Introduction
Cervical cancer is the second most common
gynecological malignancy among women worldwide, and there are an
estimated 530,000 cases of cervical cancer and 275,000 deaths from
the disease per year (1,2). The mechanisms of cervical carcinoma
formation remain unclear. Cervical carcinoma emerges from a defined
series of preneoplastic lesions with increasing cellular dysplasia
referred to as cervical intra-epithelial neoplasia (CIN) grade I,
II and III. The development and progression of cervical carcinoma
have been demonstrated associated with various genetic and
epigenetic events, especially alterations in the cell cycle
checkpoint machinery. However, the steps of the progression from
low-grade CIN to carcinoma remain elusive. Epidemiological studies
have established a causal relationship between cervical
carcinogenesis and infection of high risk HPV types (HR-HPV, such
as HPV 16 and 18) (3). It is
explicit that integration of HR-HPV DNA into the host cell genome
resulting in persistent over-expression HPV E6 and E7 oncoproteins,
subsequently induce immortalization of cells and allow virus to
replicate through their inhibitory effects on the tumor suppressor
proteins p53 and pRb, respectively (4–6).
However the E6-p53 and E7-Rb model is not sufficient to inevitably
produce cervical carcinoma (7).
Other factors must have contributed to the initiation of the
cervical cancer (8,9). Recent studies suggested a strong
association between HR-HPV types and cell cycle regulators
(10). It is well-known that the
cell cycle is regulated by a family of cyclins (cyclins A, B, D,
E), cyclin dependent kinases (CDKs, CDK1, CDK2, CDK4, CDK6) and
their inhibitors (CDKIs) through activating and inactivating
phosphorylation events. Attention has been focused on altered
expression of G1 cyclins and Cdks because the major regulatory
events leading to cell proliferation and differentiation occur
within the G1 phase of the cell cycle (11–13).
However, it is unclear how and when cell cycle factors that are
innate to the HPV-infected cells, including genetic aberrations
launch the host cell into an irreversible progression to
cancer.
BLCAP is a small 87-amino acid, evolutionary
conserved protein with no homology to any known protein in
mammalians (14). Studies have
shown BLCAP gene exerts tumor suppressor function in bladder
cancer, osteosarcoma, tongue carcinoma, renal cancer, breast cancer
and other malignant tumors (15,16).
Our previous studies found that BLCAP protein putatively includes
two trans-membrane domains, cytoplasmic domains at the N and C
terminals, a phosphorylation site that might bind DNA. We found
that BLCAP mRNA could be detected in normal cervical tissue,
but it was absent or reduced in cervical cancer tissue.
Overexpression of BLCAP could play its function as a tumor
suppressor gene to inhibit the growth of cervical cancer cell line
in vitro (17). However,
little was known about the regulation and function of BLCAP
protein. It seems likely that BLCAP might play a role not only in
regulating cellular proliferation but also coordinating the cell
cycle and apoptosis via a novel way independent of p53 and NF-κB as
previously reported by us (18).
By analyzing the signal peptides of BLCAP protein,
we found a PXXP (proline-X-X-proline) and an SPXX (Ser-Pro-X-X)
motif located within it (17). In
addition, using NetPhos and KinasePhos program analysis we
identified several putative phosphorylation sites in the BLCAP
protein: Ser66, Ser71 (Ataxia Telangiectasia Mutated
phosphorylation site), Ser73 (cdc2 phosphorylation site) and Ser78
(casein kinase II phosphorylation site) (19–22).
Through a computer-based search BLCAP was identified as a
target of adenosine to inosine (A-to-I) by RNA editing.
In this study, we determined the molecular targets
of BLCAP by protein interaction technology, combined with the cell
cycle signal path analysis and apoptosis induction by regulating
expression of key molecules to explore the molecular mechanism of
BLCAP gene.
Materials and methods
Cell culture and reagents
Human cervical cancer cell line (HeLa) was used in
this study. HeLa cells were maintained in RPMI-1640 supplemented
with 10% fetal bovine serum (FBS, Invitrogen) at 37°C in a
humidified 5% Co2 atmosphere. The sources of antibodies
were as follow: antibodies against pRB1, E2F were from Cell
Signaling Technology. Antibodies against cyclin D1, cyclin B1,
CDK4, CDK6, caspase-3 and p53 were from Abcam, Inc. Antibodies
against β-actin were obtained from Santa Cruz Biotechnology. Goat
peroxidase (HRP), conjugated secondary antibodies, and protease
inhibitor were from Roche. Chemiluminescence substrate was obtained
from Pierce.
Plasmid construction
The pRNA-U6.1/Hygro vector was purchased from
GenScript (Piscataway, NJ, USA) to use to construct shRNA plasmids
of BLCAP. DNA template corresponding to BLCAP gene
(GenBank accession no. NM006698) was used for design of shRNA.
shRNAs of BLCAP gene were designed and synthesized through
database of https://www.genscript.com. The
sequences of shRNA are shown in Table
I.
 | Table IThe sequences of shRNA targeting
BLCAP. |
Table I
The sequences of shRNA targeting
BLCAP.
| Name | Sequence of
shRNA |
|---|
| pRNA-B1 |
5′-GGATCCCGTTGTGCAAGGCTTCCGTTCCATTGATATCCGTGGAACGGAAGCCTTGCACAA-3′ |
| pRNA-B2 |
5′-GGATCCCGTGCAAGGCTTCCGTTCCAGGATTGATATCCGTCCTGGAAGCCTTGCA-3′ |
| pRNA-B3 |
5′-GGATCCCGAATCGGAGCAGTGGTACAGGTTGATATCCGCCTGTACCACTGCTCCGATTC-3′ |
| NS |
5′-GCGAGATCTGTGCCGCTCCTCATCATCCATGTTCAAGAGACATGGATGATGAGGAGCGGCA-3′ |
BamHI/HindIII fragments from the
sequences were subcloned into the same sites of pRNA-U6.1/Hygro to
generate the pshRNA-B1, pshRNA-B2, pshRNA-B3 and pRNA-NS (a
non-specific shRNA) plasmid, respectively. The recombinant vectors
were confirmed by digestion analysis of restriction endonuclease
and DNA sequencing. The full length cDNA of BLCAP was
acquired from recombinant pCD-3.1(-)-BLCAP plasmid (it was
stored by our group) and subcloned into pEgFP-N1 and pEF-CARD-3x
Flag plasmids to generate recombinant pEgFP-BLCAP and
pEF-BLCAP-3xFlag expression plasmids. The pEgFP-BLCAP
plasmid is co-transfected with shRNA plasmids of BLCAP to
detect the effects of RNAi with the help of alteration of EgFP in
cells. The Flag protein of pEF-BLCAP-3xFlag was used to
detect the BLCAP protein in immunoprecipitation reactions.
