Introduction
Cancer development results from the interaction
between genetic factors, environment and dietary factors (1). Breast cancer accounts for 522,000
deaths and is the most frequently diagnosed cancer among women,
with 1.7 million cases worldwide in 2012 (2). Even though new strategies for breast
cancer therapy are being developed, unfortunately cancer frequently
recurs in patients after first-line treatment. Tumor progression is
commonly associated with disorders in the regulation of the cell
cycle. The Aurora kinase A (AURKA) and Polo-like kinases 1 (PLK1)
proteins play key roles in cell cycle phase transition as they
co-regulate multiple processes such as mitotic spindle formation,
sister chromatid resolution, chromosome spindle connections and
cytokinesis. Moreover, AURKA and breast cancer 1 susceptibility
(BRCA1) proteins may interplay to control the cell cycle,
chromosome polyploidy, and tumorigenesis. The silencing of AURKA
was found to suppress cell cycle progression mainly by blocking
G1-S and G2-M transition, while the disruption of BRCA1 was found
to promote cell cycle progression through induction of G1-S and
G2-M transition (3). On the other
hand, targeted inhibition of PLK1 can produce antitumor effects,
while its constitutive expression in mammalian cells leads to
malignant transformation suggesting that aberrant PLK1 expression
is oncogenic (4). Because of its
important role in cancer, diverse pharmacologic inhibitors have
been developed for AURKA (MLN8054, ENMD-2076, Hesperidin and VX680)
and PLK1 (BI2536, GSK-461364, Poloxin, CYC-800 and HMN-241)
proteins (5). However, resistance
to these developed inhibitory molecules is frequently observed in
clinical practice; thus alternative therapeutic approaches are
needed to overcome resistance and improve patient survival and
outcome.
The use of phytochemicals from vegetables, fruits,
spices, teas, herbs and medicinal plants, represents one of the
most feasible means of cancer chemoprevention (6). Resveratrol
(3,5,4′-trihydroxy-trans-stilbene) is a phytoalexin found in
grapes, berries, peanuts, chocolate, red wine, herbs and plants,
and its concentration increases during environmental stress and
pathogen invasion. Notably, this natural polyphenol possesses
cancer chemopreventive and chemotherapeutic activites by inhibiting
diverse cellular events associated with tumor development (7). In fact, resveratrol represents a
promising class of anticancer drug, since it acts by targeting
multiple proteins in cancer cells having limited toxic effects on
normal cells. It also has a potential antitumor effect against the
progression of various types of cancer such as prostate, breast,
liver, skin and lung cancer (8).
Resveratrol exhibits antitumor activity in breast cancer through
suppression of migration, invasion and cell cycle progression. It
also activates apoptosis leading to chemosensitization of tumor
cells in vitro (9).
Importantly, resveratrol triggers numerous intracellular pathways
leading to cell growth and cell cycle arrest (10). In this study, we evaluated the
changes in gene expression profiles induced by resveratrol
treatment of MDA-MB-231 breast cancer cells to identify modulated
genes that may represent potential therapeutic targets.
Interestingly, we found that resveratrol regulates cell cycle
progression by targeting AURKA, PLK1 and BRCA1. Our data highlight
the potential use of resveratrol in combination with antineoplastic
drugs as adjuvant therapy in breast cancer.
Materials and methods
Cell cultures
MCF-7 and MDA-MB-231 breast cancer cell lines were
obtained from the American Type Culture Collection (ATCC; Manassas,
VA, USA). Cells were maintained in Dulbecco's modified Eagle's
minimal essential medium (DMEM), supplemented with 10% fetal bovine
serum and antibiotics (100 U/ml penicillin and 100 U/ml
streptomycin) at 37°C in a humidified atmosphere of 5%
CO2.
Reagents
Resveratrol was purchased from Sigma-Aldrich (St.
Louis, MO, USA), and dissolved at a concentration of 80 mmol/l in
ethanol, stored at −20°C and diluted with DMEM to a 100-µM
working concentration.
DNA microarrays and validation
Total RNA was processed and hybridized into the
GeneChip Human Gene 1.0 ST (Affymetrix Inc., Santa Clara, CA, USA),
following the manufacturer's recommendations. Microarray quality
assessment, condensing of the probe sets, data normalization, and
filtering were conducted using the Transcriptome Analysis Console
version 2.0 (Affymetrix). To identify those biological processes
that show differentially expressed genes, we used KEGG and the
Database for Annotation, Visualization and Integrated Discovery
(DAVID) v6.7 for functional annotation. To validate the microarray
data, real-time quantitative PCR (RT-qPCR) was performed for 13
selected genes using specific primers (Table I). The SYBR-Green reaction was
carried out using a QuantiTect SYBR Green PCR reagents kit (Qiagen)
following the manufacturer's recommendations. GADPH expression was
used as control. Samples were tested in triplicate and data were
analyzed using the 2−∆∆Ct method (11).
 | Table IPrimer sequences for quantitative
reverse-transcription polymerase chain reaction. |
Table I
Primer sequences for quantitative
reverse-transcription polymerase chain reaction.
| Gene | Forward primer
5′-3′ | Reverse primer
5′-3′ | Reference
sequence |
|---|
| AURKA |
CGTGTTCTCGTGACTCAGCA |
TGGTTGCCTGCAATTGCTTC | NM_003600 |
| FUT11 |
CAACGGCTTCGAGTGTTTCG |
CTCTTTCCAACTGTCATTCTCAGG | NM_173540 |
| HK2 |
TCCAACCTTAGGCTTGCCATT |
CTTGGACATGGGATGGGGTG | NM_000189 |
| UBASH3B |
AAATTCCCGGACTGCTAGGC |
CTCTGACGCTTCCTGGTGAG | NM_032873 |
| CPA4 |
CCTGCAGGCCCTTTTAGACA |
CGGCCGGTTTTCAAACGAAT | NM_016352 |
| DSCC1 |
CAAGCTGTGCTGTGCAGTAA |
TGGGTCTACGTCTTCTTAATTCCC | NM_024094 |
| NEIL3 |
TGGAAGTGCAGCTCACCAAA |
AGCACATCACCTAGCATCCG | NM_018248 |
| PEG10 |
GAGAACAGCGGAGAAGGTCC |
TCAAAACCCGCTTATTTCGCA | NM_001172437 |
| DDR2 |
CCTACAAGTTGCCTGGGGTT |
AGGCTACAGTCTCCCTCTGG | NM_001014796 |
| MSH2 |
GGAGGTGAGGAGGTTTCGAC |
GTATAGAAGTCGCCCCGGTC | NM_000251 |
| UGT2B11 |
TGCCAAGATCCCACAAAAGGT |
TGGAATGCCCACCATAGGGA | NM_001073 |
| CYP1B1 |
AGTTCTTGAGGCACTGCGAA |
GTGATAGTGGCCGGTACGTT | NM_000104 |
| BRCA1 |
ACGGAAAAGCGCGGGAATTA |
TCTTCAACGCGAAGAGCAGA | NM_007294 |
| GAPDH |
CCCCACCACACTGAATCTCC |
GTACATGACAAGGTGCGGCT | NM_001289746.1 |
Cell cycle analysis
MDA-MB-231 and MCF-7 cells were plated at a density
of 5×104 cells/ml for 24 h, and then treated with
resveratrol (100 µM) for 24 and 48 h. Cells were harvested
by trypsinization and fixed in ice-cold 70% methanol overnight at
−20°C. Cells were centrifuged at 1,200 rpm for 5 min and incubated
with a propidium iodide (PI) working solution (100 µg/ml PI
and 100 µg/ml RNase A) for 30 min at 37°C. Cell cycle
distribution was analyzed using a FACScan flow cytometer
(FACSCalibur; Becton-Dickinson).
