Introduction
Colorectal cancer ranks third in worldwide incidence
of malignant tumors, and fourth in terms of the cancer-related
mortality rate (1). The 5-year
survival rate of patients with colorectal cancer ranges between 50
and 60% (2). Several key genes and
signaling pathways were found to play an important role in the
pathogenesis of colorectal cancer, for example, EGFR,
Wnt, TGFβ, p53 and DNA-mismatch repair pathway
(3). Nonetheless, we have yet to
gain a complete understanding of the molecular genetics of
colorectal cancer. In the future, we also need to investigate the
molecular mechanisms as well as the corresponding genetic
alterations in colorectal cancer, to determine the pathophysiology
and potential diagnostic markers and therapeutic targets.
The sirtuin family (SIRT1-7) includes an
NAD+-dependent histone deacetylase, deacetylase and ADP
ribosyltransferases playing an important role in pressure
resistance, genomic stability, energy metabolism and aging
(4). To date, almost all of the
SIRT family members have been considered to play an important role
in the development of cancer (5).
In regards to SIRT1, which is the most extensively studied member
of the SIRT family, the deacetylation protein group and a series of
non-histone protein substrates affect the corresponding
tumor-related genes including apoptosis of FOXO proteins, tumor
suppressor p53, and DNA mismatch repair protein Ku70 (6,7),
SIRT1, SIRT3, SIRT6 and SIRT7 have been shown to play the role of
oncogenes or tumor-suppressor genes in colorectal cancer (8–11).
However, SIRT2, SIRT4 and SIRT5 have yet to be investigated in
colorectal cancer.
SIRT2 is an NAD+-dependent deacetylase
located in the cytoplasm (12), and
catalyzes substrates such as H4K16 (13), H3K56 (14), FOXO1 (15) and p53 (16). Kim et al (17) found that SIRT2 regulates mitosis,
and SIRT2 knockout leads to the emergence of sex-specific tumors in
mice, female breast cancer and male hepatocellular carcinoma.
SIRT4 is an ADP-dependent NAD+
transferase located in the mitochondria (18). SIRT4 regulates insulin secretion and
fatty acid oxidation and other cellular metabolic functions
(18–20). Recent studies have indicated that
SIRT4 functions as a tumor-suppressor gene by regulating the
metabolism of glutamine (21,22).
SIRT5 is also located in the mitochondria,
catalyzing deacetylation of carbamoyl-phosphate synthetase 1
(CPS1). The initial reaction of the urea cycle results in the
removal and degradation of ammonia in cells (23–25).
Lu et al (26) showed that
SIRT5 expression in human non-small cell lung cancer was elevated
and SIRT5 knockdown inhibits the growth and metastasis of lung
cancer cells in vitro and in vivo. Therefore, we
sought to determine whether SIRT2, SIRT4 and SIRT5 also play a role
in colorectal cancer.
Materials and methods
Patients and colorectal cancer
specimens
Tissue specimens from 16 colorectal cancer patients
(age range, 45–78 years; average age, 58 years) were used in the
PCR analysis. Patients underwent radical surgery for colorectal
cancer at Shanghai First People's Hospital from January 2013 to May
2013. No patient received neoadjuvant chemotherapy. Patients were
pathologically diagnosed with only a single primary lesion of
colorectal cancer. The patients included in the present study
provided written (signed) informed consent. The research protocol
was approved by the Ethics Committee of Shanghai First People's
Hospital. Colorectal cancer and normal colorectal tissues were
removed from a 5 cm tumor edge, frozen with liquid nitrogen and
stored at −80°C until further use.
Reverse transcription-RT-PCR
Total tissue RNA was purified using the TRIzol kit
(Invitrogen, Carlsbad, CA, USA) following the manufacturer's
protocol. Total cDNA (500 ng) was synthesized using reverse
transcription kit (PrimeScript™ RT Master Mix; Takara, Japan). The
cDNA was diluted three times using an RT-PCR kit (SYBR®
Premix Ex Taq™ II; Takara) in the RT-PCR reaction apparatus (DNA
Engine Opticon 2 system; Bio-Rad, Hercules, CA, USA). GAPDH was
selected as the reference gene. Primers for each gene were as
follows: SIRT2 forward primer, ATAACCCACACCCAGCGTAG and reverse
primer, AATGTCTTCTGCCCATCCAG; SIRT4 forward primer,
GATGACTTGGCGTGTCTGAA and reverse primer, TTGAATGGGAACTGGAATCTG;
SIRT5 forward primer, TTGAATGGGAACTGGAATCTG and reverse primer, TTG
AATGGGAACTGGAATCTG; and GAPDH forward primer,
CGGAGTCAACGGATTTGGTCGTAT and reverse primer,
AGCCTTCTCCATGGTGGTGAAGAC. The PCR reaction conditions were as
follows: 2 min at 94°C and then 30 sec at 94°C, 30 sec at 57°C, 1
min at 72°C for 40 cycles, and 5 min at 72°C and maintained at 4°C.
After the loop, melting curve was analyzed to ensure uniformity of
the PCR product. The data were converted using the
2−ΔΔCt method.
Analysis of SIRT2, SIRT4 and SIRT5
expression using an online microarray database
The human colorectal cancer expression microarray
data downloaded from the The Cancer Genome Atlas (TCGA) website
(http://cancergenome.nih.gov/) were used
to analyze the mRNA expression of SIRT2, SIRT4 and SIRT5 between
normal colorectal and colorectal cancer tissues.
Tissue microarray
Tissue microarray was obtained from a commercial
chip Co. (Superchip Inc., Shanghai, China) using 89 cases of
patient samples, each containing colorectal cancer and the
corresponding normal colorectal tissue specimen at each point. The
point diameter was 1.5 mm, and all points were overlaid with
paraffin wax. No patient received neoadjuvant chemotherapy or
radiotherapy. Surgeries were conducted between January 2009 and
October 2009. The follow-up time ranged from 4.65 to 5.3 years,
ending May 2014. The total survival time was defined as the time
until death following radical surgery. Clinicopathological
parameters included age, gender, tumor size, growth mode, degree of
differentiation, tumor invasion depth and scope, lymph node and
distant metastases, the Union for International Cancer Control
(UICC) stage and post-operative overall survival time (OS)
(Table II).
 | Table IICorrelation between the
clinicopathologic variables and SIRT4 expression in colorectal
cancer. |
Table II
Correlation between the
clinicopathologic variables and SIRT4 expression in colorectal
cancer.
| Clinicopathological
parameters | All cases | SIRT4 expression
| χ2 | P-valuea |
|---|
| Low | High |
|---|
| Age (years) | | | | 1.435 | 0.263 |
| ≤65 | 41 | 16 | 25 | | |
| >65 | 48 | 13 | 35 | | |
| Gender | | | | 0.829 | 0.377 |
| Male | 46 | 17 | 29 | | |
| Female | 43 | 12 | 31 | | |
| Tumor size
(cm) | | | | 0.113 | 0.820 |
| ≤5 | 53 | 18 | 35 | | |
| >5 | 36 | 11 | 25 | | |
|
Differentiation | | | | 5.791 | 0.031 |
| Well-moderate | 75 | 23 | 52 | | |
| Poor | 14 | 9 | 5 | | |
| Stage (T) | | | | 3.308 | 0.326 |
| T1 | 3 | 0 | 3 | | |
| T2 | 10 | 3 | 7 | | |
| T3 | 49 | 16 | 33 | | |
| T4 | 27 | 13 | 14 | | |
| Stage (N) | | | | 1.800 | 0.475 |
| N0 | 58 | 22 | 36 | | |
| N1 | 23 | 6 | 17 | | |
| N2 | 8 | 4 | 4 | | |
| Stage (M) | | | | 0.009 | 1.000 |
| M0 | 86 | 31 | 55 | | |
| M1 | 3 | 1 | 2 | | |
| UICC stage | | | | 2.381 | 0.478 |
| I | 11 | 2 | 9 | | |
| II | 45 | 19 | 26 | | |
| III | 30 | 10 | 20 | | |
| IV | 3 | 1 | 2 | | |
Immunohistochemistry (IHC)
IHC was performed as previously described (27). SIRT4 immunoreactivity at each tissue
point was evaluated in terms of staining intensity (0, no staining;
1, weak staining; 2, medium staining; and 3, strong staining), and
staining area (0, <5%; 1, 5–25%; 2, >25–50%; 3, >50–75%;
and 4, >75%). The staining intensity score was multiplied with
the staining area score to obtain the final staining score. The
tissue points were divided into two groups based on the final
staining score: low, 0–4; high, 6–12. In case of inconsistencies,
the scoring was reevaluated by two researchers using a multi-headed
microscope until a conclusion or consensus was reached.