Plasmid transfection and stable
selection
HeLa cells (1×105) were seeded in 6-well
plates and subsequently transfected with recombinant plasmid
containing selection marker using Lipofectamine 2000 (grand Island,
Ny, uSA) according to the manufacturer's protocols. Transfectant
cells were selected with 50 µg/ml of G418 or 8 µg/ml
of hygromycin (Sigma, USA) for two weeks, respectively. Single
clones were isolated and expanded for an additional one months in
media containing selection antibiotics. The stable transfectants
were named HeLa-wt (transfected with wild-type of BLCAP
gene), HeLa-M1 (transfected with BLCAP AXXA type), HeLa-M2
(transfected with BLCAP SAXX type) and HeLa-M3 (transfected
with BLCAP Ala type). The stable transfectants of shRNA in
HeLa were HeLa-B1 (transfected with pshRNA-B1), HeLa-B2
(transfected with pshRNA-B2), HeLa-B3 (transfected with pshRNA-B3)
and HeLa-NS (transfected with pshRNA-NS), respectively.
Detection of apoptosis
Apoptosis analysis was performed by Annexin V-FITC
Apoptosis Detection kit (BD Biosciences, San Jose, CA, USA)
following the manufacturer's instructions. Briefly, the kit
includes Annexin V conjugated to FITC and propidium iodide. For
each sample, 1×105 cells were harvested and washed twice
with cold PBS buffer. All cells were gently suspended in 100
µl of binding buffer, and 5 µl Annexin V-FTIC and 5
µl propidium iodide was added. Then, cells were gently
vortex and incubated for 15 min at room temperature (RT) (25°C) in
the dark. Finally, cells were analyzed with a flow cytometer. At
least 10,000 cells were counted per analysis.
RNA extraction and cellular signal
pathway chip analysis
To determine the role that BLCAP play in the signal
transduction of cell cycle and to determine the target molecules
for interaction, signal pathway chip experiment was performed using
human signal pathway gene Oligo chips (CapitalBio Corp., Beijing,
China). This chip contains 897 genes related to signal
transduction. All cells were divided into two cell groups, one
group includes HeLa cells and HeLa BLCAPwt (transfected with
wild-type BLCAP plasmid, pCD-3.1(-)-BLCAP) cells. The
other includes HeLa-BLCAPwt and
HeLa-BLCAPwt-siRNA(HeLa- BLCAPwt cells transfected
with siRNA against BLCAP gene) cells. Total RNA was isolated
with NucleoSpin RNA II kit according to the manufacturer's
instructions. Procedures for cDNA synthesis, labeling and
hybridization were carried out according to the manufacturer's
protocol. Sequences of oligo genes were obtained from database of
human genome oligo (Operon). The arrays were hybridized, washed and
scanned according to the standard protocol. The gene chips were
scanned with a luxScan 10KA (CapitalBio Corp.). Data analysis was
performed using GenePix Pro 4.0 software (Axon Instruments). After
background correction, we performed normalization for each array
and gene. Gene activity was considered to differ between HeLa cells
and wild-type BLCAP or siRNA plasmid-transfected HeLa cells
(P<0.01) when compared by the unpaired Student's t-test using
multiple testing correction. Classification of differentially
expressed genes was also analyzed. All assays were performed in 2–4
independent experiments run in triplicates.
Western blotting
Approximately 1×105 cells were harvested
and washed with PBS buffer, and lysed using RIPA buffer in the
presence of protease inhibitor according to the manufacturer's
protocols. BCA protein assay was used to measure the protein
concentration of the lysates. Equivalent amounts of protein were
resolved and boiled in loading buffer. Then, total proteins were
fractionated by SDS-PAGE and electrophoretically transferred onto
PVDF (polyvinylidene difluoride) membrane for western blotting.
Subsequently, the membranes were blocked with 5% non-fat milk and
then incubated with the primary antibodies at 4°C overnight,
following washed with TBST buffer, and incubated again with an
appropriate HRP-conjugated secondary antibody at room temperature
for 1 h. Quantification of blotting was done using
chemiluminescence detection. The detected bands were quantitated
with laser densitometry.
Co-immunoprecipitation
Immunoprecipitations from extracts of HeLa cells
were performed according to Mehta and Ticku (23). First, recombinant
pEF-3xFlag/BLCAP plasmid containing full length BLCAP
gene and three copies of Flag tag was constructed to express
BLCAP-Flag fusion protein. The Flag protein in fusion protein is a
tag to detect the BLCAP protein in immunoprecipitation reaction due
to the lack of special antibody of BLCAP protein. After transient
transfection with Lipofectamine 2000 reagent (Invitrogen), HeLa
cells were incubated with ice-cold lysis buffer for 30 min and
homogenized with a Pyrex glass homogenizer. The cell lysate were
centrifuged at 10,000 × g for 10 min at 4°C. The super-natants were
incubated with 50% protein A/g plus agarose (Santa Cruz
Biotechnology) at 4°C for 2 h. Anti-Flag antibody (Santa Cruz
Biotechnology) was added to the reaction mixture and incubated at
4°C for 4 h. Then the mixture was incubated with 50% protein A/g
plus agarose on a rocking platform at 4°C overnight. Subsequently,
protein A/g agarose complexes were collected by centrifugation at
1000 × g for 3 min, washed three times with ice-cold
phosphate-buffered saline, eluted with 2X SDS sample buffer, and
finally separated by SDS-polyacrylamide gel electrophoresis and
subjected to immunoblot analysis.
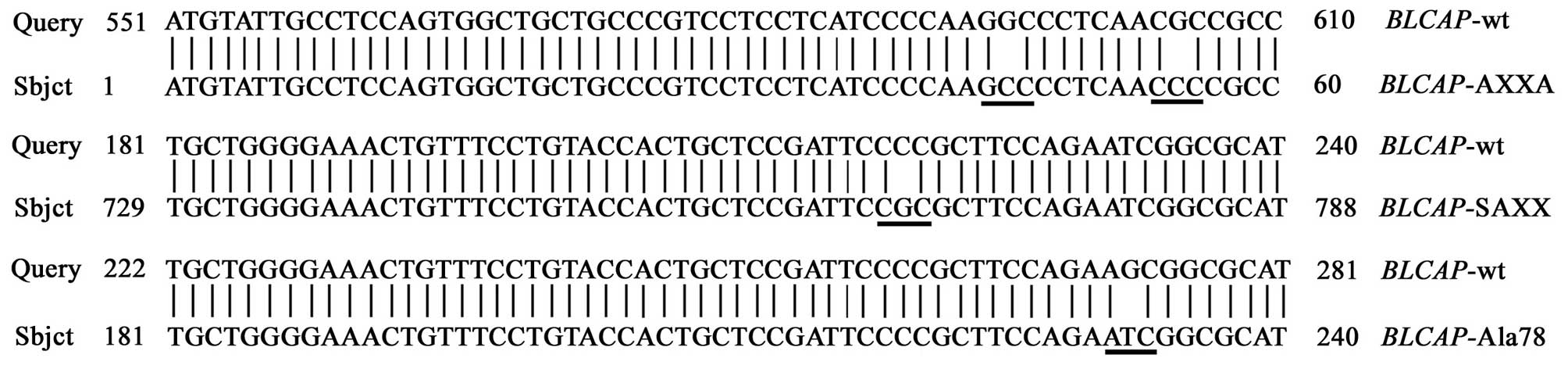
Site-specific mutagenesis
Site-specific mutagenesis has been extensively used
to study gene function. Three highly conserved amino acid positions
which may potential interact with other proteins or genes were
singled out to identify the importance of amino acid residues.
