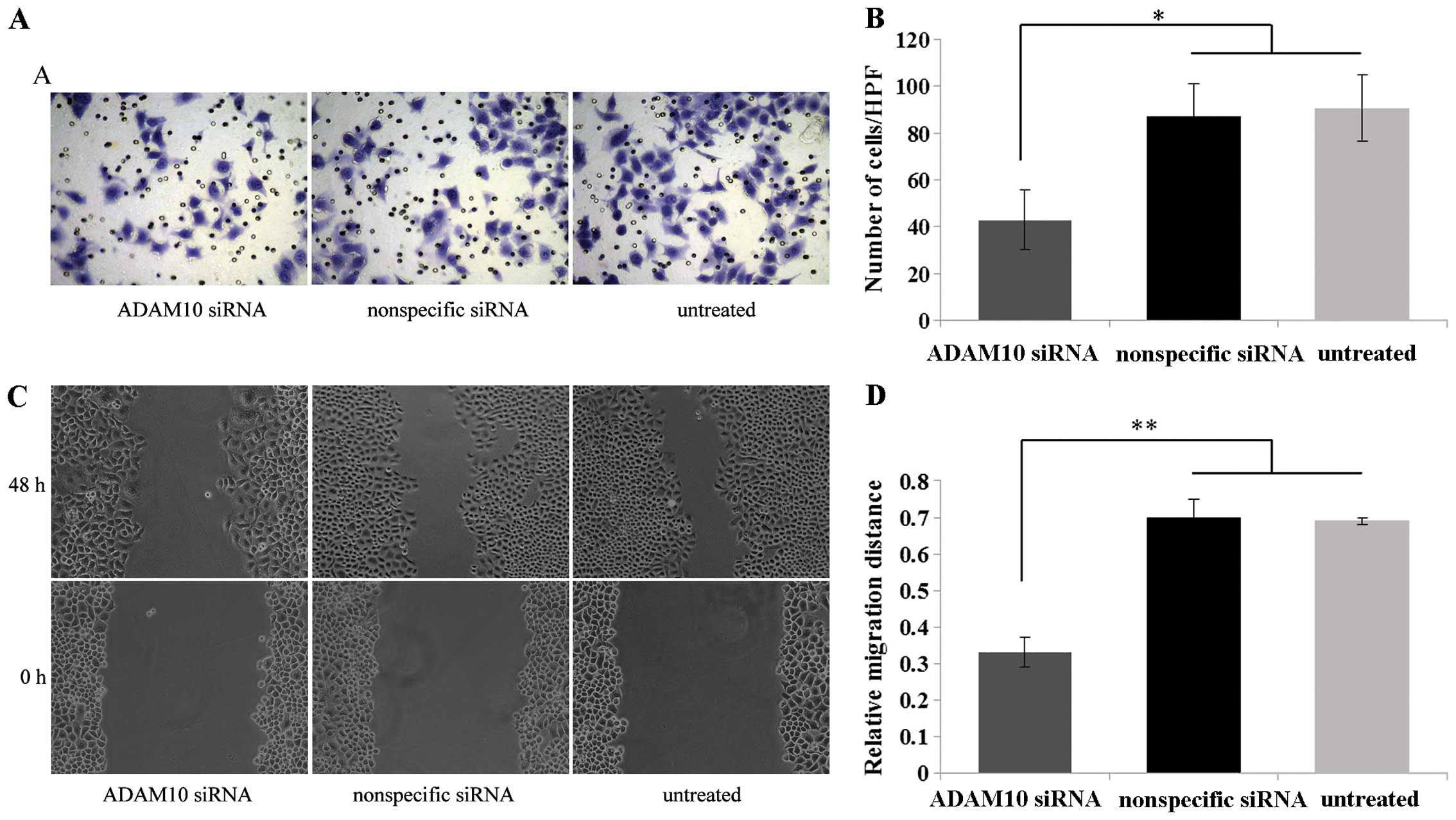
We examined the effect of ADAM10 knockdown on EC-1
cell invasion and migration using a Matrigel invasion and scratch
wound healing assays. Matrigel invasion assay demonstrated that
EC-1 cells transfected with the ADAM10 siRNA exhibited a
significant decrease in the number of cells that traversed the
membrane toward a chemoattractant as compared with the control
nonspecific siRNA-transfected and untreated cells (P<0.05)
(Fig. 5A and B). Furthermore, the
scratch wound healing assay demonstrated that the EC-1 cells
transfected with the ADAM10 siRNA resulted in a significant
decrease in the tumor cell migration distance in comparison to the
control nonspecific siRNA-transfected and untreated cells
(P<0.01) (Fig. 5C and D). These
data indicated that downregulation of ADAM10 inhibited the invasion
and migration of the EC-1 cells in vitro.