Vector and virus production
A lentivirus for SIRT4 overexpression was purchased
from HanBio (Shanghai, China). The virus vector was pHBLV-CMVIE-Zs
Green-T2A-Puro. The final virus titer of the overexpressing
lentivirus and the negative control virus was 2×108
PFU/ml.
Cell lines and culture conditions
Human colorectal cancer cell lines RKO and HT29 were
purchased from Shanghai Institute of Cell Biology, Chinese Academy
of Sciences. The cells were maintained in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% fetal bovine serum
(FBS) and penicillin/streptomycin (Gibco, Grand Island, NY, USA)
and incubated at 37°C and 5% CO2. The stable cell line
with SIRT4 overexpression was transfected with the lentivirus and
screened with puromycin (2 µg/ml) for two weeks. The other
reagents used in the cell experiments were: DMEM without glucose,
DMEM (both from Gibco) without glutamine, DM-KG (349631), 2-DG
(Klamar; 154-17-6) and 5-fluorouracil (5-FU) (F6627) (all from
Sigma, St. Louis, MO, USA). None of the culture media contained
sodium pyruvate.
Cell proliferation activity and
toxicity
Cells were seeded at 1,000/well for cell
proliferation activity and 5,000/well for cell proliferation
toxicity into a 96-well plate. For detection, each well was
supplemented with 10 µl Cell Counting Kit-8 (CCK-8)
(Dojindo, Japan) solution, and the absorbance was read at 450 nm
after culturing in a CO2 incubator for 2 h. The cell
proliferation activity in media without glutamine or glucose was
tested by changing the media to the corresponding experimental
conditions on the second day after seeding the cells. For cell
proliferation toxicity, the media were replaced the next day with
different doses of 5-FU. The cell proliferation toxicity was
calculated dynamically: cell viability (%) = A450 of
treated cells/A450 of untreated cells. Statistical
analysis of cell proliferation toxicity was carried out using the
cell viability (%) in three independent experiments.
Clone formation assay
The cells were seeded at 200/well in 6-well plates,
changing the liquid every other day. After culturing for 2 weeks,
the number of clones was counted directly with the naked eye after
fixing with methanol and staining by Giemsa.
Flow cytometric analysis of apoptosis and
cell cycle
Cells were harvested by trypsinization, pelleted by
centrifugation and resuspended in phosphate-buffered saline (PBS)
containing 3% FBS. Early cellular apoptosis was measured by flow
cytometry (C6) using Annexin V-APC and 7-AAD staining with Accuri
C6 software (all from BD, USA). The survival rate was calculated
using unstained APC or 7-AAD and found to be 100%. Cell cycle was
measured with PI/Nase kit (BD) according to the manufacturer's
instructions. The cell cycle results were analyzed using the
software ModFit (Verity Software House, Topsham, ME, USA).
Western blotting
Cells were lysed with RIPA lysis buffer supplemented
with protease inhibitor cocktail (both from Beyotime, China).
Protein concentrations were determined using the BCA protein
concentration reagent kit (Beyotime). Cell lysates were separated
by SDS-PAGE and transferred to PVDF membranes. Antibodies used
were: rabbit anti-human SIRT4 polyclonal antibody (HPA029692;
Sigma), goat anti-rabbit antibody (ab97200) and rabbit anti-human
β-actin polyclonal antibody (ab11971) (both from Abcam, Cambridge,
UK).
Xenograft tumorigenesis
Eight 4-week-old male BALB/c nude mice were obtained
from Shanghai SLAC Laboratory Animal Co., Ltd. (SLAC; China) and
bred under specific pathogen-free conditions. All animal studies
were conducted in accordance with the NIH animal use guidelines and
current Chinese regulations and standards for laboratory animal
use. Vector and SIRT-OE RKO cells were resuspended in DMEM
containing 10% FBS. The cell suspension was pre-cooled on ice
before bilateral inguinal subcutaneous injection, into each mouse
with an equal number of RKO cells (5×106), in a volume
of ~200 µl, on the left side of the negative vector group,
and the right side in the SIRT-OE group. Two months after
injection, the mice were sacrificed and the tumor was weighed.
Statistical analysis
Statistical analysis was performed using the SPSS
20.0 version of the statistical software. PCR analysis of 16 paired
human colorectal cancer was followed by t-test comparing adjacent
normal colorectal tissues and the proliferation toxicity of the
colorectal cancer cells to 5-FU. Card and Fisher's exact tests were
used to analyze the SIRT4 expression in the tumor and matched
non-tumor tissues, and the SIRT4 expression in relation to clinical
and pathological parameters in colorectal cancer. Kaplan-Meier
analysis (the log-rank test) was used for single factor analysis.
Cox proportional hazards regression model was used to identify
independent prognostic factors. Other experiments were analyzed by
the non-paired t-test. A P-value of <0.05 (two-tailed) was
considered statistically significant.