Wild-type and three mutation types of BLCAP gene were
subcloned into PCI-neo eukaryotic expression vector, respectively.
Each recombination plasmid was confirmed by PCR, digestion analysis
of restriction endonuclease and DNA sequencing.
According to the results of bioinformatics analysis
of BLCAP protein, the mutation of proline of PXXP and SPXX motifs
into Alanine, and the mutation of Serine78 into Alanine78 were
performed with the PCR-based DpnI-treatment. The primer sequences
of the BLCAP gene were designed with Primer 5.0 software,
and the coding region of wild-type BLCAP gene was amplified
by PCR using pfu high fidelity polymerase. The forward primer:
5′-CTAGTCTA GATTAGGTGCCCACAACGC-3′, and reverse primer:
5′-GCAGAATTCATGTATTGCCTCCAGTG-3′ were generated to amplify the
wild-type BLCAP gene. Three mutation type BLCAP genes
were amplified using site-specific mutagenesis PCR, the primers
were as follows. For AXXA gene type, forward primer:
5′-ACAGGGCGGCGTTGAGGGCC TTGGGGATG-3′, reverse primer:
5′-CATCCCCAAGGC CCTCAACgCCgCCCTgTg-3′; For SAXX gene type, forward
primer: 5′-GCTCCGATTCCGCGCTTCCAGAA-3′, reverse primer:
5′-GATTCTGGAAGCGCGGAATCGG AgC-3′; For Ala gene type, forward
primer: 5′-CgCTT CCAGAAGCGGCGCATGATCC-3′, reverse primer:
5′-gATCATgCgCCggTTCTggAAgC-3′. Finally, recombination plasmids were
confirmed by PCR, digestion analysis of restriction endonuclease
and DNA sequencing.
Statistical analysis
Statistical analyses were performed using SPSS 16.0
software. Student t-test was used for the comparison between two
samples. The results were considered statistically significant at
P<0.05.
Results
BLCAP might increase apoptosis in HeLa
cells
To investigate the potential role of BLCAP
gene as an inhibitor of cell proliferation, we assessed effects of
wild-type BLCAP gene and its siRNA in HeLa cells by use of a
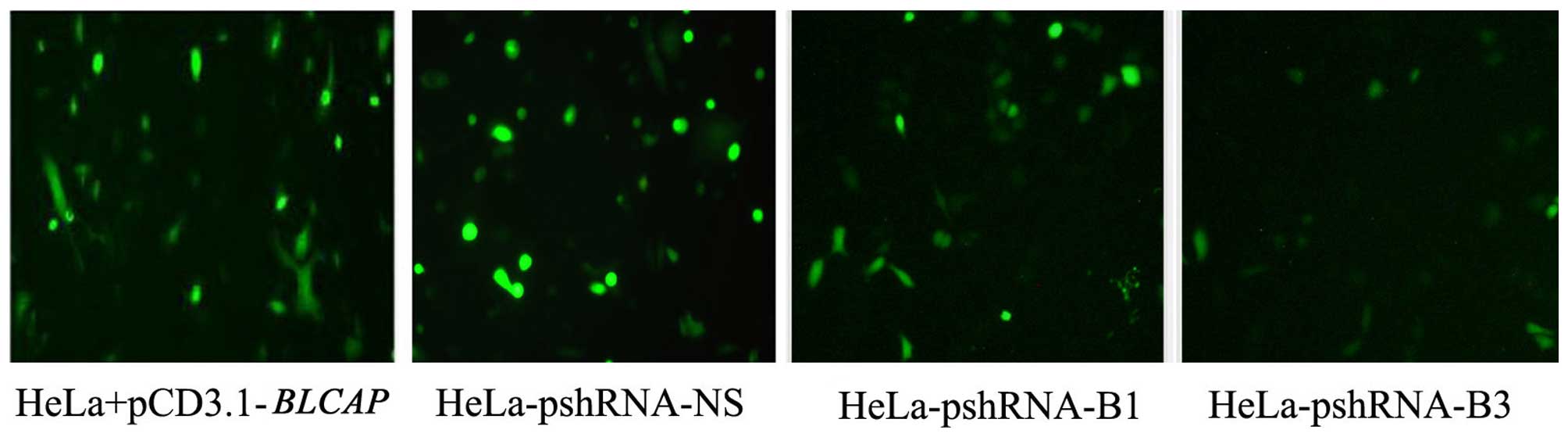
fluorescence microscope and flow cytometry. The shRNAs of
BLCAP gene (pshRNA-B1, pshRNA-B2, and pshRNA-B3) and
pshRNA-NS (a negative control shRNA) was transfected into the HeLa
cells. The efficiency of silencing and expression level of BLCAP
protein was measured with expression of green fluorescent proteins.
We found shRNAs of BLCAP did decrease the levels of BLCAP
protein, and pshRNA-B3 had the best effect in silencing (Fig. 1).
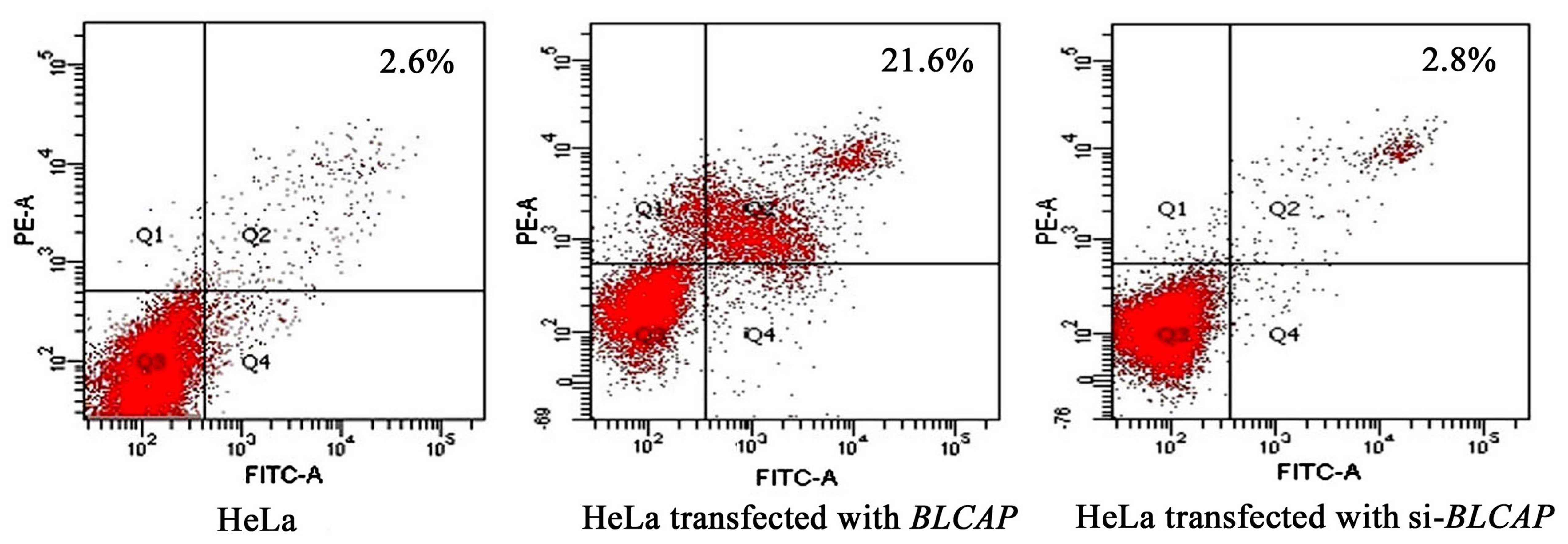
To elucidate the mechanism of induced apoptosis by
BLCAP, we assessed the effect of BLCAP knockdown on the cellular
apoptosis with flow cytometry. The results showed that the
apoptosis rate was increased to 21.6% after transfection with the
wild-type BLCAP gene while in control HeLa cells it was only
2.6%. When the expression of BLCAP was silenced by
pshRNA-B3, the apoptosis rate of cells returned to 2.8%. This
result suggested that BLCAP can increase apoptosis in HeLa cells
(Fig. 2).