Knockdown of ADAM10 inhibits
proliferation of EC-1 cells in vitro
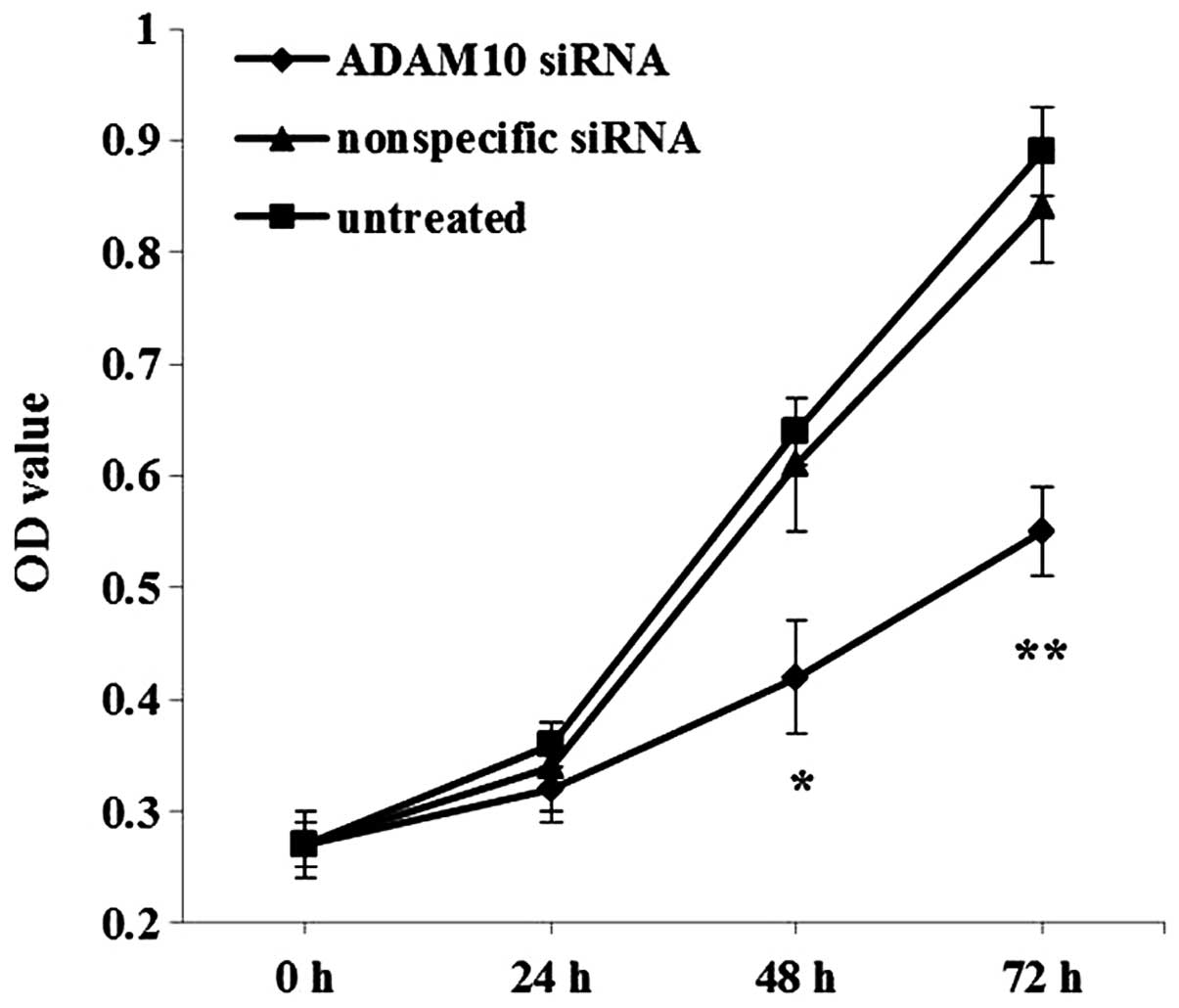
To evaluate the effect of ADAM10 knockdown on EC-1
cell proliferation, CCK-8 assay was carried out. Compared with the
control nonspecific siRNA-transfected and untreated cells, the EC-1
cells transfected with the ADAM10 siRNA showed significantly
decreased OD values after incubation for 48 (P<0.05) and 72 h
(P<0.01) (Fig. 6), suggesting
that downregulation of ADAM10 inhibited EC-1 cell growth in
vitro.
Knockdown of ADAM10 suppresses E-cadherin
expression in EC-1 cells in vitro
As described previously, expression levels of ADAM10
and E-cadherin were inversely correlated in the ESCC tissues. To
further characterize the intrinsic relationship between ADAM10 and
E-cadherin expression and explore the possible impact that ADAM10
exerts on the invasion and metastasis of ESCC, the expression
changes in E-cadherin mRNA and protein in the EC-1 cells
transfected with ADAM10 siRNA were determined by quantitative
real-time PCR and western blot analysis assays. As shown in
Fig. 7, downregulation of ADAM10 by
ADAM10 siRNA dramatically increased E-cadherin mRNA and protein in
the ADAM10 siRNA-transfected EC-1 cells as compared with the
control nonspecific siRNA-transfected and untreated cells
(P<0.05). These results demonstrated that ADAM10 is essentially
involved in E-cadherin cleavage in EC-1 cells in vitro.
Discussion
The role of ADAM10 has been clarified in a variety
of important biological processes, such as cell adhesion (33), cell migration (38), immunoreaction regulation (39,40)
and apoptosis control (41).
However recently, it has emerged as a major and key player in human
diseases. ADAM10 is considered to contribute to Alzheimer's disease
(42) and chronic inflammatory
diseases (43) such as lung
inflammation (44) and eczematous
dermatitis (45); most importantly,
it is overexpressed in many cancers and participates in tumor
progression (18–30). Guo et al (19) demonstrated that ADAM10 protein
expression is significantly increased in human NSCLC tissues,
particularly in metastatic tissues; furthermore, downregulation of
ADAM10 by ADAM10 shRNA reduced the invasion and migration of NSCLC
cells. Consistent with this result, Gaida et al (28) also found that ADAM10 mRNA and
protein were overexpressed in pancreatic carcinoma compared to
normal pancreatic tissues, and ADAM10 silencing markedly reduced
the invasiveness and migration of the tumor cells. However, the
role of ADAM10 in ESCC is not yet well understood.