Results
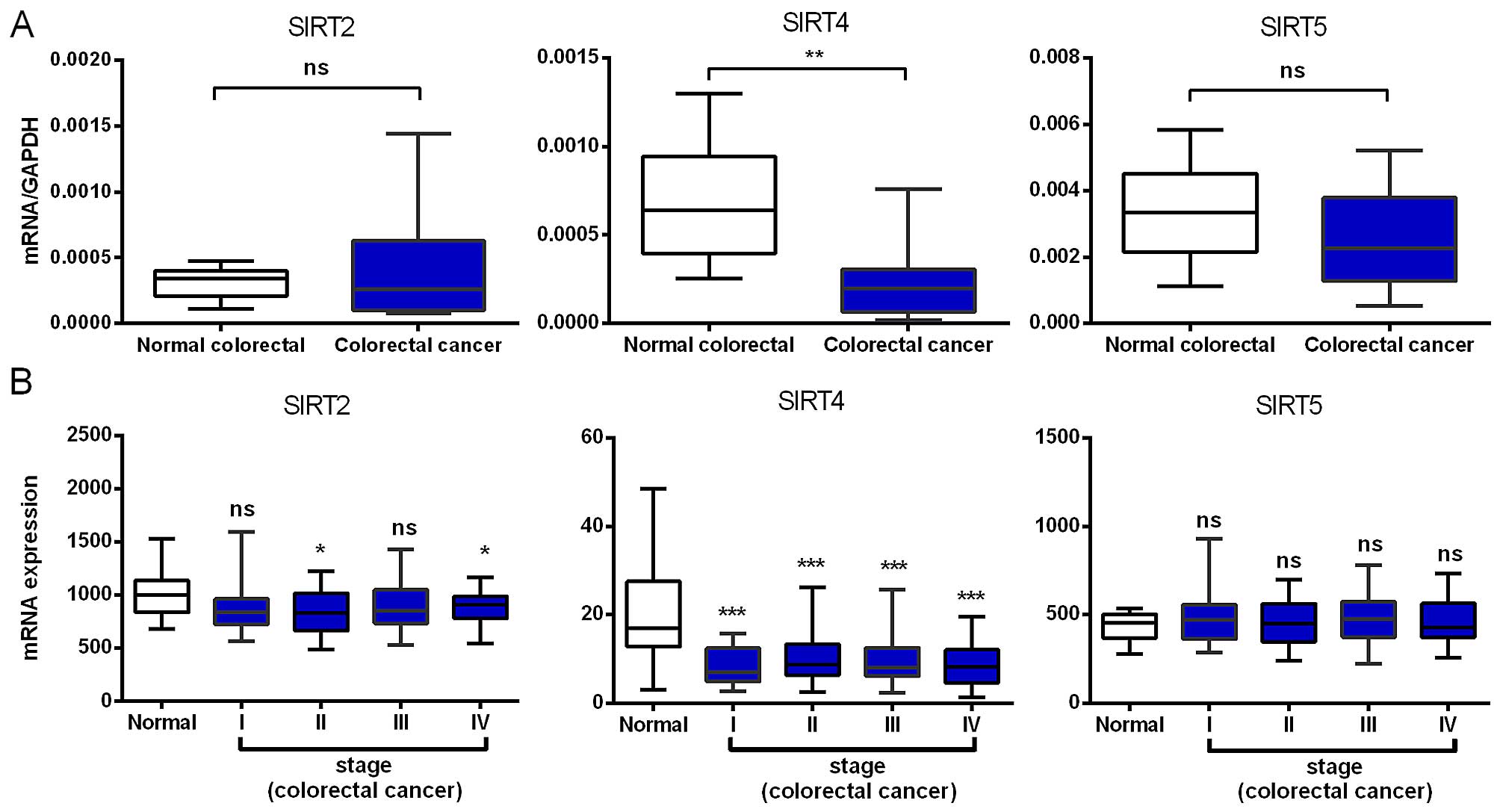
Decreased SIRT4 mRNA expression
We compared the mRNA expression levels of SIRT2,
SIRT4 and SIRT5 in 16 paired colorectal cancer and adjacent normal
tissues and found that SIRT4 was significantly reduced in
colorectal cancer, while SIRT2 and SIRT5 showed no significant
change (Fig. 1A). Although not
statistically significant, the expression of SIRT5 in colorectal
cancer showed a downward trend.
To validate the above results, we further analyzed
the expression profile of SIRT2, SIRT4 and SIRT5 using microarray
data, with 236 cases of colorectal cancer and 22 normal colorectal
tissue samples from the TCGA database. Consistent with our RT-PCR
results, we found that SIRT4 was downregulated in early stages and,
importantly, its low expression was maintained during cancer
progression, indicating that SIRT4 downregulation may be required
for both tumor initiation and maintenance. The SIRT2 mRNA
expression was downregulated in stages II and IV with no
significant changes in stages I and III. SIRT5 showed no
significant change in any colorectal cancer stage (Fig. 1B). In brief, the SIRT4 mRNA
expression in colorectal cancer tissues was significantly
decreased.
SIRT4 expression correlates with
pathological differentiation and prognosis
We next evaluated SIRT4 protein expression in tissue
microarray analysis of 89 colorectal cancer patients by
immunohistochemistry. We observed that SIRT4 was expressed in the
cytoplasm (Fig. 2). We then divided
the samples into two groups defined as low and high expression
based on the staining results. We found that in normal colorectal
tissues, 91.01% (81/89) of the SIRT4 segment was highly expressed
and 8.99% (8/89) were low. By contrast, in colorectal cancer
tissues, these numbers were 64.04% (57/89) and 35.96% (32/89),
respectively. The difference was statistically significant
(P<0.001; Table I).
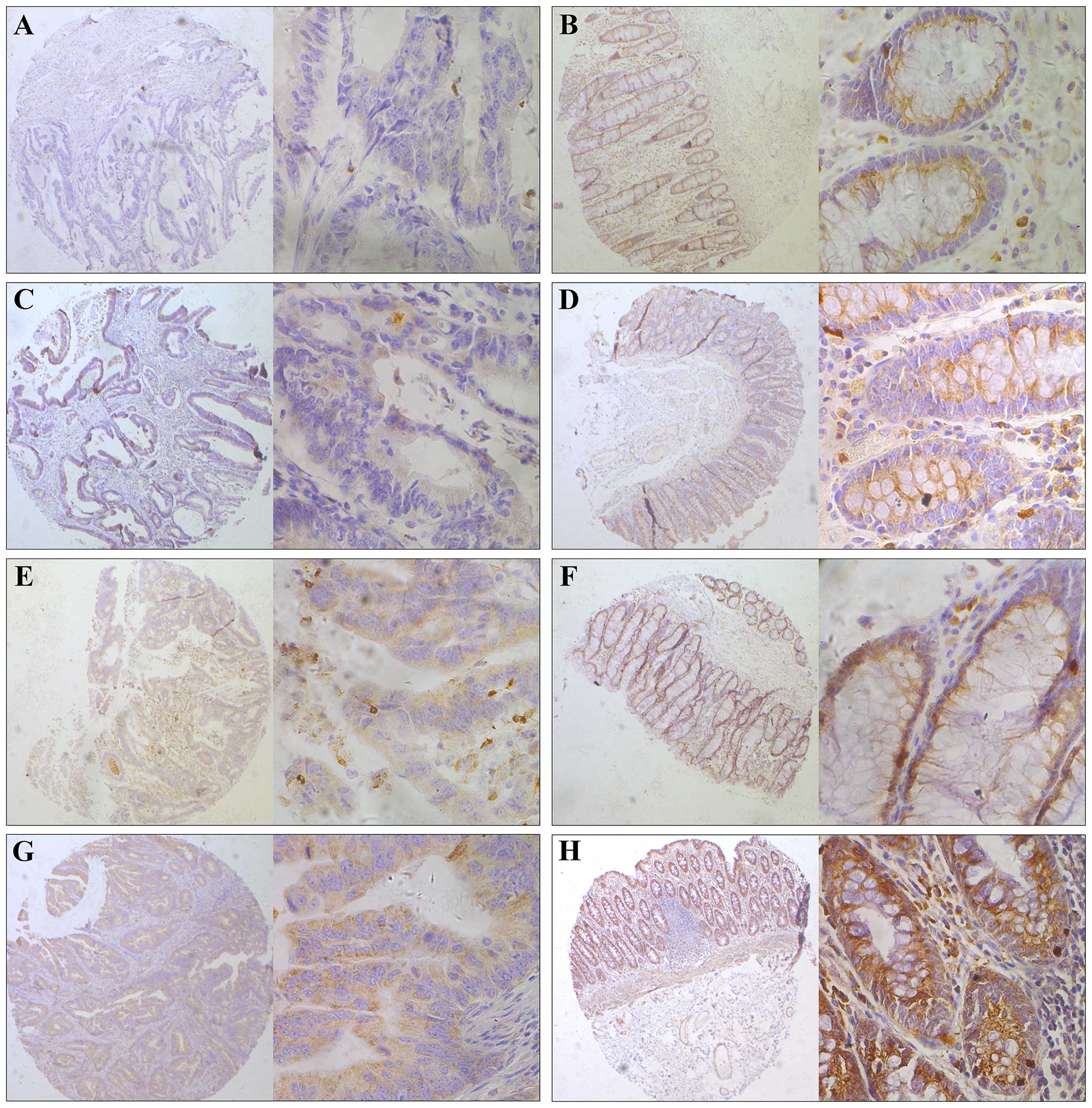 | Figure 2Representative immunohistochemical
staining of SIRT4 in human colorectal cancer tissues. SIRT4
expression in the cytoplasm was significantly lower in tumor
tissues compared with that observed in the adjacent normal
colorectal tissue. The micrographs showed negative (A), weak (C),
medium (E) and strong (G) expression of SIRT4 in colorectal cancer
tissues The relevant SIRT4 expression in corresponding adjacent
normal colorectal tissues of cases in A, C, E, and G is shown in B,
D, F and H, respectively (magnification: left panel, ×100; right
panel, ×400). |
 | Table ISIRT4 protein expression in
colorectal cancer and adjacent normal colon tissues. |
Table I
SIRT4 protein expression in
colorectal cancer and adjacent normal colon tissues.