BLCAP regulates the expression of
genes
In order to detect whether BLCAP protein may
modulate signaling pathways in HeLa cells, we subjected the
differently treated HeLa cells (HeLa-BLCAPwt,
HeLa-BLCAPwt-siRNA-B3) to gene expression analyses using
human signal pathway gene oligo chips. A comparison of the
expression profiles in the BLCAPwt treatment vs. control
(HeLa without treatment) revealed 46 genes which were significantly
up- or down-regulated with a mean change ≥2-fold. A total of 11
genes were significantly downregulated in HeLa-BLCAPwt cell
lines, while remaining 35 genes were upregulated. Furthermore, the
blcapwt treatment vs. HeLa-BLCAP-siRNA revealed that there
were 61 genes which were significantly up- or down-regulated. Ten
genes were significantly downregulated while 51 genes were
upregulated in HeLa-BLCAP-siRNA cells.
BLCAP regulates G1 to S phase of the cell
cycle
From gene expression analyses using human signal
pathway gene Oligo chips, we found 30 differential expression genes
in HeLa cells (Table II), and at
least half of them played important roles in the cell cycle, growth
or proliferation. Among them at least 7 genes were related with G1
to S phase of the cell cycle, followed by cell signal pathway,
protein synthesis or transcription factors (Table III).
 | Table IIThe characteristics of 30
differential expression genes by go analysis. |
Table II
The characteristics of 30
differential expression genes by go analysis.
| Acession code | Unigene. ID | Gene symbol | Gene name | FC | Dir | CC | Cg | CS | CF |
|---|
| NM_006609 | Hs.28827 | MEKK2 | Mitogen-activated
protein kinase kinase 2 | 4.7291 | ↑ | 1 | 0 | 1 | 0 |
| NM_001786 | Hs.334562 | cdk1
(cdc2) | Cell division cycle
2, G1 to S and G2 to M | 4.3154 | ↑ | 1 | 0 | 0 | 0 |
| NM_005721 | Hs.380096 | ARP3 | ARP3 actin-related
protein 3 homolog (yeast) | 3.9601 | ↑ | 0 | 0 | 1 | 1 |
| NM_002734 | Hs.183037 | PRKAR1A | Protein kinase,
cAMP-dependent, regulatory, type I | 3.7921 | ↑ | 1 | 0 | 1 | 0 |
| NM_005722 | Hs.393201 | ARP2 | ARP2 actin-related
protein 2 homolog (yeast) | 3.4129 | ↑ | 0 | 0 | 1 | 1 |
| NM_003670 | Hs.171825 | l-Dec | Basic
helix-loop-helix domain containing, class B, 2 | 3.3637 | ↑ | 0 | 0 | 0 | 1 |
| NM_000321 | Hs.75770 | Rb1 | Retinoblastoma 1
(including osteosarcoma) | 3.0727 | ↑ | 1 | 0 | 0 | 0 |
| NM_003816 | Hs.2442 | ADAM9 | A disintegrin and
metalloproteinase domain 9 | 3.0339 | ↑ | 0 | 0 | 1 | 0 |
| NM_001892 | Hs.283738 | CSNK1A1 | Casein kinase 1,
α1 | 2.6738 | ↑ | 0 | 1 | 1 | 0 |
| NM_004156 | Hs.80350 | PPP2CB | Protein phosphatase
2, β isoform | 2.6287 | ↑ | 0 | 1 | 1 | 0 |
| NM_001752 | Hs.76359 |
Catalase | Catalase | 2.6148 | ↑ | 0 | 0 | 0 | 1 |
| NM_031966 | Hs.23960 | cyclin
B1 | Cyclin B1 | 2.5594 | ↑ | 1 | 1 | 0 | 0 |
| NM_005655 | Hs.82173 | TIEG | TgF-β inducible
early growth response | 2.4786 | ↑ | 0 | 1 | 0 | 1 |
| NM_016026 | Hs.179817 | ARSDR1 | CgI-82 protein | 2.4469 | ↑ | 0 | 1 | 0 | 0 |
| NM_002592 | Hs.78996 | PCNA | Proliferating cell
nuclear antigen | 2.3775 | ↑ | 1 | 1 | 0 | 0 |
| AF059531 | Hs.152337 | PRMT3 | Protein arginine
N-methyltransferase 3-like 3 | 2.3432 | ↑ | 0 | 0 | 0 | 1 |
| AF073310 | Hs.143648 | IRS2 | Insulin receptor
substrate 2 | 2.3229 | ↑ | 0 | 1 | 1 | 0 |
| NM_005359 | Hs.75862 | DPC4 | MAD, mothers
against decapentaplegic homolog 4 | 2.2822 | ↑ | 1 | 0 | 1 | 0 |
| NM_005171 | Hs.36908 |
ATF-1/TREB | Activating
transcription factor 1 | 2.2552 | ↑ | 0 | 0 | 0 | 1 |
| AB036063 | Hs.94262 | p53R2 | Ribonucleotide
reductase M2 B (TP53 inducible) | 2.1887 | ↑ | 0 | 0 | 0 | 1 |
| NM_005917 | Hs.75375 | MDH1 | Malate
dehydrogenase 1, NAD (soluble) | 2.1807 | ↑ | 0 | 0 | 0 | 1 |
| NM_002884 | Hs.865 | RAP1A | RAP1A, member of
RAS oncogene family | 2.1792 | ↑ | 1 | 0 | 0 | 0 |
| NM_002227 | Hs.50651 | JAK1 | Janus kinase 1 (a
protein tyrosine kinase) | 2.0847 | ↑ | 0 | 0 | 1 | 0 |
| NM_004329 | Hs.2534 | ALK-3 | Bone morphogenetic
protein receptor, type IA | 2.0835 | ↑ | 0 | 0 | 1 | 0 |
| X68560 | Hs.154295 | Sp3 | Sp3 transcription
factor | 2.0498 | ↑ | 0 | 0 | 0 | 1 |
| NM_001239 | Hs.514 | cyclin
H | Cyclin H | 2.0299 | ↑ | 1 | 0 | 0 | 0 |
| NM_002467 | Hs.79070 | c-myc | V-myc
myelocytomatosis viral oncogene homolog | 2.015 | ↑ | 1 | 1 | 0 | 0 |
| NM_022740 | Hs.236131 | HIPK2 | Homeodomain
interacting protein kinase 2 | 0.4943 | ↓ | 0 | 0 | 1 | 1 |
| S76638 | Hs.73090 | NFKB2 | Nuclear factor of
kappa light polypeptide gene | 0.4135 | ↓ | 0 | 0 | 0 | 1 |
| NM_001955 | Hs.2271 | EDN1 | Endothelin 1 | 0.2809 | ↓ | 0 | 0 | 1 | 1 |
 | Table IIISignificant signal pathway analysis
through human signal pathway gene oligo chips. |
Table III
Significant signal pathway analysis
through human signal pathway gene oligo chips.