Western blotting
Protein extracts (50 µg) were separated by
12% SDS-PAGE and electrotransferred to a nitrocellulose membrane
(Bio-Rad). Membranes were blocked with 5% nonfat milk in PBS with
0.1% Tween-20 and then probed with primary antibodies as follows:
anti-AURKA (1:1,000; Cell Signaling Technology), anti-PLK1 (1:500),
anti-cyclin D1 (1:1,000), anti-cyclin B1 (1:1,000) and anti-BRCA1
(1:1,000) (all from Santa Cruz Biotechnology), followed by
secondary antibodies, anti-rabbit (1:2,500) or anti-mouse (1:2,500)
(both from Santa Cruz Biotechnology). The membranes were probed
with anti-α-tubulin (1:1,000; Sigma-Aldrich) or actin (1:1,000;
Abcam) as a loading control. Immunoreactive bands were developed
using the ECL chemiluminescence system (Amersham Pharmacia
Biotech).
Statistical analysis
A two-way ANOVA was performed to identify
differentially expressed genes of the microarray data. Only genes
with statistically significant differences in expression levels
(P-value <0.05) and a fold change criteria of ≥1.5 were included
in the final set of differentially expressed genes. RT-qPCR results
were analyzed using the Student's test. Differences of P<0.05
were considered statistically significant. Values are presented as
the mean ± standard deviation of three independent experiments.
Results
Resveratrol induces changes in the
transcriptome of human MDA-MB-231 breast cancer cells
Resveratrol is able to inhibit cell viability
through apoptosis activation in diverse types of cancer cells
(12,13). Here, the effects of resveratrol (100
µM) on cell viability and apoptosis of triple-negative
MDA-MB-231 breast cancer cells were evaluated. Results of the MTT,
Annexin V and TUNEL assays confirmed that resveratrol treatment was
able to significantly (P<0.05) inhibit cell viability and induce
apoptosis after 24 and 48 h treatments (data not shown). These
results concur with previous reports on diverse types of cancer
cells (14–17). Then, in order to obtain insight
concerning the cellular transcripts modulated by resveratrol that
may explain its anticancer effects, we performed a genome-wide
analysis of the transcriptome of MDA-MB-231 breast cancer cells
after treatment with resveratrol (100 µM) for 24 and 48 h
using DNA microarrays. Data from two biological replicates were
analyzed, normalized, and raw P-values adjusted. Only genes with a
significant fold change (FC>1.5; P<0.05) were included in
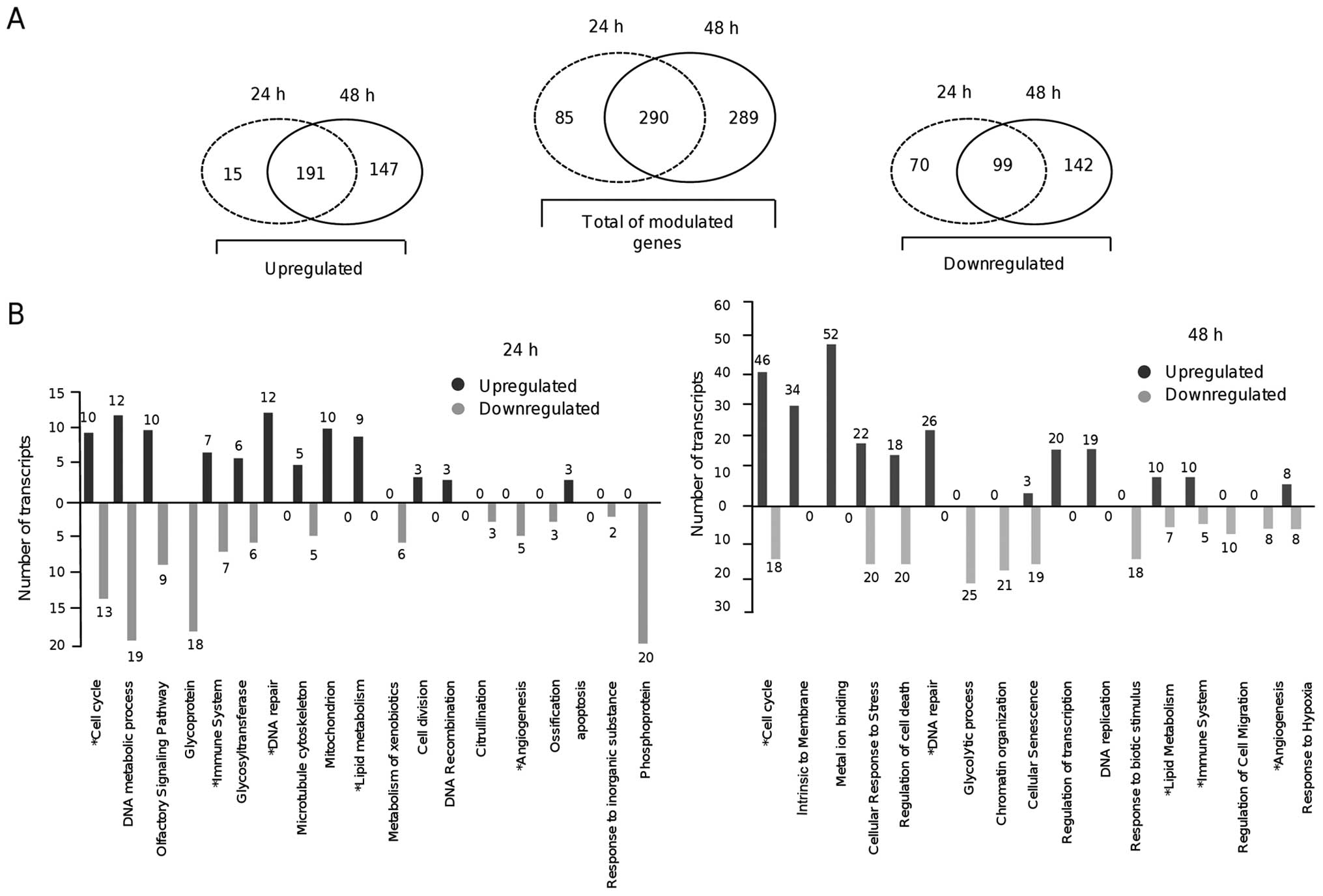
this analysis. Transcriptional profiling showed that 375 genes (206
upregulated and 169 downregulated) were significantly modulated at
24 h (Table II). After a 48-h
treatment with resveratrol, changes in the expression of 579 genes
were detected (338 upregulated and 241 downregulated). Of these,
290 genes (30%) were modulated in common at 24 and 48 h (Fig. 1A).
 | Table IIDeregulated genes and cellular
processes modulated by resveratrol treatment at 24 h. |
Table II
Deregulated genes and cellular
processes modulated by resveratrol treatment at 24 h.