In the present study, we first examined the
expression levels of ADAM10 mRNA and protein in human ESCC tissues
and cells. As expected, ADAM10 mRNA and protein were both
dramatically elevated in the ESCC tissues and cells in comparison
to these levels in the normal esophageal epithelial mucosa tissues
and cells, suggesting that aberrant activation of ADAM10 plays a
fundamental role in the carcinogenesis of ESCC. Subsequently, the
correlation of ADAM10 with tumor invasion and metastasis was
analyzed. In ESCC tissues, ADAM10 expression was positively
correlated with depth of invasion, lymph node metastasis and high
TNM stage of the cancers; in addition, in ESCC cells, EC-1 with
high ADAM10 expression exhibited higher invasion and migration
ability, whereas TE-1 with low ADAM10 expression exhibited poor
invasion and migration ability. Since EC-1 cells possessed a higher
level of ADAM10 expression and markedly enhanced invasion and
migration ability compared to the Eca109 and TE-1 cells, EC-1 cells
were chosen to perform follow-up experiments to ascertain the
effect of ADAM10 silencing on cell invasion and migration. After
the knockdown of ADAM10 using ADAM10 siRNA, the EC-1 cells
exhibited a significant decrease in the number of invasive cells,
cell migration distance and cell proliferation in comparison to the
control nonspecific siRNA-transfected and untreated cells. All
these above results demonstrated that aberrant ADAM10 is associated
with tumor cell invasion, migration, metastasis and proliferation
in ESCC in vitro and in vivo.
A large body of evidence has shown that tumor
invasion, metastasis and proliferation are associated with the
shedding activity of ADAM10. Nevertheless, the precise mechanism
ADAM10 exerts in the course of ESCC progression is still poorly
understood. ADAM10 promotes tumor progression through the cleavage
of its substrates, such as adhesion molecules (22). E-cadherin, an adhesion molecule, has
been known as an ADAM10 substrate (33). ADAM10 has been implicated in the
shedding of E-cadherin in response to Helicobacter (H.)
pylori infection in gastric cancer cell lines (46). H. pylori infection was found
to correlate with increased ADAM10 expression in gastric cancer
patient samples and to induce ADAM10 expression in gastric cancer
cell lines (47). Indeed, chemical
inhibition or shRNA-mediated knockdown of ADAM10 resulted in
decreased soluble E-cadherin generation in response to H.
pylori infection, suggesting that induction of ADAM10 by H.
pylori in gastric cancer promotes E-cadherin cleavage (46). In the present study, we analyzed the
correlation of ADAM10 with E-cadherin in ESCC tissues and found a
significantly negative correlation between them, suggesting that
ADAM10 promotes the progression of ESCC via E-cadherin shedding.
Accordingly, we next detected the effect of ADAM10 silencing on the
shedding of E-cadherin in EC-1 cells in vitro. The data
showed that the knockdown of ADAM10 dramatically promoted
E-cadherin mRNA and protein in the ADAM10 siRNA-transfected cells
as compared with the control nonspecific siRNA-transfected and
untreated cells, indicating that ADAM10 is critically involved in
the proteolytic processing of E-cadherin in EC-1 cells in
vitro and ADAM10-mediated cleavage of E-cadherin represents a
potent mechanism for regulating invasion, migration, and
proliferation of EC-1 cells. ADAM10 could shed the extracellular
domain of E-cadherin, and allow β-catenin to translocate to the
nucleus (33). Nuclear
translocation of β-catenin further contributes to the activation
and transcription of genes involved in cell proliferation such as
c-myc and cyclin D1 (48). In
addition to enhanced cell proliferation, E-cadherin shedding was
found to simultaneously weaken cadherin-mediated cell-cell adhesion
and promote dissociation of potentially invasive and metastatic
cells located in primary cancers; moreover, such dissociation could
directly place malignant cells on their pathway to metastasis
(10).
In conclusion, the present study suggests that
ADAM10 is a key regulator in the invasion and metastasis of ESCC,
and may control invasion and metastasis at least in part through
the regulation of E-cadherin activity. Disruption of ADAM10
expression may provide a possible route for therapeutic
intervention for ESCC.
Acknowledgments
This project was financially supported by the
Education Program of Henan Province, China (grant no.
12A310014).