| All cases | SIRT4 expression
| χ2 | P-valuea |
|---|
| Low (%) | High (%) |
|---|
| Tissue type | | | | 18.574 | 0.000 |
| Normal | 89 | 8 (8.99) | 81 (91.01) | | |
| Cancer | 89 | 32 (35.96) | 57 (64.04) | | |
We next analyzed the relationship between SIRT4
expression and the clinicopathological parameters and found that
patients expressing low SIRT4 levels manifested increasingly
adverse pathological grade (P=0.031). However, we did not find any
statistical relationship between SIRT4 expression with other
parameters, including age, gender, tumor size, tumor invasion depth
(T), lymph node positive number (N), distant metastasis (M) and
UICC stage (P>0.05). The relationship between SIRT4 expression
and the clinicopathological is summarized in Table II.
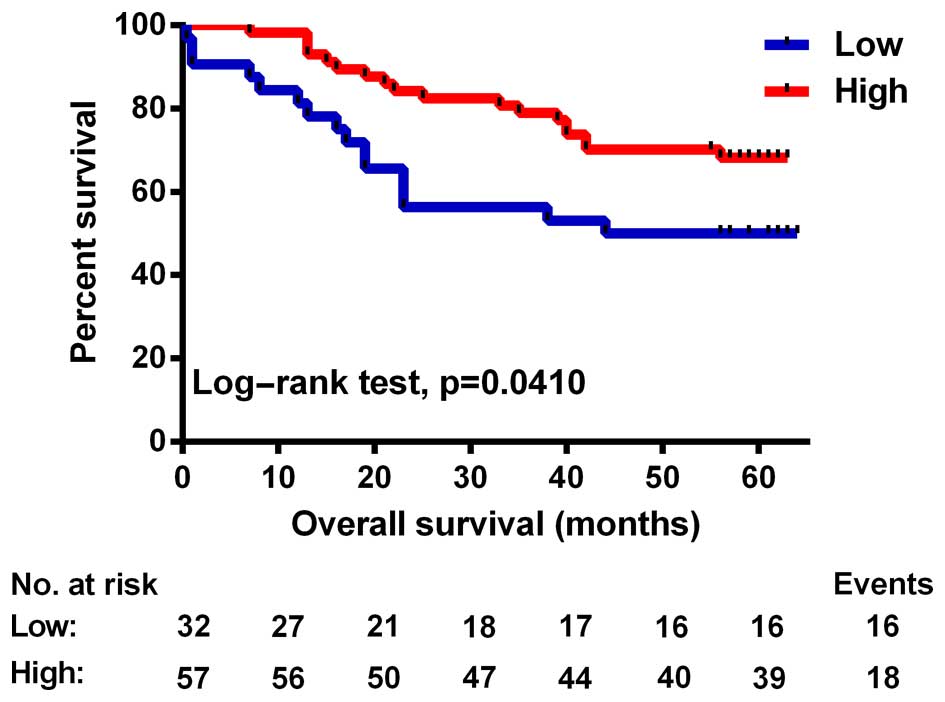
To further explore the prognostic value of SIRT4 in
colorectal cancer, we first performed univariate analysis. The
results showed that SIRT4 and tumor size, tumor differentiation,
lymph node and tumor distant metastasis, and UICC stage were
related to OS time post-operatively (Table III). The OS of patients with low
SIRT4 expression was significantly lower than that noted in
patients with high SIRT4 expression (P=0.041, test log-rank;
Fig. 3). Next, using COX regression
analysis adjusted for the prognostic factors established in the
univariate analysis, we found a significant correlation between low
SIRT4 expression and worse OS time of the colorectal cancer
patients (P=0.003, HR=0.339; Table
IV). Together, these results suggest that SIRT4 expression is
associated with a worse pathological grade and is an independent
prognostic factor for OS in patients with colorectal cancer.
 | Table IIIUnivariate analysis of SIRT4
expression and clinicopathological variables in 89 patients with
colorectal cancer. |
Table III
Univariate analysis of SIRT4
expression and clinicopathological variables in 89 patients with
colorectal cancer.
| Variable | All cases | Overall survival
(months)
| P-valuea |
|---|
| Mean | Median |
|---|
| Age (years) | | | | 0.531 |
| ≤65 | 41 | 46.6 | NR | |
| >65 | 48 | 49.3 | NR | |
| Gender | | | | 0.368 |
| Male | 46 | 49.3 | NR | |
| Female | 43 | 46.7 | NR | |
| Tumor size
(cm) | | | | 0.005 |
| ≤5 | 53 | 54.2 | NR | |
| >5 | 36 | 38.9 | 38 | |
|
Differentiation | | | | 0.003 |
| Well-moderate | 75 | 51.1 | NR | |
| Poor | 14 | 30.6 | 17 | |
| T stage | | | | 0.247 |
| T1-T2 | 13 | 52.6 | NR | |
| T3-T4 | 76 | 47.0 | NR | |
| N stage | | | | 0.000 |
| N0 | 58 | 53.7 | NR | |
| N1-N2 | 31 | 37.1 | 39 | |
| M stage | | | | 0.002 |
| M0 | 86 | 49.1 | NR | |
| M1 | 3 | 18.3 | 17 | |
| UICC stage | | | | 0.000 |
| I-II | 56 | 54.9 | NR | |
| III-IV | 33 | 36.0 | 25 | |
| SIRT4
expression | | | | 0.041 |
| Low | 32 | 40.3 | 44 | |
| High | 57 | 51.7 | NR | |
 | Table IVCox multivariate analyses of
prognostic factors on overall survival. |
Table IV
Cox multivariate analyses of
prognostic factors on overall survival.
| Variables | HR | 95% CI | P-valuea |
|---|
| Tumor size (cm) (≤5
vs. >5) | 2.781 | 1.384–5.590 | 0.004 |
| Differentiation
(Well/moderate vs. poor) | | | NS |
| N stage (N0 vs.
N1/N2) | | | NS |
| M stage (M0 vs.