| Process | No. of genes | Gene name |
|---|
| Cell cycle | 22 | TXNIP,
CCNB1, FAM83D, CDCA8, HJURP,
PLK1, CENPA, DLGAP5, BUB1,
AURKA, ASPM, WEE1, CDC6, DSN1, E2F8, MIS12,
SESN3, CCNE2, CCNE1, PSMC3IP, MNS1, DSCC1 |
| DNA metabolic
process | 31 | HIST1H2AB,
H1F0, HIST1H2BB, HIST1H3J, HIST1H2AE,
HIST1H2BM, H2BFS, HIST1H2BK, HIST1H3A,
HIST1H3B, HIST1H2AH, HIST1H3C,
HIST1H3D, HIST1H2AJ, HIST1H3E,
HIST1H3F, HIST1H3G, HIST1H3H, HIST1H3I,
CLSPN, CDC6, GINS2, DNAH14, FAM175A, DTL, GINS3, RNASEH2A, MCM6,
CCNE2, FANCL, PRIM1 |
| Olfactory signaling
pathway | 19 | OR13C4,
OR5P3, OR5P2, ARHGAP11A, ARHGAP11B,
SLC6A6, SLC5A3, SLC14A1, SLC2A3,
HK2, C5, SAA2, CCL20, PRICKLE1, KITLG, DKK1, DKK1, NAPEPLD,
MAP2 |
| Glycoprotein | 18 | SLC5A3,
NOG, TNC, IGHM, OR13C4, SLC2A3,
CLEC2B, IGHA1, OR5P3, OR5P2,
IL24, IGHV3-11, EYS, GPR110,
SLC6A6, STC1, TM4SF1, SLC14A1 |
| Immune system | 14 | IGHG1,
IGHV311, IGV330, IGV348, SCFV,
IGHA1, IGHM, C5, IL18, ICAM2, TRIM68, IL18, CLU,
C5 |
|
Glycosyltransferase | 12 | ALOX5AP,
GPSM2, ARHGAP11B, ARHGAP11A, DEPDC1,
ERRFI1, UNG, AMY1C, AMY2A, AMY1B, AMY1A, GBA |
| DNA repair | 12 | BRIP1, UNG, BRCA2,
MLH1, FANCL, BRCA1, EXO1, CLSPN, BLM, ESCO2, DNA2, POLN |
| Microtubule
cytoskeleton | 10 | CCNB1,
FAM83D, KIF14, CDCA8, KIF20A, CDC6,
ROCK1P1, DNAH14, MAP2, DNAH6 |
| Mitochondrion | 10 | UNG, KIAA0101,
ASAH2, SLIT3, TIMM8A, TST, ASAH2C, ASAH2B, SLC25A19, SLC27A2 |
| Lipid
metabolism | 9 | ACADSB, NAPEPLD,
LIPK, ASAH2C, ASAH2B, SLC27A2, ASAH2, GBA, AADAC |
| Metabolism of
enobiotics by cytochrome P450 | 6 | UGT2B17,
CYP1B1, UGT2B11, UGT2B10, UGT2B15,
UGT2B7 |
| Cell division | 3 | CDC6, DSN1,
MIS12 |
| DNA
recombination | 3 | EXO1, BLM,
PSMC3IP |
| Citrullination | 3 | HIST1H2AH,
HIST1H3D, HIST1H3E |
| Angiogenesis | 5 | JUN,
VEGFA, LOX, ANGPTL4, PTGS2 |
| Ossification | 3 | NOG,
STC1, RUNX2 |
| Induction of
apoptosis by intracellular signals | 3 | TP53I3, CCNE1,
SESN3 |
| Response to
inorganic substance | 2 | TXNIP,
NDRG1 |
| Phosphoprotein | 20 | PRR11,
CDCA8, H2BFS, HIST1H2BK, ANKZF1,
NFIL3, ERRFI1, ASPM, KIF14, ND2,
TMEM71, DEPDC1, SLITRK6, PFKFB3,
GNE, TNC, LPXN, GPSM2, NDRG1,
BHLHE40 |
Overview of the modulated genes by
resveratrol in MDA-MB-231 breast cancer cells
To evaluate the impact of resveratrol on cellular
processes and pathways of MDA-MB-231 cells, a bioinformatic
analysis of the DNA microarray data set was performed. We used the
Database for Annotation, Visualization and Integrated Discovery
(DAVID) and Reactome Database for functional annotation and
visualization of the expression data in the KEGG biological pathway
context. Gene ontology category analysis indicated that a number of
genes involved in cancer-related processes were modulated by
resveratrol at both times (Fig.
1B). Of these, 47 suppressed genes are well-known oncogenes,
which highlight the potential effects of resveratrol on the
suppression of cancer-related processes. Notably, a significant
decrease in the expression of genes involved in the cell cycle, DNA
metabolic processes, cytoskeleton organization, metabolism of
xenobiotics by cytochrome P450, and angiogenesis was detected at 24
h after resveratrol treatment (Table
II). Several of these genes, including SLC6A6, HK2, GPR110,
IGHG1, ALOX5AP, CCNB1, CYP1B1, JUN, VEGFA, ANGPTL4 and histones,
are clinical therapeutic targets in several types of cancers
(18–20). Remarkably, after a 24-h resveratrol
treatment we identified the overexpression of a set of DNA repair
genes including BRIP1, UNG, BRCA2, MLH1, FANCL, BRCA1, EXO1, CLSPN,
BLM, ESCO2, DNA2, POLN (Table
II).