| Pathway name | P-value | Number of genes
found in the pathway | Gene name and
accession number |
|---|
|
Hs_Cell_cycle_KEGG | 0.00316 | 7 | CCNB1
NM_031966 |
| | | CDC2
NM_001786 |
| | | SMAD4
NM_005359 |
| | | RB1
NM_000321 |
| | | PRKAR1A
NM_002734 |
| | | CCNH
NM_001239 |
| | | PCNA
NM_002592 |
|
Hs_G1_to_S_cell_cycle_Reactome | 0.02107 | 5 | CCNB1
NM_031966 |
| | | RB1
NM_000321 |
| | | CCNH
NM_001239 |
| | | MYC
NM_002467 |
| | | PCNA
NM_002592 |
Interaction of BLCAP and Rb1
proteins
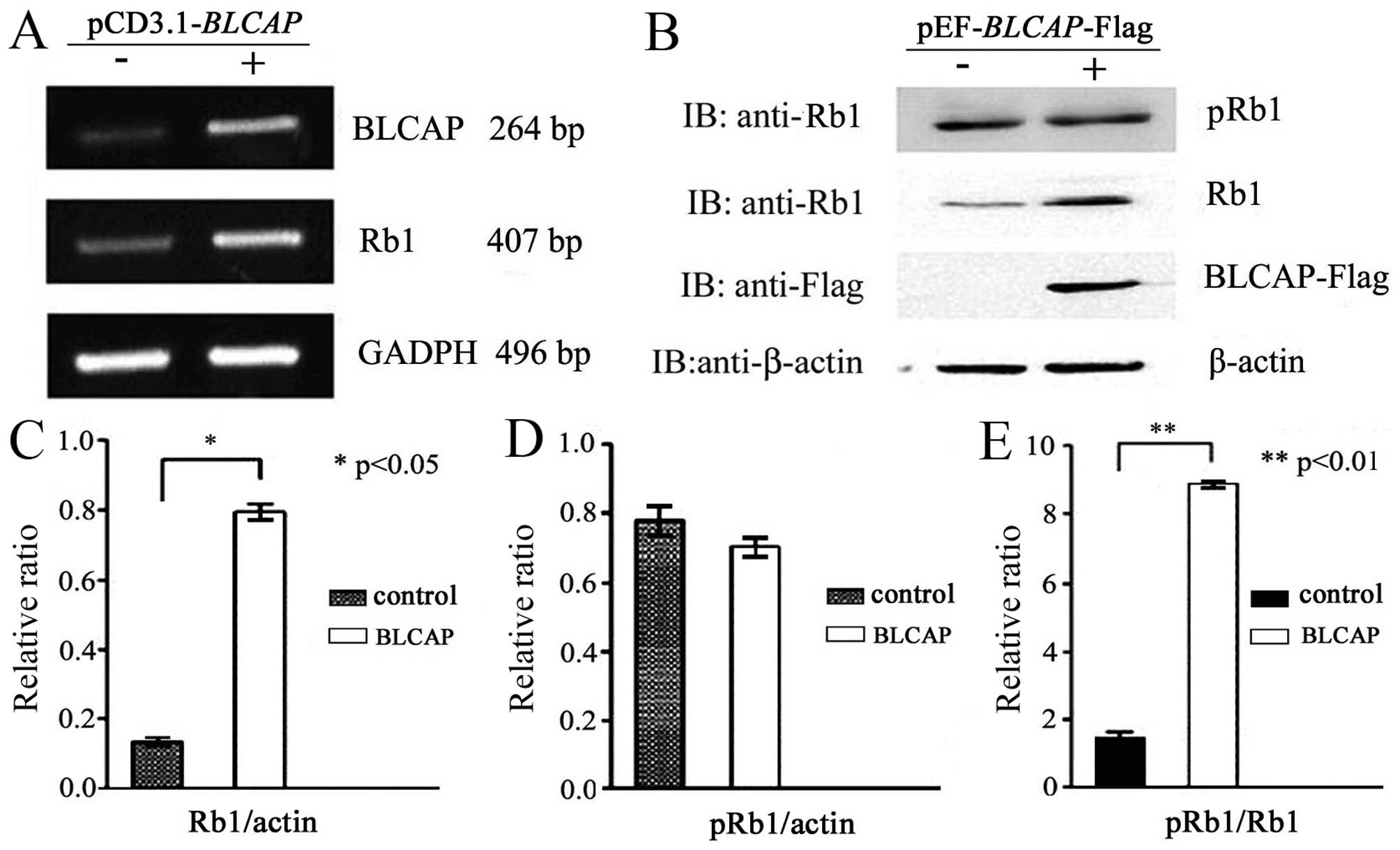
We performed RT-PCR, western blotting and Co-IP
assays to confirm the exact association between BLCAP and Rb1
proteins. Firstly, the candidate protein Rb was identified to
participate in BLCAP signal pathway with chip analysis (Table III). We prepared HeLa cell line
expressing flag-tagged BLCAP and examined the phosphorylation
status of Rb1. As shown in Fig. 3A and
B, upregulation of BLCAP proteins could specifically increase
RB1 protein expression level, but did not phosphorylate Rb1
proteins. The pRb1/Rb1 was significantly decreased (Fig. 3C, D and E). These data implied that
almost all of the Rb1 protein was phosphorylated in HeLa cells, but
only half of Rb1 protein was phosphorylated in HeLa cells
transfected with BLCAP expression plasmid. The interaction between
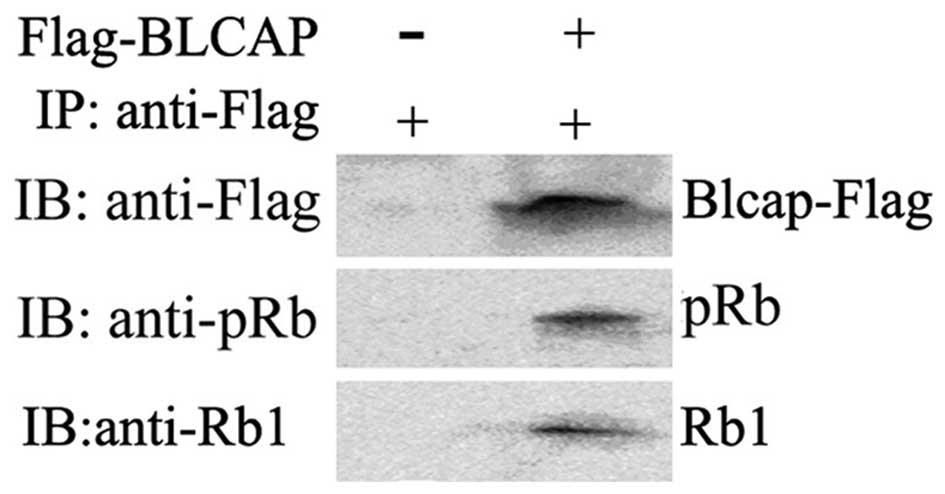
BLCAP and Rb1 or pRb1 was clearly observed with
co-immunoprecipitation analysis (Fig.