M1) | | | NS |
| UICC stage (I/II
vs. III/IV) | 4.555 | 2.201–9.426 | 0.000 |
| SIRT4 expression
(Low vs. High) | 0.339 | 0.165–0.695 | 0.003 |
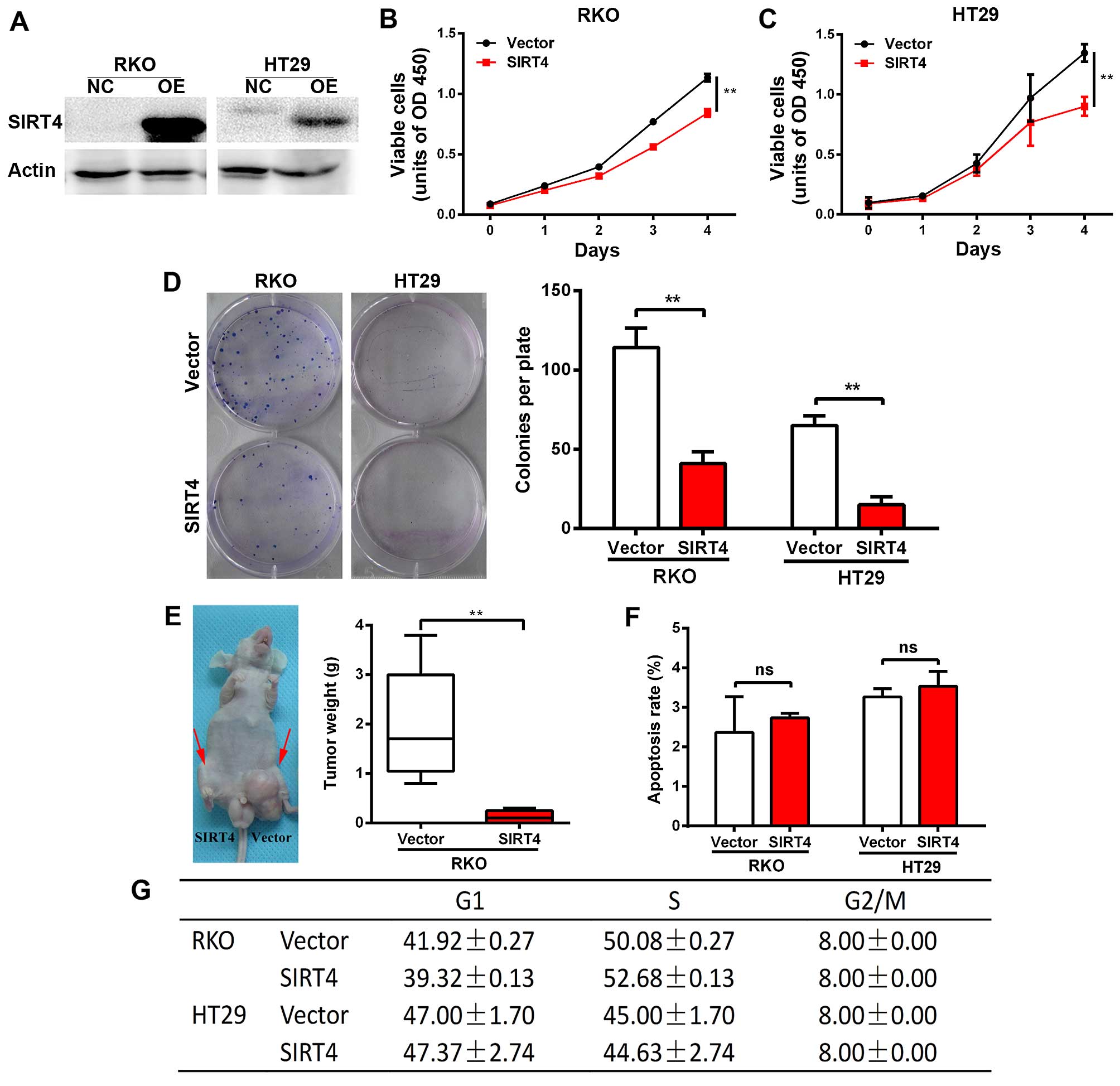
SIRT4 inhibits the growth of human
colorectal cancer cells
We constructed a stable cell line overexpressing
SIRT4 in colorectal cancer lines RKO and HT29 using the lentivirus,
and verified the results by western blotting (Fig. 4A). We found that SIRT4
overexpression significantly reduced the proliferation of RKO and
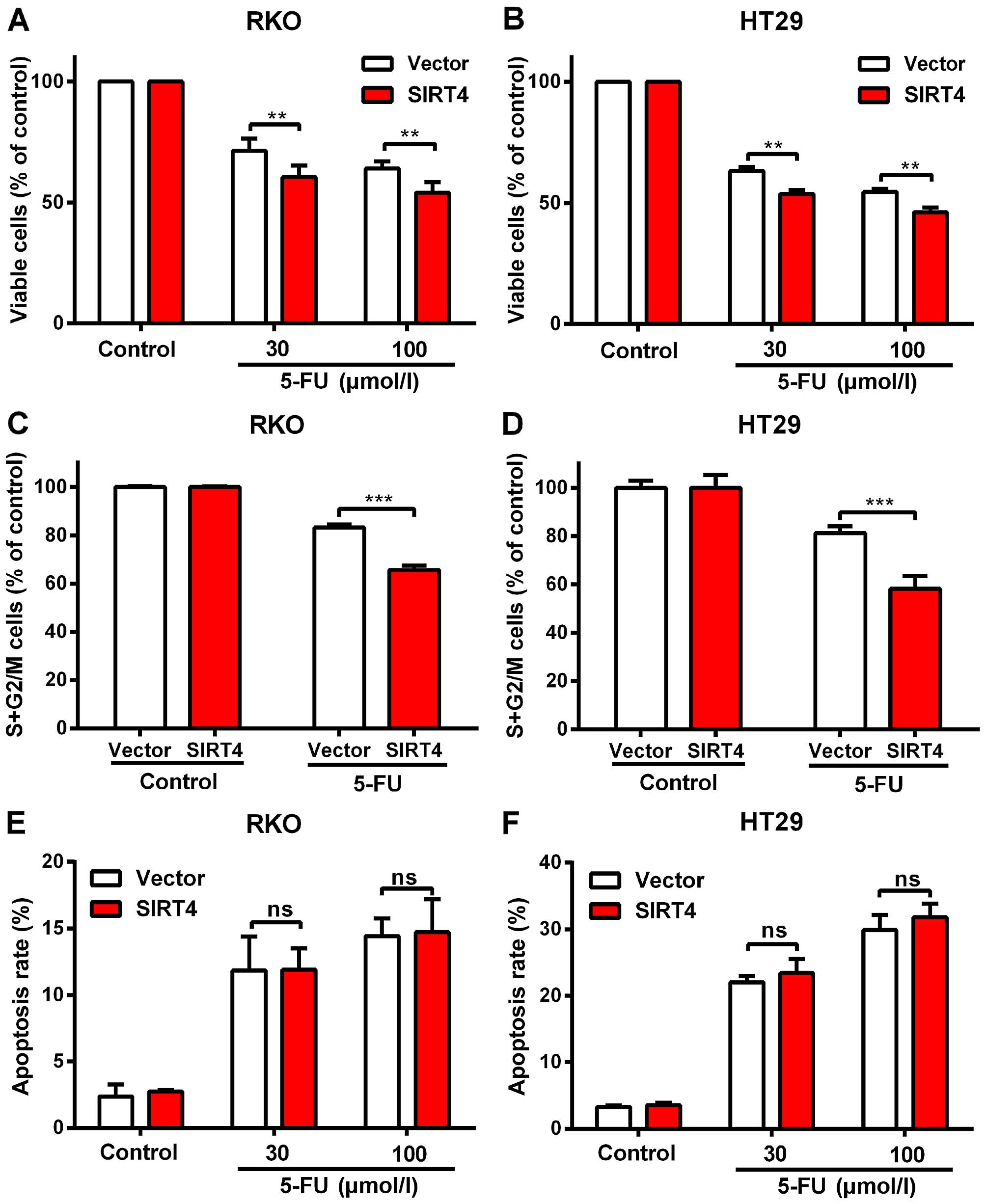
HT29 cells (Fig. 4B and C).