In contrast, at 48 h after resveratrol treatment we
detected the upregulation of genes involved in cellular response to
stress, regulation of cell death, glycolytic process, chromatin
organization and cellular senescence. Notably, DNA repair genes
involved in homologous recombinational repair (RAD54L, EXO1, RECQL,
BRCA1, BRCA2, BLM, DNA2, RBBP8, POLN), mismatch repair (MLH1,
MSH2), Fanconi anemia pathway (FANCB, FANCD2, FANCL), base excision
repair (UNG), DNA double-strand break repair (BRIP1, FAM175A) and
repair of DNA inter-strand crosslinks (EME1) were upregulated at 48
h (Table III). Moreover, several
genes involved in cell death, such as TP53I3, CCNE1 and SESN3, were
overexpressed at 48 h. Remarkably, 22 and 63 genes involved in the
regulation of cell cycle progression were modulated at 24 and 48 h,
respectively (Tables II and
III). Repressed cell cycle genes
at 24 h included AURKA, PLK1, TXNIP, CCNB1, FAM83D, CDCA8, HJURP,
CENPA, DLGAP5, BUB1, ASPM, and WEE1; whereas at 48 h of treatment,
AURKA, PLK1, JUN, ARHGAP18, KIF20A, SH3KBP, EDN1 and PSMD11 were
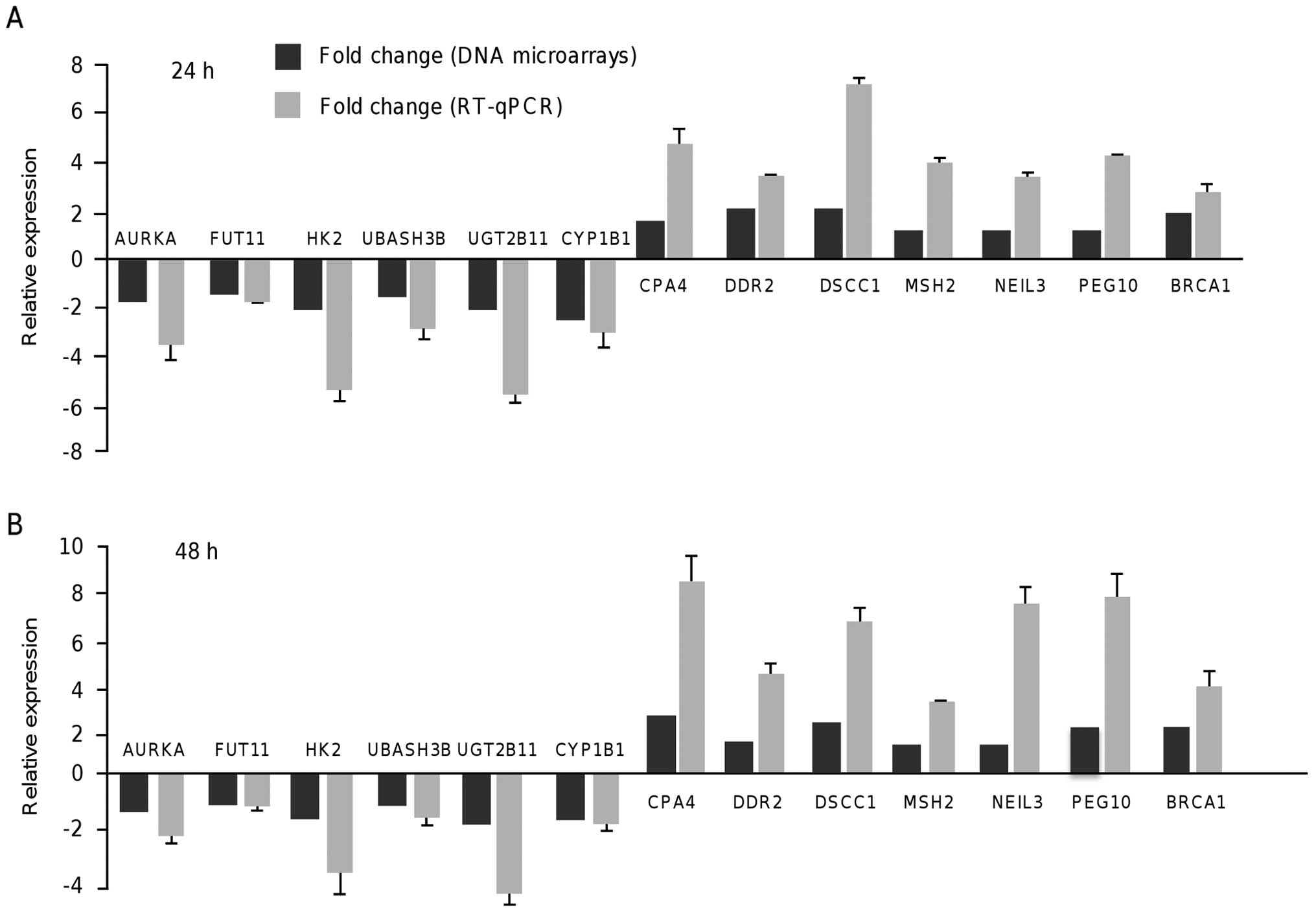
suppressed. To validate the DNA microarray data, we analyzed using
RT-qPCR the expression of 13 modulated genes by resveratrol
including six oncogenes at 24 and 48 h. In all cases, the RT-qPCR
data were similar to those obtained by the DNA microarray analyses
(Fig. 2).
 | Table IIIDeregulated genes and cellular
processes modulated by resveratrol treatment at 48 h. |
Table III
Deregulated genes and cellular
processes modulated by resveratrol treatment at 48 h.
| Process | No. of genes | Gene name |
|---|
| Cell cycle | 64 | AURKA,
PLK1, HIST1H3F, HIST1H2AJ, JUN,
ARHGAP18, KIF20A, SH3KBP, EDN1,
HIST1H2BH, HIST1H2AH, HIST1H1E,
HIST1H2BM, H1F0, HIST1H1C, H2BFS,
HIST1H2BB, PSMD11, MSH2, ARL3, DSN1, E2F1, E2F8,
FBXO43, HAUS4, MPHOSPH9, MTBP, PSMC31B, TSPYL2, AVPI1, CASP8AP2,
CDC6, CDT1, CLSPN, CCN1, CCN2, ESCO2, EXO1, MNS1, MND1, MLH1,
RBBP8, SESN3, TIMELESS, DSNI, MASTL, CDT1, BLM, PRIM1, PRKACA,
CENP, CENP1, RRM2, MLF1IP, RFC5, RFC4, TUBB4B, PSMC3IP, DNA2, CDC6,
EMD, TPH1, ALDH7A1, ACADSB |
| Intrinsic to
membrane | 34 | MFGE8, MOXD1,
OR5L1, PIGX, PLCE1, PSTP1P2, P4HB, RTN3, STAM2, SVIP, SLC16A12,
SLC27A2, SLC35AS, SLC38A4, SLC40A1, SLC5A10, ATRNL1, B3GAT3, CLGN,
C14orf21, DCXR, ENTPD3, EMD, EDNRB, EPHXI, GBA, GBP2, TMEM232,
ITGB3, MGAT4A, TSPANI2, TSPANI3, TMEMIO6C, UCP2 |
| Metal ion
binding | 51 | PEG10, CPA4, ARL3,
ATP8A1, EFCAB5, EFEMP1, FBX043, FANCL, GPR98, NAPEPLD, RFESD,
KDMIB, AMY1A, AMY1C, AMY1B, AMY2A, AMY2B, BIRC3, B3GAT3, CAPS2,
CLGN, C4orf21, ENTPD3, EME1, FBNI, GUCYIB3, IDH2, MATN2, MOXD1,
PLCE1, PRICKLE1, SLC38A4, SLC40A1, SLC5A10, TCF19, TRIM68, TPH1,
UBR7, VSNL1, ZC3H6, ZNF367, ZNF443, ZNF594, ZNF665, ZNF678, ZNF682,
ZNF701, ZNF724P, ZNF730, ZNF799, ZNF845 |
| Cellular response
to stress | 42 | ALOX5AP,
PTGS2, CYP1B1, PLA2G4A, SLC2A3,
TNC, LDHA, KIF204, PSMD11,
H2BFS, SH3KBP1, APOBEC3G, BHLHL40,
UGT2B10, UGT2B11, UGT2B15, UGT2B17,
CYP1B1, GPI, HK2, BRIP1, ATAD5, FAM175A,
CCL20, CLU, C5, CFB, ITGB3, PTX3, SAA2, SAA4, TFPI2, DCXR, TP53I3,
VNN2, EPHXI, GGH, MFGE8, SLIT3, RPS6KA5, P4HB, FBN1 |
| Regulation of cell
death | 38 | HIST1H1E,
PSMD11, H1F0, HIST1H1C, TNFRSF10D,
BNIP3, MMP9, PHLDA1, SERPINB2,
TNFRSF10D, IL24, DDIT4, NOG,
EDN1, OR5P2, OR5P3, ALG3,
PSMD11, SLC2A3, TMEM87B, PSMD11, H1F0,
HIST1H1C, TNFRSF10D, BNIP3, PHLDA1, SERPINB2, TNFRSF10D, IL24,
DDIT4, NOG, EDN1, OR5P2, OR5P3, ALG3, PSMD11, SLC2A3, TMEM87B |
| DNA repair | 26 | NEIL3, BRIPI, BLM,