4A and B). These above results further suggested that
overexpression of BLCAP protein might interact and inhibit the
phosphorylation of Rb1 in HeLa cells.
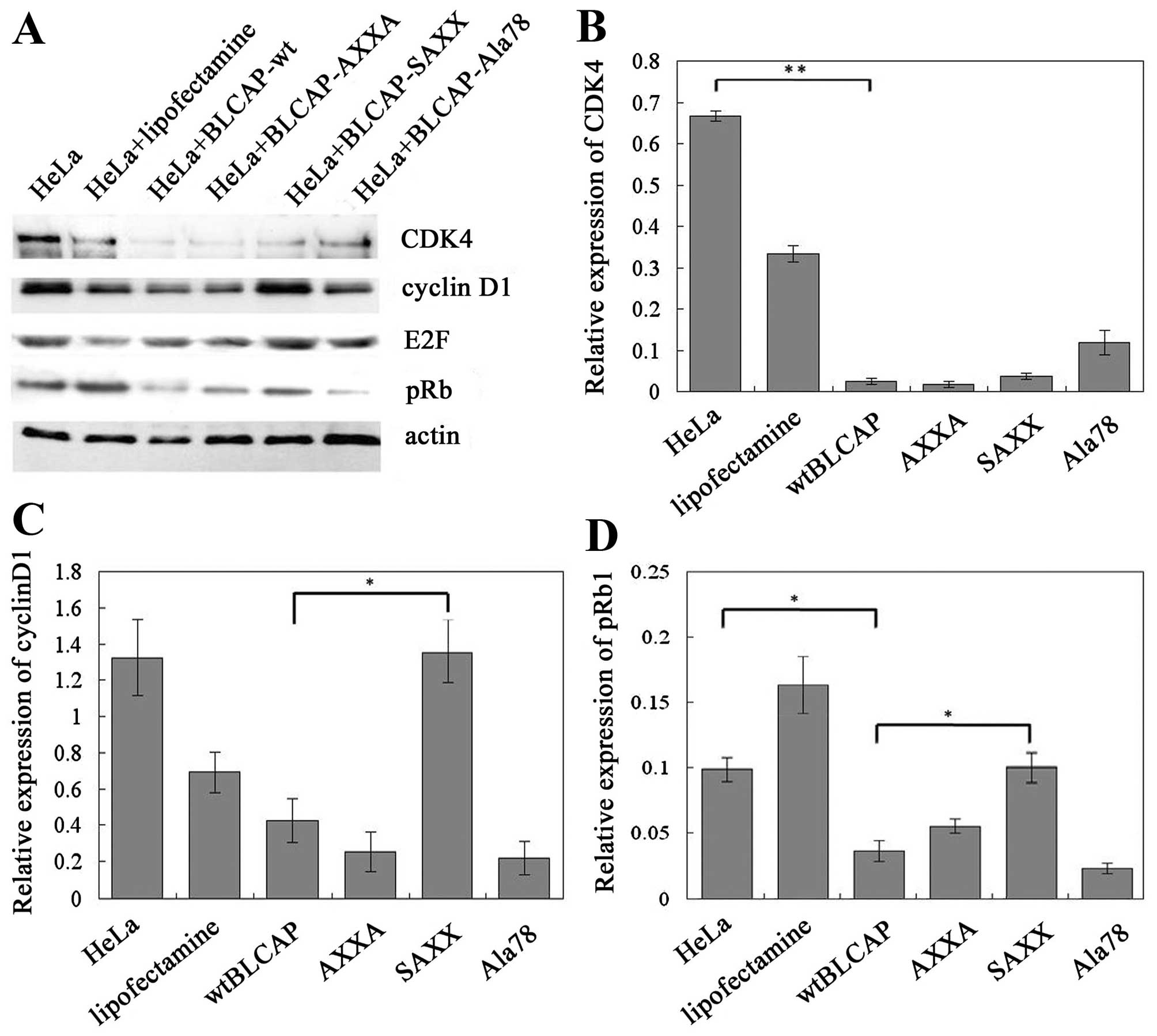
SAXX mutation of BLCAP promotes the
expression of pRb1 protein
In this study, we targeted three highly conserved
amino acid positions within the BLCAP protein that potentially
correspond to amino acids predicted to directly interact with other
proteins or genes, and combined site-specific mutagenesis to
identify amino acid residues important for BLCAP (Fig. 5). The effects of the mutants in
cells were tested by protein expression analyses for a potential
BLCAP target gene. We found that both AXXA and Ala78 mutation of
BLCAP could inhibit the expression of cell cycle G1/S regulators
such as cyclin D1 and CDK4 proteins similarly to wild-type BLCAP in
cells. Thus, AXXA and Ala78 motifs of BLCAP protein were not the
key regions in terms of BLCAP structure-function relationships.
However, SAXX mutation of BLCAP significantly suppresses the BLCAP
inhibition of expression of cyclin D1 and promotes the expression
of pRb1 proteins in HeLa cells. (Fig.
6). As mentioned above, SPXX (Ser-Pro-X-X) motif located in
many regulatory proteins could attend the regulation of gene
expression by the phospholated site in BLCAP protein. These results
provided novel information regarding the role of these residues in
BLCAP function, and how BLCAP regulates expression of genes
involved in the cell cycle and apoptosis.
Discussion
It is widely recognized that cervical carcinogenesis
is related to various genetic and epigenetic events, especially the
alterations in cell cycle checkpoint. BLCAP gene located on
chromosome 20 is regarded as a tumor-suppression gene and was
identified in human bladder carcinoma (19,24–26).
until now, the exact function and mechanisms have been obscure.
By bioinformatics analysis BLCAP was found to
contain SPXX, PXXP sequences and the phospholation site was located
at 78th amino acid. A number of studies have shown that these
domains of protein usually play a role in cell signaling pathways.
PXXP is often involved in the multi-protein interactions, such as
the activation of transcription and signal transduction. SPXX
participated in regulating BLCAP function of gene expression by the
phospholated site in BLCAP protein (21,22).
Therefore, we hypothesized that BLCAP may play an important role in
growth, reproduction, or malignant transformation of cervical cells
through signal transduction pathway. In order to confirm this
hypothesis, we used the cell signal transduction chip to analyze
the alteration of gene profiles in HeLa cells which were
transfected with wild-type BLCAP gene and siRNA targeted
BLCAP gene. BLCAP was found to exert anti-tumor activity in
cervical cancer cells. We identified at least 30 up- or
down-regulated genes that might represent potential target genes
for BLCAP in HeLa cells using microarray assay combined with GO
pathways analysis. Among the potential targets, seven genes belong
to the regulation of cell cycle, including RB1, cyclin D1,
CDC2.