Furthermore, SIRT4 overexpression significantly reduced the number
and size of the clones of RKO and HT29 cells (Fig. 4D). Next, we found that SIRT4
overexpression significantly reduced the tumorigenic potential of
RKO cells in nude mice (Fig. 4E).
Together, these results indicate that SIRT4 inhibits the growth of
colorectal cancer cells.
We found no significant change in the apoptosis rate
and cell cycle of the RKO and HT29 cells following SIRT4
overexpression (Fig. 4F and G).
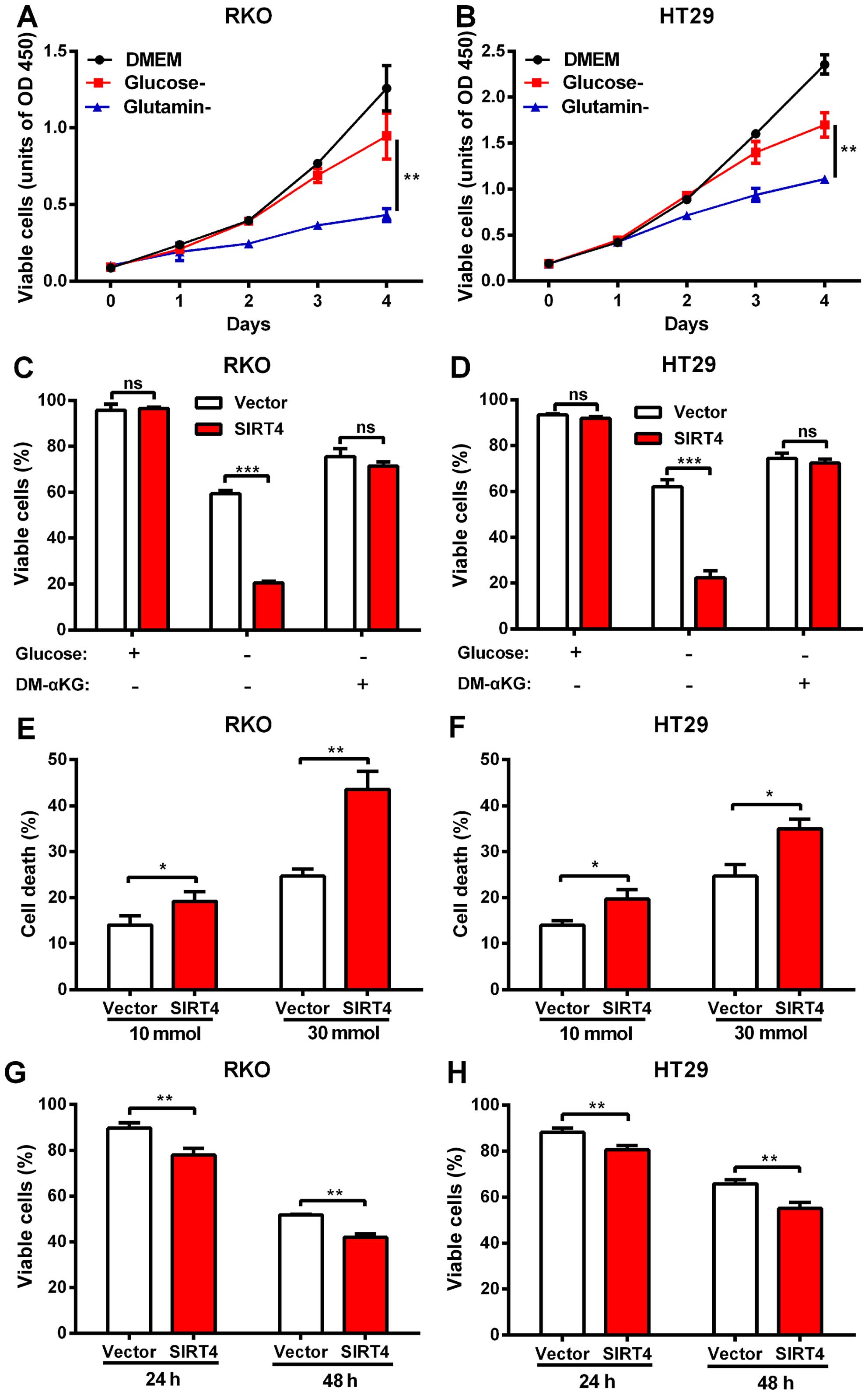
SIRT4 inhibits glutamine metabolism and
synergistically with glycolytic inhibition induces cell death in
colorectal cancer
Studies indicate that SIRT4 inhibits tumor growth
via inhibition of mitochondrial glutamine metabolism (21,22).
We investigated whether SIRT4 inhibited the growth of colon cancer
cells by inhibiting glutamine metabolism. We found that RKO and
HT29 cells still maintained growth in media in the absence of
glucose, but the growth rate was significantly weak in the absence
of glutamine (Fig. 5A and B)
suggesting that glutamine metabolism plays an important role in the
growth of human colorectal cancer cells.
We tested whether SIRT4 inhibited the utilization of
glutamine in colorectal cancer cells. We deprived RKO and HT29
cells of glucose, and forced the cells to switch to glutamine to
maintain growth. The results showed that SIRT4 overexpression
significantly reduced the survival rate of the RKO and HT29 cells
in glucose-deprivation. However, when cell-permeable DM-KG was
added, the mortality difference disappeared (Fig. 5C and D). We found that in
glucose-deprived media, the magnitude of the decrease in
proliferation of the colorectal cancer cells caused by SIRT4
overexpression was larger compared with glucose-supplemented media
(data not shown). These results indicate that SIRT4 overexpression
reduced glutamine dependence of colorectal cancer cells, suggesting
that SIRT4 inhibition of glutamine metabolism mediated the
inhibition of proliferation of the colorectal cancer cells.
Blocking tumor cells in the metabolic pathway is a
new treatment strategy. Since SIRT4 inhibited glutamine metabolism,
we further explored whether SIRT4 overexpression increased the
sensitivity of colorectal cancer cells to glucose metabolic
inhibitors. Consistent with the previous glucose deprivation
experiments, we found that SIRT4 overexpression sensitized
colorectal cancer cells to 2-deoxyglucose (2-DG)-induced cell death
(Fig. 5E and F) further supporting
the role of SIRT4 in glutamine metabolism and survival of
colorectal cells and indicating that SIRT4 overexpression and
glucose metabolism inhibitors induced a synergistic effect on the
colorectal cancer cells.
In addition to inhibiting glutamine metabolism, the
role of SIRT4 in glucose metabolism is still unclear. We found that
SIRT4 overexpression significantly reduced the survival rates of
both colorectal cancer cell lines in glutamine-deprived media
(Fig. 5G and H) suggesting that
SIRT4 affected glucose metabolism as well.
SIRT4 increases the sensitivity of
colorectal cancer cells to 5-FU by delaying the cell cycle
5-FU is the most commonly used chemotherapeutic
agent for the treatment of colorectal cancer. We found that SIRT4
overexpression increased the inhibitory effect of 5-FU on the
proliferation of colorectal cancer cells (Fig. 6A and B).
We found that SIRT4 overexpression significantly
decreased the S and G2/M rates in both colorectal cancer cell lines
after 5-FU treatment (Fig. 6E), but
had no influence on the apoptotic rates under these conditions
(Fig. 6F) suggesting that SIRT4
increased the sensitivity of colorectal cancer cells to 5-FU by
delaying mitosis.