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
2
|
Zhu YH, Fu L, Chen L, Qin YR, Liu H, Xie
F, Zeng T, Dong SS, Li J, Li Y, et al: Downregulation of the novel
tumor suppressor DIRAS1 predicts poor prognosis in esophageal
squamous cell carcinoma. Cancer Res. 73:2298–2309. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Parkin DM, Bray FI and Devesa SS: Cancer
burden in the year 2000. The global picture. Eur J Cancer. 37(Suppl
8): S4–S66. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsai ST, Wang PJ, Liou NJ, Lin PS, Chen CH
and Chang WC: ICAM1 is a potential cancer stem cell marker of
esophageal squamous cell carcinoma. PLoS One. 10:e01428342015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shimada H, Shiratori T, Okazumi S,
Matsubara H, Nabeya Y, Shuto K, Akutsu Y, Hayashi H, Isono K and
Ochiai T: Have surgical outcomes of pathologic T4 esophageal
squamous cell carcinoma really improved? Analysis of 268 cases
during 45 years of experience. J Am Coll Surg. 206:48–56. 2008.
View Article : Google Scholar
|
|
7
|
Radisky ES and Radisky DC: Matrix
metalloproteinase-induced epithelial-mesenchymal transition in
breast cancer. J Mammary Gland Biol Neoplasia. 15:201–212. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pisani P, Parkin DM, Bray F and Ferlay J:
Estimates of the worldwide mortality from 25 cancers in 1990. Int J
Cancer. 83:18–29. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Primakoff P and Myles DG: The ADAM gene
family: surface proteins with adhesion and protease activity.
Trends Genet. 16:83–87. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Duffy MJ, Mullooly M, ODonovan N, Sukor S,
Crown J, Pierce A and McGowan PM: The ADAMs family of proteases:
new biomarkers and therapeutic targets for cancer? Clin Proteomics.
8:92011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wolfsberg TG, Primakoff P, Myles DG and
White JM: ADAM, a novel family of membrane proteins containing a
disintegrin and metalloprotease domain: multipotential functions in
cell-cell and cell-matrix interactions. J Cell Biol. 131:275–278.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Walker JL, Fournier AK and Assoian RK:
Regulation of growth factor signaling and cell cycle progression by
cell adhesion and adhesion-dependent changes in cellular tension.
Cytokine Growth Factor Rev. 16:395–405. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mazzocca A, Coppari R, De Franco R, Cho
JY, Libermann TA, Pinzani M and Toker A: A secreted form of ADAM9
promotes carcinoma invasion through tumor-stromal interactions.
Cancer Res. 65:4728–4738. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mochizuki S and Okada Y: ADAMs in cancer
cell proliferation and progression. Cancer Sci. 98:621–628. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Klein T and Bischoff R: Active
metalloproteases of the a disintegrin and metalloprotease (ADAM)
family: biological function and structure. J Proteome Res.
10:17–33. 2011. View Article : Google Scholar
|
|
16
|
Lu X, Lu D, Scully M and Kakkar V: ADAM
proteins - therapeutic potential in cancer. Curr Cancer Drug
Targets. 8:720–732. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hartmann D, de Strooper B, Serneels L,
Craessaerts K, Herreman A, Annaert W, Umans L, Lübke T, Lena Illert
A, von Figura K, et al: The disintegrin/metalloprotease ADAM 10 is
essential for Notch signalling but not for alpha-secretase activity
in fibroblasts. Hum Mol Genet. 11:2615–2624. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lendeckel U, Kohl J, Arndt M, Carl-McGrath
S, Donat H and Röcken C: Increased expression of ADAM family
members in human breast cancer and breast cancer cell lines. J
Cancer Res Clin Oncol. 131:41–48. 2005. View Article : Google Scholar
|
|
19
|
Guo J, He L, Yuan P, Wang P, Lu Y, Tong F,
Wang Y, Yin Y, Tian J and Sun J: ADAM10 overexpression in human
non-small cell lung cancer correlates with cell migration and
invasion through the activation of the Notch1 signaling pathway.