DNA2, FANCB, FANCD2, FANCL, RAD54L, RECQL, BRCA1, BRCA2, CLSPN,
EME1, ESCO2, EXO1, FAM175A, MLH1, MSH2, POLN, RFC5, RBBP8, UNG,
MNS1, PIGX, MGAT4A, ST6GALNA |
| Glycolytic
process | 25 | PFKB3,
TPI1P1, GPI, LDHA, PGK1, ND2,
FUT11, WDR74, BHLHB9, DDX21,
MLKL, ANK2F1, ERRFI1, CHORDC1,
LTV1, TM45F19, PFKFB3, TKTL2,
LPXN, RAB43, TMEM87B, NDRG1,
HSPA8, RPS24 |
| Chromatin
organization | 22 | PLK1,
RARA, NEDD4, VARS, HIST1H2BB,
RPS24, PSMD11, HIST1H2BM, RUNX2,
H2BFS, HIST1H3F, HIST1H2AJ, HIST1H2BH,
HIST1H2AH, HIST1H1E, HIST1H2BM, ALG3,
PLA2G4A, HK2, NEDD4, RPS24,
HAS2 |
| Senescence | 22 | HIST1H3F,
JUN, HIST1H2AJ, HIST1H2BH, HIST1H2AH,
HIST1H1E, HIST1H2BM, H1F0, HIST1H1C,
H2BFS, PSMD11, APOBC3G, SLC2A3,
VEGFA, PLA2G4A, EDN1, MMP3, BP1,
HAS1, E2F1, CCNE1, CCNE2 |
| Regulation of
transcription | 20 | NRIPI, CBX5, CREG1,
CDCA2L, TSPYL2, PSMC3IP, MLF1IP, E2F8, ATAD2, PER3, TCF19, ZNF845,
ZNF799, ZNF367, ZNF443, ZBF594, ZNF678, ZNF682, ZNF701, ZNF730 |
| DNA
replication | 19 | DSCC1, CDT1,
CCDC111, POLN, RNASEH2A, TK1, KIF24, FANCD2, BLM, RFC4, RFC5, RRM2,
KITLF, DNA2, PRIM1, TSPYL2, CSF2, ATAD2, ATAD5 |
| Response to biotic
stimulus | 18 | SLC5A3,
SLC14A1, NEDD4, SLC2A3, HK2,
HMGCS1, HLADRA, IFITM3, ORM1,
RUNX2, NDRG1, BHLHE40, IGLON5,
VWC2L, SLITRK6, PLK1, PLOD2, SORD2 |
| Metabolism of
lipids and lipoproteins | 17 | HMGCS1,
ARSJ, GPR110, ERRF11, SLC16A6,
MT1F, MTIJP, TSPYL2, MNS1, MGAT4A, PIGX, SLC27A2,
GBA, P4HB, ELOVL7, UBR7, FBX016 |
| Immune system | 15 | KIF20A,
PSMD11, SH3KBP1, IFITM3, EDD4, IL18,
GBP2, CFB, CSF2, SAMHD1, BIRC3, TUBB4B, RPS6KA5, CD226, TRIM68 |
| Regulation of cell
migration | 12 | NEXN,
PLOD2, ARSJ, PTGS2, TNC,
SH3KBP1, ARHGAP18, ORM1, S100A11,
GPR110 |
| Angiogenesis | 8 | ANGPTL4,
EDN1, GPI, JUN, ROBO4, VEGFA,
LOX, PTGS2 |
| Response to
hypoxia | 8 | SHISA3,
DDIT4, PLOD2, VARS, LDHA, RARA,
KIF20A, SLC6A6 |
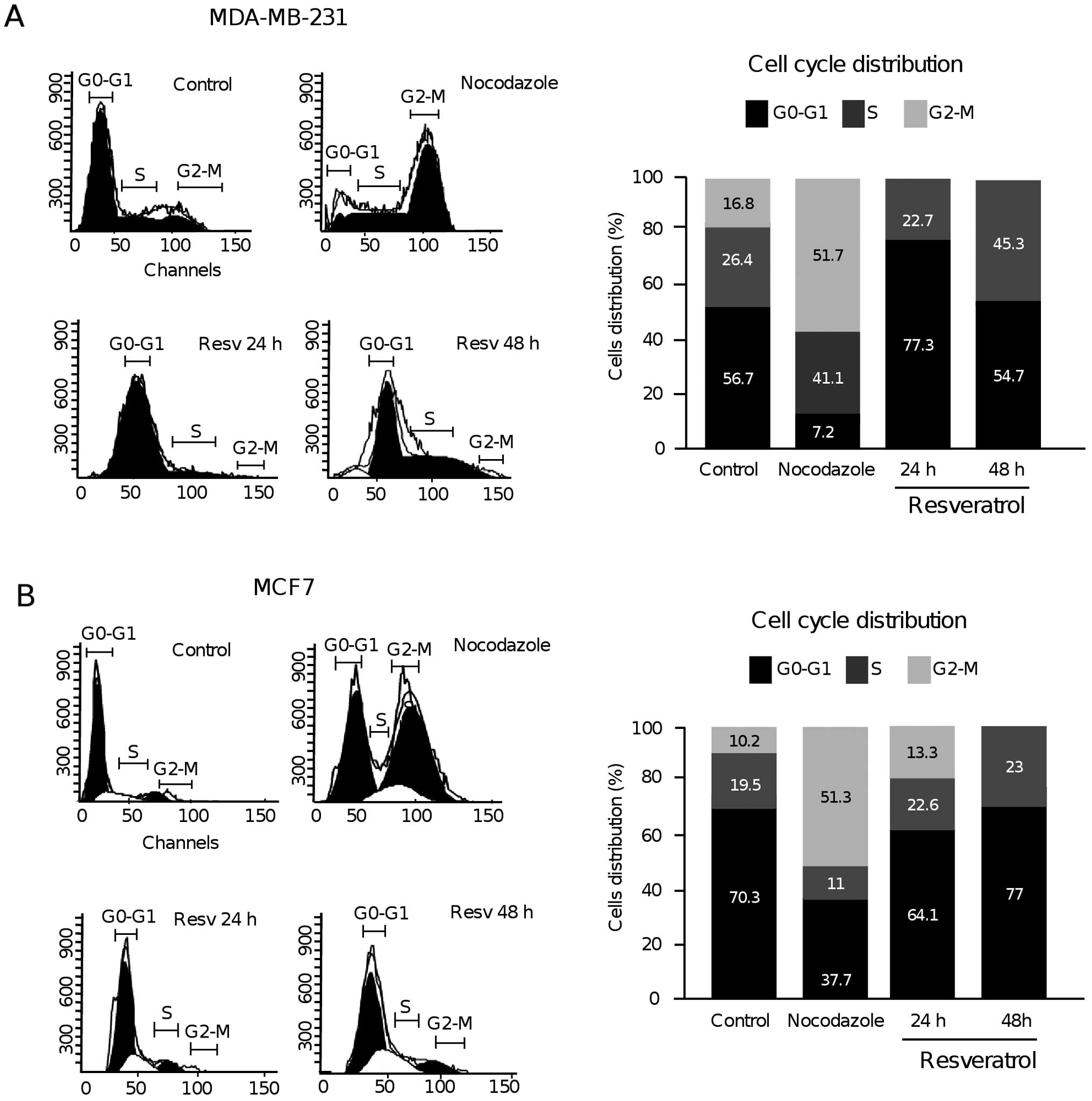
Resveratrol inhibits the G1/S phase
transition in breast cancer cells
In our genome-wide analysis of gene expression in
MDA-MB-231 cells treated with resveratrol at 24 and 48 h, we
identified 63 modulated genes involved in the cell cycle (Tables II and III). Thus, we decided to confirm the
effects of resveratrol (100 µM) on cell cycle progression in
the MDA-MB-231 and MCF-7 breast cancer cell lines. The cells were
treated with resveratrol and the distribution of cell cycle phases
was determined by flow cytometric analysis as described in
Materials and methods. The results showed that the percentage of
cells in the G0–G1 phase increased from 56.7 to 77.3% at 24 h after
resveratrol treatment, but retained a normal level at 48 h (54.7%)
in comparison to the control (ethanol vehicle) in the MDA-MB-231
cells. This change was accompanied by an increment in the
proportion of cells in the S phase from 26.4 in the control to
45.3% in the resveratrol-treated cells at 48 h, but not at 24 h
(Fig. 3A). However, we did not
observed arrest in the G2-M phase in cells treated with resveratrol
at 24 and 48 h, in comparison with the control assays using
nocodazole a known blocker of G2-M phase transition. In contrast,
in the MCF-7 cells the percentage of cells in the G0–G1 phase did
not significantly change after 24 h of resveratrol treatment in
comparison with the control, whereas at 48 h we observed a slight
but significant increase in cells in the G0–G1 phase (Fig. 3B). Taken altogether, these data
indicate that resveratrol inhibits cell cycle progression and that
its effects are cell line-specific.