Discussion
In the present study, we found that decreased SIRT4
expression in human colorectal cancer was associated with poor
pathologic differentiation and worse prognosis. In vitro and
in vivo experiments demonstrated that SIRT4 decreased the
proliferation activity, the number of cells and tumor formation in
nude mice injected with colorectal cancer cells. We found that
glutamine plays an important role in the growth of colorectal
cancer cells, and SIRT4 weakened the ability of colorectal cancer
cells in glutamine utilization and enhanced cell death caused by
glucose metabolism inhibitor 2-DG. Finally, we found that SIRT4
increased the sensitivity of colorectal cancer cells to
chemotherapeutic agents by delaying the cell cycle. Our research
has uncovered the clinical significance of SIRT4 in human colon
cancer.
According to the present study, multiple SIRT family
members are involved in different tumors, which may depend on the
specific tissue and tumor type (28). For instance, SIRT1 expression levels
are elevated in gastric (29),
colorectal (30), prostate
(31) and skin cancers (32), suggesting that it promotes tumor
formation. In addition, studies have shown that SIRT1 may act as a
tumor suppressor. For example, SIRT1 was downregulated in breast
cancer (33) and inhibited the
formation of intestinal tumors in APC (Min/+) mouse
models (34). Similarly, SIRT2 was
found to be downregulated in breast (17), glioma (35) and skin cancers (36), but upregulated in acute myeloid
leukemia (37) and prostate cancer
(38). Jeong et al (21,39)
found that SIRT4 inhibited the growth of HeLa cells and MYC-induced
B-lymphoma cells. SIRT4-knockout MEF cells in nude mice formed
larger tumors. SIRT4-knockout mice spontaneously developed cancers
of the lung, liver, breast and lymphoma. Csibi et al
(22) also found that SIRT4
inhibited the growth of human colorectal cancer DLD-1 cells and
human prostate cancer DU145 cells. Our previous studies suggested
that SIRT4 expression is decreased in gastric cancer tissues and is
correlated with gastric cancer pathology (27). In the present study, we found that
SIRT4 was downregulated in human colorectal cancer tissues and
inhibited the growth of colorectal cancer cells in vitro and
in vivo. Our results suggest that SIRT4 plays a
tumor-suppressor role in human colorectal cancer, and that SIRT4 is
a tumor-suppressor gene.
We found no significant difference in SIRT2 and
SIRT5 mRNA levels between colorectal cancer and normal colorectal
tissues. Since the role of SIRT family members is tissue-specific,
they do not play a role in the development of colorectal cancer. In
contrast, their expression in colorectal cancer may be regulated by
post transcriptional modification. SIRT1, which has a higher level
of protein expression in hepatocellular carcinoma tissues compared
with normal liver tissues, showed no difference in mRNA expression
between HCC tissues and adjacent normal liver tissues (40). The next step will be to study the
protein expression in colorectal cancer.
Jeong et al (21) and Csibi et al (22) found that SIRT4 inhibited tumor
growth by inhibiting mitochondrial glutamine metabolism. However,
recent studies show that SIRT4 inhibits pyruvate dehydrogenase
(41). Since pyruvate dehydrogenase
is a key enzyme in the tricarboxylic acid cycle, SIRT4 may also
play a role in glucose metabolism. The present study found that
SIRT4 increased the cell death of colorectal cancer cells in
glucose-deprived culture media, and the addition of glutamine to
downstream metabolite DM-KG abrogated this effect suggesting that
SIRT4 inhibited glutamine metabolism. However, we also found that
overexpression of SIRT4 reduced the survival rate of the colorectal
cancer cells in glutamine-deprived media, indicating that SIRT4 may
also inhibit glucose metabolism in colorectal cancer cells.
Targeting glucose metabolism using glucose
inhibitors has been used as a therapeutic strategy (42–44).
However, tumor cells activate other metabolic pathways, such as
glutamine metabolism to survive, since mitochondrial glutamine
metabolism can substitute for the lack of glucose recharge
mitochondrial tricarboxylic acid cycle (45,46).
Our experiments found that colorectal cancer cells under glucose
deprivation still maintained growth, and SIRT4 overexpression
increased the death associated with glucose deprivation in
colorectal cancer. SIRT4 overexpression and glucose inhibitors 2-DG
synergistically acted to significantly increase colorectal cancer
cell death. These results indicate the therapeutic potential of
SIRT4 targeting in metabolism, particularly in treating tumors with
a glucose metabolism inhibitor.
Previous research has shown that SIRT4 delays the
cell cycle in damaged DNA (21).
The present study found that 5-FU increased the sensitivity of
colorectal cancer cells to chemotherapy drugs by delaying the cell
cycle. The present study reveals the potential of SIRT4 in
chemotherapy.
In summary, our results indicate that SIRT4 plays a
tumor-suppressor role and is an independent prognostic factor in
colorectal cancer. SIRT4 for the treatment of colorectal cancer,
particularly in conjunction with metabolic and cytotoxic
chemotherapy, is a promising strategy. The present study reveals
the clinical significance of SIRT4 in human colorectal cancer. Our
results suggest that SIRT4 is a potential diagnostic and
therapeutic target in colorectal cancer.
Acknowledgments
We thank Yueqin Tang for technical assistance in the
experimental study of nude mice. We also thank Huamei Tang for
assistance with the immunohistochemistry studies. The present study
was funded by the National High Technology Research and Development
Program (SS2014AA020803), the National Natural Science Foundation
of China (81220108021), the Project of Shanghai Science and
Technology Commission (14411950502), the Joint Research Projects of
Shanghai Municipal Hospital (SHDC12012105), the Project of Shanghai
JiaoTong University (YG2012ZD01), and the Nutriology of the Medical
Support Discipline of Zhejiang Province.
References
|
1
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar
|
|
2
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar
|
|
4
|
Finkel T, Deng CX and Mostoslavsky R:
Recent progress in the biology and physiology of sirtuins. Nature.
460:587–591. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yuan H, Su L and Chen WY: The emerging and
diverse roles of sirtuins in cancer: A clinical perspective. Onco
Targets Ther. 6:1399–1416. 2013.PubMed/NCBI
|
|
6
|
Chen WY, Wang DH, Yen RC, Luo J, Gu W and
Baylin SB: Tumor suppressor HIC1 directly regulates SIRT1 to
modulate p53-dependent DNA-damage responses. Cell. 123:437–448.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brunet A, Sweeney LB, Sturgill JF, Chua
KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et
al: Stress-dependent regulation of FOXO transcription factors by
the SIRT1 deacetylase. Science. 303:2011–2015. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu H, Ye W, Wu J, Meng X, Liu RY, Ying X,
Zhou Y, Wang H, Pan C and Huang W: Overexpression of sirt7 exhibits
oncogenic property and serves as a prognostic factor in colorectal
cancer. Clin Cancer Res. 20:3434–3445. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu C, Huang Z, Jiang H and Shi F: The
sirtuin 3 expression profile is associated with pathological and
clinical outcomes in colon cancer patients. Biomed Res Int.
2014:8712632014.PubMed/NCBI
|
|
10
|
Sebastián C, Zwaans BM, Silberman DM,
Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber
D, et al: The histone deacetylase SIRT6 is a tumor suppressor that
controls cancer metabolism. Cell. 151:1185–1199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kabra N, Li Z, Chen L, Li B, Zhang X, Wang
C, Yeatman T, Coppola D and Chen J: SirT1 is an inhibitor of
proliferation and tumor formation in colon cancer. J Biol Chem.
284:18210–18217. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
North BJ, Marshall BL, Borra MT, Denu JM
and Verdin E: The human Sir2 ortholog, SIRT2, is an
NAD+-dependent tubulin deacetylase. Mol Cell.
11:437–444. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vaquero A, Scher MB, Lee DH, Sutton A,
Cheng HL, Alt FW, Serrano L, Sternglanz R and Reinberg D: SirT2 is
a histone deacetylase with preference for histone H4 Lys 16 during
mitosis. Genes Dev. 20:1256–1261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Das C, Lucia MS, Hansen KC and Tyler JK:
CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature.