Oncol Rep. 28:1709–1718. 2012.PubMed/NCBI
|
|
20
|
You B, Shan Y, Shi S, Li X and You Y:
Effects of ADAM10 upregulation on progression, migration, and
prognosis of naso-pharyngeal carcinoma. Cancer Sci. 106:1506–1514.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jones AV, Lambert DW, Speight PM and
Whawell SA: ADAM 10 is over expressed in oral squamous cell
carcinoma and contributes to invasive behaviour through a
functional association with αvβ6 integrin. FEBS Lett.
587:3529–3534. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ko SY, Lin SC, Wong YK, Liu CJ, Chang KW
and Liu TY: Increase of disintergin metalloprotease 10 (ADAM10)
expression in oral squamous cell carcinoma. Cancer Lett. 245:33–43.
2007. View Article : Google Scholar
|
|
23
|
Shao Y, Sha XY, Bai YX, Quan F and Wu SL:
Effect of a disintegrin and metalloproteinase 10 gene silencing on
the proliferation, invasion and migration of the human tongue
squamous cell carcinoma cell line TCA8113. Mol Med Rep. 11:212–218.
2015.
|
|
24
|
Yue Y, Shao Y, Luo Q, Shi L and Wang Z:
Downregulation of ADAM10 expression inhibits metastasis and
invasiveness of human hepatocellular carcinoma HepG2 cells. BioMed
Res Int. 2013:4345612013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang W, Liu S, Liu K, Wang Y, Ji B, Zhang
X and Liu Y: A disintegrin and metalloprotease (ADAM)10 is highly
expressed in hepatocellular carcinoma and is associated with tumour
progression. J Int Med Res. 42:611–618. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang YY, Ye ZY, Li L, Zhao ZS, Shao QS and
Tao HQ: ADAM 10 is associated with gastric cancer progression and
prognosis of patients. J Surg Oncol. 103:116–123. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gavert N, Sheffer M, Raveh S, Spaderna S,
Shtutman M, Brabletz T, Barany F, Paty P, Notterman D, Domany E, et
al: Expression of L1-CAM and ADAM10 in human colon cancer cells
induces metastasis. Cancer Res. 67:7703–7712. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gaida MM, Haag N, Günther F, Tschaharganeh
DF, Schirmacher P, Friess H, Giese NA, Schmidt J and Wente MN:
Expression of a disintegrin and metalloprotease 10 in pancreatic
carcinoma. Int J Mol Med. 26:281–288. 2010.PubMed/NCBI
|
|
29
|
Fu L, Liu N, Han Y, Xie C, Li Q and Wang
E: ADAM10 regulates proliferation, invasion, and chemoresistance of
bladder cancer cells. Tumour Biol. 35:9263–9268. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Doberstein K, Pfeilschifter J and Gutwein
P: The transcription factor PAX2 regulates ADAM10 expression in
renal cell carcinoma. Carcinogenesis. 32:1713–1723. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Turner SL, Blair-Zajdel ME and Bunning RA:
ADAMs and ADAMTSs in cancer. Br J Biomed Sci. 66:117–128.
2009.PubMed/NCBI
|
|
32
|
Woods N, Trevino J, Coppola D, Chellappan
S, Yang S and Padmanabhan J: Fendiline inhibits proliferation and
invasion of pancreatic cancer cells by interfering with ADAM10
activation and β-catenin signaling. Oncotarget. 6:35931–35948.