Rb protein has profound effect on multiple cellular
processes and has been reported to regulate the expression of genes
involved in cell cycle progression, differentiation, development,
proliferation and apoptosis (27,28).
Rb protein molecular weight is about 110 kDa, with localization in
the nucleus. The most important structure domains of Rb protein is
the A/B pocket. A variety of protein such as viral oncogene
proteins, SV40 large T antigen, adenovirus E1A, HPV E7 protein and
cellular E2F protein was able to combine with A/B pocket of Rb
protein (29). Rb protein function
was regulated by the phosphorylation state through a cascade of
cell cycle dependent kinases, and the binding transcription factor
E2F, to determine cell entry into S-phase of the cell cycle
(30,31).
Diverse essential molecular processes within a cell
are carried out by a large number of protein components organized
by protein-protein interactions. Co-immunoprecipitation technology
is a classic method for the study of protein-protein interactions.
In this study, co-immunoprecipitation and immuno blotting were
applied to detect the interactions between BLCAP and other
biological macromolecules. We constructed the recombinant
eukaryotic expression plasmid pEF BLCAP-3xFlag as a
commercial BLCAP monoclonal antibody is not available. The Flag
protein in fusion protein is a tag to detect the BLCAP protein in
immunoprecipitation reaction and Western Blot detection after HeLa
transient transfection with plasmids. We found that BLCAP could
co-immunoprecipitate with Rb1 and pRb1 in physiological conditions.
The results were further confirmed in HeLa cells transfected with
wild-type BLCAP expression plasmid showing that the expression
levels of Rb1 were significantly up regulated, and pRb1 level was
significantly downregulated. It suggested that BLCAP can inhibit
phosphorylation of Rb1, which blocks the cells from going through
the G1/S checkpoint of the cell cycle. The cell proliferation is
inhibited and apoptosis induction follows the overexpression of
BLCAP. Our results indicate the important role that BLCAP plays in
its biological function through Rb1 pathway, and provide a novel
way for clinical treatment of malignant tumors such as cervical
cancer.
Next we designed a working model of BLCAP to mutate
the potential functional motif of BLCAP including AXXA, SPXX and
Ser78 site. The recombination plasmids were transfected into HeLa
cells to investigate the function model of BLCAP protein in
vitro. We identified that motif SPXX was a key region for the
function of BLCAP, and SPXX motif showed a significant effect on
inhibition of BLCAP function. As mentioned above, SPXX
(Ser-Pro-X-X) was the phosphorylation site in BLCAP protein. When
the SPXX (Ser-Pro-X-X) motif was mutated to SAXX (Ser-Ala-X-X) by
site-specific mutagenesis in our study, the expression of cyclin D1
and pRb1 proteins were significantly upregulated. In contrast, AXXA
mutation and Ala78 mutation of BLCAP did not show these effects in
the same assay.
Transcription factor E2F family members
played a major role in regulating cell cycle process by promoting
the timely expression of genes required for DNA synthesis at the
G1/S phase transition and their altered expression contribute to a
number of human diseases, including cancer (32). During cancer cells growth and
progression, E2F promotes tumor cell proliferation, but inhibits
apoptosis. It was clear that E2F activity was controlled by the
Rb1. Commonly Rb1 binds to E2F which represses E2F activity,
affecting the G1/S phase of the cell cycle, while Rb phosphorylated
by CDK4/cyclin D complexes, then E2F was released to activate its
target genes. Cyclin D1, CDK4 and Rb1 played important roles in the
cell cycle G1/S regulation in several human tumors (33,34).
Overexpression of cyclin D1 and CDK4 is a commonly
observed alteration in tumors. Some reports suggest that the
overexpression of cyclin D1 may serve as a driving force through
its cell cycle regulating function (35). Our results indicated that BLCAP
could induce overexpression of pRb1 and promote CDK4/cyclin D
activity, resulting in increased Rb phosphorylation and thus E2F
accumulation, eliciting its potential tumor-suppression effects.
Based on our studies, we concluded that wild-type BLCAP protein
plays its biological function through regulating the expression of
cyclin D1, CDK4 and pRb1 proteins. In cervical cancer, the role of
cyclin D1 and CDK4 in cervical carcinogenesis was not clearly
understood and controversial results have been described. The
inactivation of the E2F repressor resulting in increased E2F
activity was a key step for cervical carcinogenesis (9). Our work provided detailed information
regarding the role of BLCAP, and novel insight into how BLCAP
regulates gene expression involving the cell cycle and apoptosis in
Hela cells. Further, our studies suggested that BLCAP might be a
new Rb1 activator or a potential E2F repressor. BLCAP is
suggested as a prospective biomarker and a possible new therapeutic
target of cervical cancer (36).
Our results showed that knock down of BLCAP
by the use of siRNA or mutagenesis was useful to inhibit the cell
growth and induce apoptosis of HeLa cells in vitro. This
study strongly suggested that interaction between BLCAP and Rb1 was
a frequent event in HeLa cells leading to cell growth inhibition
and apoptosis induction. Therefore, BLCAP played an important role
in the pathogenesis of cervical cancer, which might be due to the
regulation of Rb expression.
Acknowledgments
We thank Professor Guo Deyin who kindly provided
pEF-CARD-3x Flag eukaryotic expression plasmid. This work was
supported by grants provided by the National Natural Science
Foundation of China (nos. 81072123 and 30571955).
References
|
1
|
Forouzanfar MH, Foreman KJ, Delossantos
AM, Lozano R, Lopez AD, Murray CJ and Naghavi M: Breast and
cervical cancer in 187 countries between 1980 and 2010: A
systematic analysis. Lancet. 378:1461–1484. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arbyn M, Castellsagué X, de Sanjosé S,
Bruni L, Saraiya M, Bray F and Ferlay J: Worldwide burden of
cervical cancer in 2008. Ann Oncol. 22:2675–2686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
zur Hausen H: Papillomaviruses in the
causation of human cancers - a brief historical account. Virology.
384:260–265. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johansson H, Bjelkenkrantz K, Darlin L,
Dilllner J and Forslund O: Presence of high-risk HPV mRNA in
relation to future high-grade lesions among high-risk HPV DNA
positive women with minor cytological abnormalities. PLoS One.