Resveratrol targets the cell cycle
regulators AURKA, PLK1, and BRCA1
Our DNA microarrays and RT-qPCR analysis indicated
that the activators of cell cycle AURKA, CCND1 and CCNB1 were
significantly repressed by resveratrol at 24 and 48 h, whereas PLK1
was only suppressed at 24 h. Moreover, the cell cycle repressor and
AURKA/PLK1 inhibitor, BRCA1, was overexpressed at 24 and 48 h after
resveratrol treatment. Thus, in order to confirm whether the
alterations in mRNA abundance of these cell cycle genes also occur
at the protein level, western blot assays using specific antibodies
were performed. Results of the immunoblot analysis indicated that
changes in the protein levels of these cell cycle genes were
correlated with changes at the mRNA level. Data showed that after
24 h of resveratrol treatment, AURKA levels were markedly decreased
(95%) in both the MDA-MB-231 and MCF-7 cells in comparison with the
control (Fig. 4A and B). In
contrast, no significant changes were detected at 48 h in both cell
lines. In addition, PLK1 was slightly but significantly decreased
by resveratrol at 24 and 48 h in the MDA-MB-231 cells, but not in
the MCF-7 cells (Fig. 4C and D). On
the other hand, BRCA1 protein levels increased at 24 h whereas no
significant changes were identified after 48 h of resveratrol
treatment in the MDA-MB-231 cells (Fig.
4A). In addition, we confirmed the suppression of CCND1 and
CCNB1 cyclins, which were previously described as two cell cycle
markers modulated by resveratrol in MCF-7 and MDA-MB-231 cells
(21).
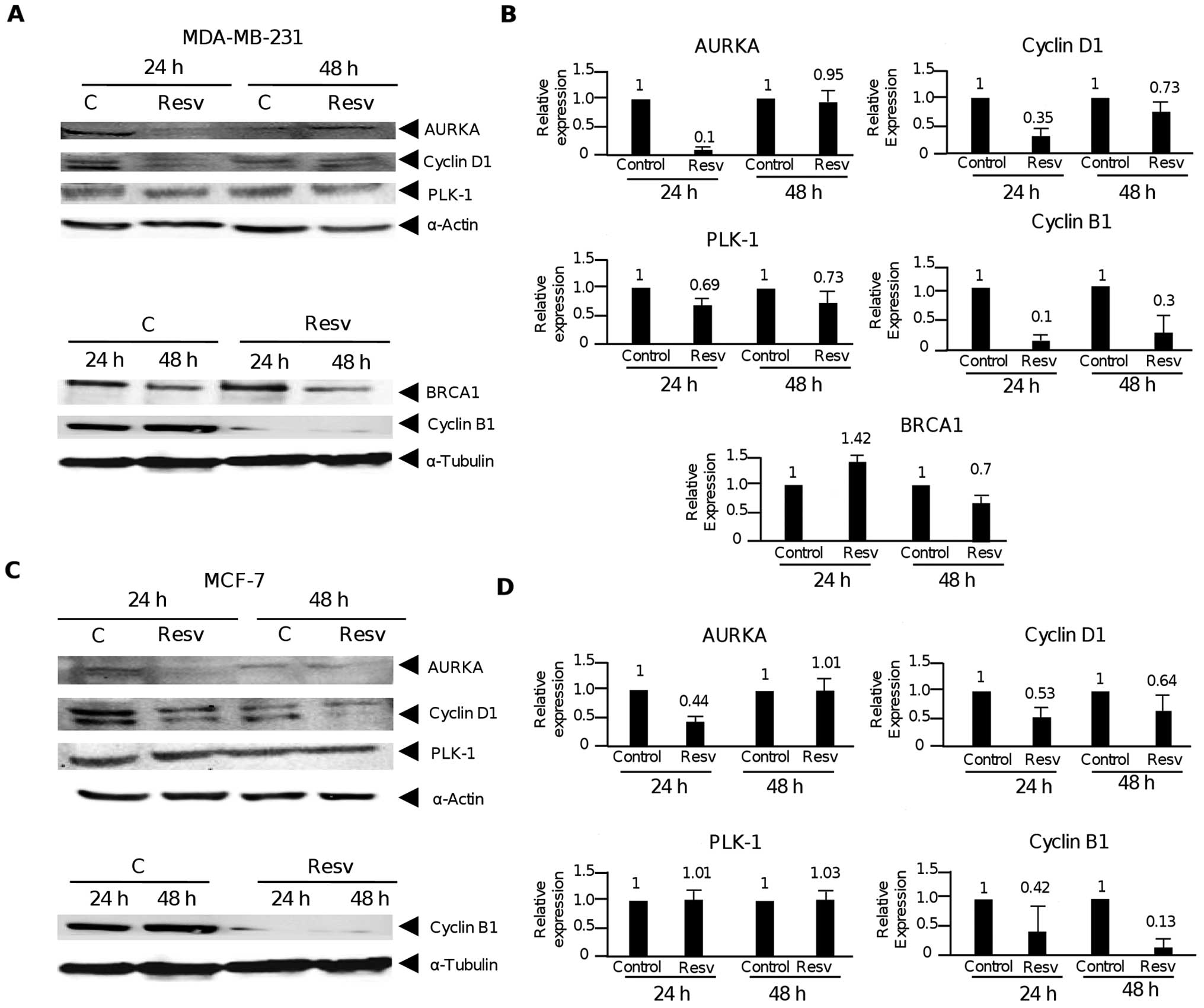 | Figure 4Resveratrol targets cell cycle
proteins. Western blot analysis of (A) MDA-MB-231 and (C) MCF-7
cells treated with resveratrol (100 µM, Resv), or
non-treated (control) during 24 and 48 h, using AURKA (1:1,000),
cyclin D1 (1:1,000); PLK1 (1:500), BRCA1 (1:1,000) and cyclin B
(1:1,000) antibodies. Actin (1:1,000) and tubulin (1:1,000)
antibodies were used as control. (B and D) Densitometric analysis
of bands in panels A and C. Data were normalized using actin or
tubulin expression. Images are representative of three independent
experiments. Resv, resveratrol. |
Discussion
Here, we identified novel targets of resveratrol
that may represent potential therapeutic targets in breast cancer.