2015.PubMed/NCBI
|
|
33
|
Maretzky T, Reiss K, Ludwig A, Buchholz J,
Scholz F, Proksch E, de Strooper B, Hartmann D and Saftig P: ADAM10
mediates E-cadherin shedding and regulates epithelial cell-cell
adhesion, migration, and beta-catenin translocation. Proc Natl Acad
Sci USA. 102:9182–9187. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dittmer A, Hohlfeld K, Lützkendorf J,
Müller LP and Dittmer J: Human mesenchymal stem cells induce
E-cadherin degradation in breast carcinoma spheroids by activating
ADAM10. Cell Mol Life Sci. 66:3053–3065. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee SB, Schramme A, Doberstein K, Dummer
R, Abdel-Bakky MS, Keller S, Altevogt P, Oh ST, Reichrath J, Oxmann
D, et al: ADAM10 is upregulated in melanoma metastasis compared
with primary melanoma. J Invest Dermatol. 130:763–773. 2010.
View Article : Google Scholar
|
|
36
|
Bánkfalvi A, Krassort M, Buchwalow IB,
Végh A, Felszeghy E and Piffkó J: Gains and losses of adhesion
molecules (CD44, E-cadherin, and beta-catenin) during oral
carcinogenesis and tumour progression. J Pathol. 198:343–351. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
38
|
Mancia F and Shapiro L: ADAM and Eph: how
Ephrin-signaling cells become detached. Cell. 123:185–187. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hasegawa-Ishii S, Shimada A, Inaba M, Li
M, Shi M, Kawamura N, Takei S, Chiba Y, Hosokawa M and Ikehara S:
Selective localization of bone marrow-derived ramified cells in the
brain adjacent to the attachments of choroid plexus. Brain Behav
Immun. 29:82–97. 2013. View Article : Google Scholar
|
|
40
|
Gibb DR, Saleem SJ, Chaimowitz NS, Mathews
J and Conrad DH: The emergence of ADAM10 as a regulator of
lymphocyte development and autoimmunity. Mol Immunol. 48:1319–1327.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schulte M, Reiss K, Lettau M, Maretzky T,
Ludwig A, Hartmann D, de Strooper B, Janssen O and Saftig P: ADAM10
regulates FasL cell surface expression and modulates FasL-induced
cytotoxicity and activation-induced cell death. Cell Death Differ.
14:1040–1049. 2007.PubMed/NCBI
|
|
42
|
Endres K and Fahrenholz F: The role of the
anti-amyloidogenic secretase ADAM10 in shedding the APP-like
proteins. Curr Alzheimer Res. 9:157–164. 2012. View Article : Google Scholar
|
|
43
|
Crawford HC, Dempsey PJ, Brown G, Adam L
and Moss ML: ADAM10 as a therapeutic target for cancer and
inflammation. Curr Pharm Des. 15:2288–2299. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dreymueller D, Uhlig S and Ludwig A:
ADAM-family metalloproteinases in lung inflammation: potential
therapeutic targets. Am J Physiol Lung Cell Mol Physiol.
308:L325–L343. 2015. View Article : Google Scholar
|
|
45
|
Maretzky T, Scholz F, Köten B, Proksch E,
Saftig P and Reiss K: ADAM10-mediated E-cadherin release is
regulated by proinflammatory cytokines and modulates keratinocyte
cohesion in eczematous dermatitis. J Invest Dermatol.
128:1737–1746. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schirrmeister W, Gnad T, Wex T,
Higashiyama S, Wolke C, Naumann M and Lendeckel U: Ectodomain
shedding of E-cadherin and c-Met is induced by Helicobacter pylori
infection. Exp Cell Res. 315:3500–3508. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yoshimura T, Tomita T, Dixon MF, Axon AT,
Robinson PA and Crabtree JE: ADAMs (a disintegrin and
metalloproteinase) messenger RNA expression in Helicobacter
pylori-infected, normal, and neoplastic gastric mucosa. J Infect
Dis. 185:332–340. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
48
|
Balzer EM and Konstantopoulos K:
Intercellular adhesion: mechanisms for growth and metastasis of
epithelial cancers. Wiley Interdiscip Rev Syst Biol Med. 4:171–181.
2012. View Article : Google Scholar
|