459:113–117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jing E, Gesta S and Kahn CR: SIRT2
regulates adipocyte differentiation through FoxO1
acetylation/deacetylation. Cell Metab. 6:105–114. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin YH, Kim YJ, Kim DW, Baek KH, Kang BY,
Yeo CY and Lee KY: Sirt2 interacts with 14-3-3 beta/gamma and
down-regulates the activity of p53. Biochem Biophys Res Commun.
368:690–695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim HS, Vassilopoulos A, Wang RH, Lahusen
T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H, et al: SIRT2
maintains genome integrity and suppresses tumorigenesis through
regulating APC/C activity. Cancer Cell. 20:487–499. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Haigis MC, Mostoslavsky R, Haigis KM,
Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos
GD, Karow M, Blander G, et al: SIRT4 inhibits glutamate
dehydrogenase and opposes the effects of calorie restriction in
pancreatic beta cells. Cell. 126:941–954. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nasrin N, Wu X, Fortier E, Feng Y, Bare'
OC, Chen S, Ren X, Wu Z, Streeper RS and Bordone L: SIRT4 regulates
fatty acid oxidation and mitochondrial gene expression in liver and
muscle cells. J Biol Chem. 285:31995–32002. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ahuja N, Schwer B, Carobbio S, Waltregny
D, North BJ, Castronovo V, Maechler P and Verdin E: Regulation of
insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase.
J Biol Chem. 282:33583–33592. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jeong SM, Xiao C, Finley LW, Lahusen T,
Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, et al:
SIRT4 has tumor-suppressive activity and regulates the cellular
metabolic response to DNA damage by inhibiting mitochondrial
glutamine metabolism. Cancer Cell. 23:450–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Csibi A, Fendt SM, Li C, Poulogiannis G,
Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T,
et al: The mTORC1 pathway stimulates glutamine metabolism and cell
proliferation by repressing SIRT4. Cell. 153:840–854. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tan M, Peng C, Anderson KA, Chhoy P, Xie
Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, et al: Lysine
glutarylation is a protein posttranslational modification regulated
by SIRT5. Cell Metab. 19:605–617. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang
H, Kim J, Woo J, Kim JH, Choi BH, et al: Sirt5 is a NAD-dependent
protein lysine demalonylase and desuccinylase. Science.
334:806–809. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakagawa T, Lomb DJ, Haigis MC and
Guarente L: SIRT5 deacetylates carbamoyl phosphate synthetase 1 and
regulates the urea cycle. Cell. 137:560–570. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu W, Zuo Y, Feng Y and Zhang M: SIRT5
facilitates cancer cell growth and drug resistance in non-small
cell lung cancer. Tumour Biol. 35:10699–10705. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang G, Cui F, Yu F, Lu H, Zhang M, Tang
H and Peng Z: Sirtuin-4 (SIRT4) is downregulated and associated
with some clinicopathological features in gastric adenocarcinoma.
Biomed Pharmacother. 72:135–139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Roth M and Chen WY: Sorting out functions
of sirtuins in cancer. Oncogene. 33:1609–1620. 2014. View Article : Google Scholar
|
|
29
|
Cha EJ, Noh SJ, Kwon KS, Kim CY, Park BH,
Park HS, Lee H, Chung MJ, Kang MJ, Lee DG, et al: Expression of
DBC1 and SIRT1 is associated with poor prognosis of gastric
carcinoma. Clin Cancer Res. 15:4453–4459. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stünkel W, Peh BK, Tan YC, Nayagam VM,
Wang X, Salto-Tellez M, Ni B, Entzeroth M and Wood J: Function of
the SIRT1 protein deacetylase in cancer. Biotechnol J. 2:1360–1368.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huffman DM, Grizzle WE, Bamman MM, Kim JS,
Eltoum IA, Elgavish A and Nagy TR: SIRT1 is significantly elevated
in mouse and human prostate cancer. Cancer Res. 67:6612–6618. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hida Y, Kubo Y, Murao K and Arase S:
Strong expression of a longevity-related protein, SIRT1, in Bowen's
disease. Arch Dermatol Res. 299:103–106. 2007. View Article : Google Scholar
|
|
33
|
Wang RH, Sengupta K, Li C, Kim HS, Cao L,
Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al: Impaired DNA damage
response, genome instability, and tumorigenesis in SIRT1 mutant
mice. Cancer Cell. 14:312–323. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Firestein R, Blander G, Michan S,
Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S,
de Cabo R, Fuchs C, et al: The SIRT1 deacetylase suppresses
intestinal tumorigenesis and colon cancer growth. PLoS One.
3:e20202008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hiratsuka M, Inoue T, Toda T, Kimura N,
Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CG, Kurimasa
A, et al: Proteomics-based identification of differentially
expressed genes in human gliomas: Down-regulation of SIRT2 gene.
Biochem Biophys Res Commun. 309:558–566. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ming M, Qiang L, Zhao B and He YY:
Mammalian SIRT2 inhibits keratin 19 expression and is a tumor
suppressor in skin. Exp Dermatol. 23:207–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dan L, Klimenkova O, Klimiankou M, Klusman
JH, van den Heuvel-Eibrink MM, Reinhardt D, Welte K and Skokowa J:
The role of sirtuin 2 activation by nicotinamide
phosphoribosyltransferase in the aberrant proliferation and
survival of myeloid leukemia cells. Haematologica. 97:551–559.
2012. View Article : Google Scholar :
|
|
38
|
Hou H, Chen W, Zhao L, Zuo Q, Zhang G,
Zhang X, Wang H, Gong H, Li X, Wang M, et al: Cortactin is
associated with tumour progression and poor prognosis in prostate
cancer and SIRT2 other than HADC6 may work as facilitator in situ.
J Clin Pathol. 65:1088–1096. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jeong SM, Lee A, Lee J and Haigis MC:
SIRT4 suppresses tumor formation in genetic models of Myc-induced B
cell lymphoma. J Biol Chem. 289:4135–4144. 2014. View Article : Google Scholar :
|
|
40
|
Chen J, Zhang B, Wong N, Lo AW, To KF,
Chan AW, Ng MH, Ho CY, Cheng SH, Lai PB, et al: Sirtuin 1 is
upregulated in a subset of hepatocellular carcinomas where it is
essential for telomere maintenance and tumor cell growth. Cancer
Res. 71:4138–4149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mathias RA, Greco TM, Oberstein A,
Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T and Cristea
IM: Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase
complex activity. Cell. 159:1615–1625. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao Y, Butler EB and Tan M: Targeting
cellular metabolism to improve cancer therapeutics. Cell Death Dis.
4:e5322013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Galluzzi L, Kepp O, Vander Heiden MG and
Kroemer G: Metabolic targets for cancer therapy. Nat Rev Drug
Discov. 12:829–846. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ahmad IM, Abdalla MY, Aykin-Burns N,
Simons AL, Oberley LW, Domann FE and Spitz DR: 2-Deoxyglucose
combined with wild-type p53 overexpression enhances cytotoxicity in
human prostate cancer cells via oxidative stress. Free Radic Biol
Med. 44:826–834. 2008. View Article : Google Scholar
|
|
45
|
Daye D and Wellen KE: Metabolic
reprogramming in cancer: Unraveling the role of glutamine in
tumorigenesis. Semin Cell Dev Biol. 23:362–369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tennant DA, Durán RV and Gottlieb E:
Targeting metabolic transformation for cancer therapy. Nat Rev
Cancer. 10:267–277. 2010. View Article : Google Scholar : PubMed/NCBI
|