Our data showed that, after 24 and 48 h of resveratrol treatment,
key genes involved in the regulation of cell cycle progression and
DNA repair processes were modulated. These data are relevant as the
development of breast cancer is frequently associated with
disorders in the regulation of the cell cycle, and enhanced DNA
repair is linked to chemotherapy resistance of tumor cells. Failure
of the quality control checkpoints or a loss of balance of
regulatory molecules plays a major role in carcinogenesis. Of the
many regulatory cell cycle checkpoints, the acquisition of
abnormalities at the G1/S checkpoint appears to be the most crucial
step in the genesis and progression of cancer. Here, we found that
resveratrol inhibited the proliferation of MDA-MB-231 and MCF-7
cells by blocking G0–G1 phase transition at 24 h. In agreement with
our data, several reports indicate that resveratrol activates
apoptosis and inhibits cell proliferation of tumor cells (22,23).
Remarkably, we identified two novel targets of resveratrol, the
AURKA and PLK1 kinases, which represent pivotal proteins related to
cell division. Previous studies indicate that overexpression of
AURKA and PLK1 promote cell proliferation through activation of
G1/S cell cycle transition in multiple types of carcinomas
(24). AURKA belongs to a small
family of serine/treonine kinases with evolutionarily conserved
structure and participates in mitosis. AURKA is upregulated in a
wide range of malignancies and it acts as an oncogene both in
vitro and in vivo, by enhancing G1-S transition,
protecting cells from apoptosis, inducing centrosome amplification,
promoting multipolar spindle formation and genomic instability, and
conferring resistance to antitumor agents. PLK1 is a checkpoint
protein that participates in the regulation of centrosome
maturation and spindle assembly, the removal of cohesins from
chromosome arms, the inactivation of anaphase-promoting
complex/cyclosome (APC/C) inhibitors, and the mitotic exit and
cytokinesis. As PLK1 activates cyclin B and stimulates cell
proliferation, its silencing results in an inactivation of cyclin
B/Cdc2 complex-mediated mitotic arrest followed by apoptosis
(25). As these kinases may
co-regulate several processes in mitosis, some overlap in the
clinical performance of drugs that target PLK1 or aurora kinases is
possible. However, the knowledge of the molecular factors that
influence sensitivity and resistance to AURKA and PLK1 inhibitors
remains limited.
On the other hand, we found that resveratrol (100
µM) treatment resulted in the upregulation of the BRCA1 gene
in MDA-MB-231 cells at 24 and 48 h. Similar results were reported
in breast cancer cell lines at low resveratrol doses (30 µM)
in previous research (26). BRCA1
is a tumor suppressor that plays critical roles in DNA repair, cell
cycle checkpoint control, and maintenance of genomic stability. It
also suppresses the cell cycle progression. Remarkably, a molecular
interplay between AURKA, PLK1 and BRCA1 proteins has been reported.
BRCA1 inhibits the kinase activity of PLK1 by modulating the
interactions of AURKA with hBora, and PLK1 (27). Our data confirm that resveratrol
suppresses the expression of both CCND1 (G1-S transition) and CCNB1
(G2-M transition) cyclins at 24 h and 48 h in MDA-MB-231 and MCF-7
cells. Similar results for CCND1 and CCNB1 were previously
described in SW-480 colorectal cancer cells and MCF-7 breast cancer
cells (28). Taken all together, we
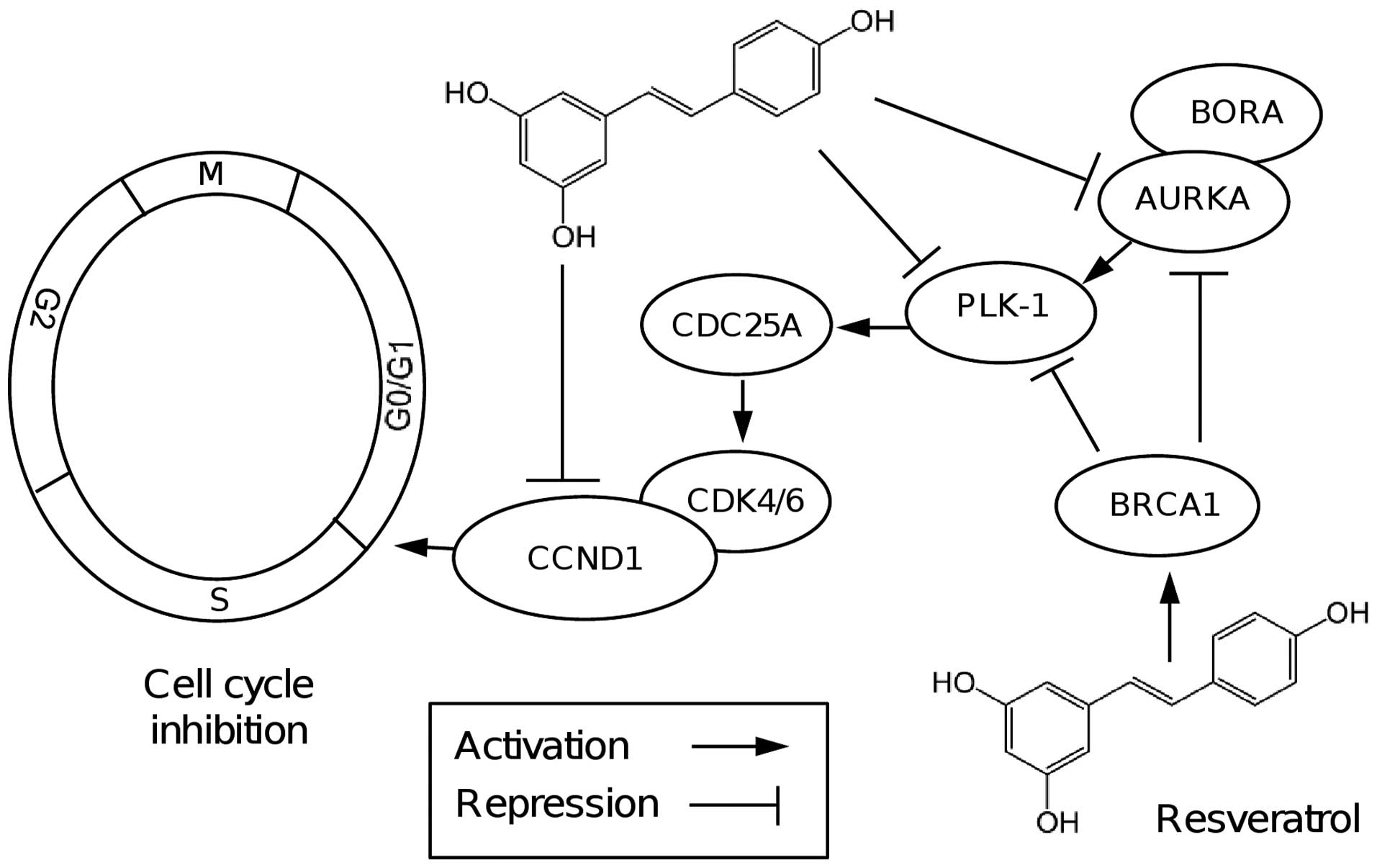
propose a working model for cell cycle regulation by resveratrol in
MDA-MB-231 cells (Fig. 5). In
conclusion, the present study shows for the first time that
resveratrol reduced mRNA and protein expression levels of AURKA and
PLK1 two potential therapeutic targets in cancer. Our findings
revealed the role of resveratrol in important cellular processes
and in the modulation of key cell cycle regulators, which
highlights its potential use as an adjuvant in breast cancer
therapy.