10:e01244602015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Naucler P, Ryd W, Törnberg S, Strand A,
Wadell G, Elfgren K, Rådberg T, Strander B, Johansson B, Forslund
O, et al: Human papillomavirus and Papanicolaou tests to screen for
cervical cancer. N Engl J Med. 357:1589–1597. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Galloway DA: Human papillomaviruses: A
growing field. Genes Dev. 23:138–142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Woodman CB, Collins SI and Young LS: The
natural history of cervical HPV infection: unresolved issues. Nat
Rev Cancer. 7:11–22. 2007. View
Article : Google Scholar
|
|
8
|
Lee HS, Yun JH, Jung J, Yang Y, Kim BJ,
Lee SJ, Yoon JH, Moon Y, Kim JM and Kwon YI: Identification of
differentially-expressed genes by DNA methylation in cervical
cancer. Oncol Lett. 9:1691–1698. 2015.PubMed/NCBI
|
|
9
|
Jensen KE, Schmiedel S, Frederiksen K,
Norrild B, Iftner T and Kjær SK: Risk for cervical intraepithelial
neoplasia grade 3 or worse in relation to smoking among women with
persistent human papillomavirus infection. Cancer Epidemiol
Biomarkers Prev. 21:1949–1955. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamashita Y, Hasegawa M, Deng Z, Maeda H,
Kondo S, Kyuna A, Matayoshi S, Agena S, Uehara T, Kouzaki H, et al:
Human papillomavirus infection and immunohistochemical expression
of cell cycle proteins pRb, p53, and p16(INK4a) in sinonasal
diseases. Infect Agent Cancer. 10:232015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Esashi F, Christ N, Gannon J, Liu Y, Hunt
T, Jasin M and West SC: CDK-dependent phosphorylation of BRCA2 as a
regulatory mechanism for recombinational repair. Nature.
434:598–604. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Plotnikova OV, Golemis EA and Pugacheva
EN: Cell cycle-dependent ciliogenesis and cancer. Cancer Res.
68:2058–2061. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Niculescu AB III, Chen X, Smeets M, Hengst
L, Prives C and Reed SI: Effects of p21(Cip1/Waf1) at both the g1/S
and the G2/M cell cycle transitions: pRb is a critical determinant
in blocking DNA replication and in preventing endoreduplication.
Mol Cell Biol. 18:629–643. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gromova I, Gromov P and Celis JE: bc10: A
novel human bladder cancer-associated protein with a conserved
genomic structure downregulated in invasive cancer. Int J Cancer.
98:539–546. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rae FK, Stephenson SA, Nicol DL and
Clements JA: Novel association of a diverse range of genes with
renal cell carcinoma as identified by differential display. Int J
Cancer. 88:726–732. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Evans HK, Weidman JR, Cowley DO and Jirtle
RL: Comparative phylogenetic analysis of BLCAP/NNAT reveals
eutherian-specific imprinted gene. Mol Biol Evol. 22:1740–1748.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zuo Z, Zhao M, Liu J, Gao G and Wu X:
Functional analysis of bladder cancer-related protein gene: A
putative cervical cancer tumor suppressor gene in cervical
carcinoma. Tumour Biol. 27:221–226. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yao J, Duan L, Fan M, Yuan J and Wu X:
Overexpression of BLCAP induces S phase arrest and apoptosis
independent of p53 and NF-kappaB in human tongue carcinoma : BLCAP
overexpression induces S phase arrest and apoptosis. Mol Cell
Biochem. 297:81–92. 2007. View Article : Google Scholar
|
|
19
|
Moreira JM, Ohlsson G, Gromov P, Simon R,
Sauter G, Celis JE and Gromova I: Bladder cancer-associated
protein, a potential prognostic biomarker in human bladder cancer.
Mol Cell Proteomics. 9:161–177. 2010. View Article : Google Scholar :
|
|
20
|
Huret JL, Ahmad M, Arsaban M, Bernheim A,
Cigna J, Desangles F, Guignard JC, Jacquemot-Perbal MC, Labarussias
M, Leberre V, et al: Atlas of genetics and cytogenetics in oncology
and haematology in 2013. Nucleic Acids Res. 41(D1): D920–D924.
2013. View Article : Google Scholar :
|
|
21
|
Suzuki M and Yagi N: Structure of the SPXX
motif. Proc Biol Sci. 246:231–235. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Suzuki M: SPXX, a frequent sequence motif
in gene regulatory proteins. J Mol Biol. 207:61–84. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mehta AK and Ticku MK: Prevalence of the
GABAA receptor assemblies containing alpha1-subunit in the rat
cerebellum and cerebral cortex as determined by
immunoprecipitation: Lack of modulation by chronic ethanol
administration. Brain Res Mol Brain Res. 67:194–199. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gromova I, Gromov P and Celis JE:
Identification of true differentially expressed mRNAs in a pair of
human bladder transitional cell carcinomas using an improved
differential display procedure. Electrophoresis. 20:241–248. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng M, Xie T, Yu J, Xu B, Song Q and Wu
X: Bladder cancer-associated protein is suppressed in human
cervical tumors. Exp Ther Med. 3:336–340. 2012.PubMed/NCBI
|
|
26
|
Galeano F, Leroy A, Rossetti C, Gromova I,
Gautier P, Keegan LP, Massimi L, Di Rocco C, O'Connell MA and Gallo
A: Human BLCAP transcript: New editing events in normal and
cancerous tissues. Int J Cancer. 127:127–137. 2010. View Article : Google Scholar :
|
|
27
|
McCormick TM, Canedo NH, Furtado YL,
Silveira FA, de Lima RJ, Rosman AD, Almeida Filho GL and Carvalho
MG: Association between human papillomavirus and Epstein-Barr virus
DNA and gene promoter methylation of RB1 and CDH1 in the cervical
lesions: A transversal study. Diagn Pathol. 10:59–65. 2015.
View Article : Google Scholar
|
|
28
|
Srinivasan SV, Mayhew CN, Schwemberger S,
Zagorski W and Knudsen ES: RB loss promotes aberrant ploidy by
deregulating levels and activity of DNA replication factors. J Biol
Chem. 282:23867–23877. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Narisawa-Saito M and Kiyono T: Basic
mechanisms of high-risk human papillomavirus-induced
carcinogenesis: Roles of E6 and E7 proteins. Cancer Sci.
98:1505–1511. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ren B, Cam H, Takahashi Y, Volkert T,
Terragni J, Young RA and Dynlacht BD: E2F integrates cell cycle
progression with DNA repair, replication, and G(2)/M checkpoints.
Genes Dev. 16:245–256. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ishida S, Huang E, Zuzan H, Spang R, Leone
G, West M and Nevins JR: Role for E2F in control of both DNA
replication and mitotic functions as revealed from DNA microarray
analysis. Mol Cell Biol. 21:4684–4699. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bertoli C, Skotheim JM and de Bruin RA:
Control of cell cycle transcription during G1 and S phases. Nat Rev
Mol Cell Biol. 14:518–528. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
McLaughlin-Drubin ME and Münger K: The
human papillo-mavirus E7 oncoprotein. Virology. 384:335–344. 2009.
View Article : Google Scholar :
|
|
34
|
Moody CA and Laimins LA: Human
papillomavirus oncoproteins: Pathways to transformation. Nat Rev
Cancer. 10:550–560. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu T, Liu Y, Bao X, Tian J, Liu Y and
Yang X: Overexpression of TROP2 predicts poor prognosis of patients
with cervical cancer and promotes the proliferation and invasion of
cervical cancer cells by regulating ERK signaling pathway. PLoS
One. 8:e758642013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gromova I, Gromov P, Kroman N, Wielenga
VT, Simon R, Sauter G and Moreira JM: Immunoexpression analysis and
prognostic value of BLCAP in breast cancer. PLoS One. 7:e459672012.
View Article : Google Scholar : PubMed/NCBI
|