Abbreviations:
|
AURKA
|
Aurora protein kinase
|
|
PLK1
|
Polo-like kinase-1
|
|
CCND1
|
cyclin D1
|
|
CCNB1
|
cyclin B1
|
Acknowledgments
CONACyT SALUD grants (233370 and 222335) supported
this work. R.M. was supported by a CONACYT doctoral fellowship.
References
|
1
|
Le Corre L, Chalabi N, Delort L, Bignon YJ
and Bernard-Gallon DJ: Resveratrol and breast cancer
chemoprevention: Molecular mechanisms. Mol Nutr Food Res.
49:462–471. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
3
|
Wang Y, Wang Z, Qi Z, Yin S, Zhang N, Liu
Y, Liu M, Meng J, Zang R, Zhang Z, et al: The negative interplay
between Aurora A/B and BRCA1/2 controls cancer cell growth and
tumorigenesis via distinct regulation of cell cycle progression,
cytokinesis, and tetraploidy. Mol Cancer. 13:942014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McInnes C and Wyatt MD: PLK1 as an
oncology target: Current status and future potential. Drug Discov
Today. 16:619–625. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun JM, Yang LN, Xu H, Chang B, Wang HY
and Yang G: Inhibition of Aurora A promotes chemosensitivity via
inducing cell cycle arrest and apoptosis in cervical cancer cells.
Am J Cancer Res. 5:1133–1145. 2015.PubMed/NCBI
|
|
6
|
Vundru SS, Kale RK and Singh RP:
β-Sitosterol induces G1 arrest and causes depolarization of
mitochondrial membrane potential in breast carcinoma MDA-MB-231
cells. BMC Complement Altern Med. 13:2802013. View Article : Google Scholar
|
|
7
|
Singh CK, George J and Ahmad N:
Resveratrol-based combinatorial strategies for cancer management.
Ann NY Acad Sci. 1290:113–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yuan L, Zhang Y, Xia J, Liu B, Zhang Q,
Liu J, Luo L, Peng Z, Song Z and Zhu R: Resveratrol induces cell
cycle arrest via a p53-independent pathway in A549 cells. Mol Med
Rep. 11:2459–2464. 2015.
|
|
9
|
Díaz-Chávez J, Fonseca-Sánchez MA,
Arechaga-Ocampo E, Flores-Pérez A, Palacios-Rodríguez Y,
Domínguez-Gómez G, Marchat LA, Fuentes-Mera L, Mendoza-Hernández G,
Gariglio P, et al: Proteomic profiling reveals that resveratrol
inhibits HSP27 expression and sensitizes breast cancer cells to
doxorubicin therapy. PLoS One. 8:e643782013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su D, Cheng Y, Liu M, Liu D, Cui H, Zhang
B, Zhou S, Yang T and Mei Q: Comparision of piceid and resveratrol
in antioxidation and antiproliferation activities in vitro. PLoS
One. 8:e545052013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-delta delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
12
|
In K, Park J and Park H: Resveratrol at
high doses acts as an apoptotic inducer in endothelial cells.
Cancer Res Treat. 38:48–53. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vanamala J, Reddivari L, Radhakrishnan S
and Tarver C: Resveratrol suppresses IGF-1 induced human colon
cancer cell proliferation and elevates apoptosis via suppression of
IGF-1R/Wnt and activation of p53 signaling pathways. BMC Cancer.
10:2382010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alkhalaf M, El-Mowafy A, Renno W, Rachid
O, Ali A and Al-Attyiah R: Resveratrol-induced apoptosis in human
breast cancer cells is mediated primarily through the
caspase-3-dependent pathway. Arch Med Res. 39:162–168. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pandey PR, Okuda H, Watabe M, Pai SK, Liu
W, Kobayashi A, Xing F, Fukuda K, Hirota S, Sugai T, et al:
Resveratrol suppresses growth of cancer stem-like cells by
inhibiting fatty acid synthase. Breast Cancer Res Treat.
130:387–398. 2011. View Article : Google Scholar
|
|
16
|
Liu B, Zhou Z, Zhou W, Liu J, Zhang Q, Xia
J, Liu J, Chen N, Li M and Zhu R: Resveratrol inhibits
proliferation in human colorectal carcinoma cells by inducing G1/S
phase cell cycle arrest and apoptosis through caspase/cyclin CDK
pathways. Mol Med Rep. 10:1697–1702. 2014.PubMed/NCBI
|
|
17
|
García-Zepeda SP, García-Villa E,
Díaz-Chávez J, Hernández-Pando R and Gariglio P: Resveratrol
induces cell death in cervical cancer cells through apoptosis and
autophagy. Eur J Cancer Prev. 22:577–584. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang I, Mitsui Y, Fukuhara S, Gill A,
Wong DK, Yamamura S, Shahryari V, Tabatabai ZL, Dahiya R, Shin DM,
et al: Loss of miR-200c up-regulates CYP1B1 and confers docetaxel
resistance in renal cell carcinoma. Oncotarget. 6:7774–7787. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pan B, Zheng S, Liu C and Xu Y:
Suppression of IGHG1 gene expression by siRNA leads to growth
inhibition and apoptosis induction in human prostate cancer cell.
Mol Biol Rep. 40:27–33. 2013. View Article : Google Scholar
|
|
20
|
Chen Y, Li D and Li S: The Alox5 gene is a
novel therapeutic target in cancer stem cells of chronic myeloid
leukemia. Cell Cycle. 8:3488–3492. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pozo-Guisado E, Alvarez-Barrientos A,
Mulero-Navarro S, Santiago-Josefat B and Fernandez-Salguero PM: The
antiproliferative activity of resveratrol results in apoptosis in
MCF-7 but not in MDA-MB-231 human breast cancer cells:
Cell-specific alteration of the cell cycle. Biochem Pharmacol.
64:1375–1386. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Meeran SM and Katiyar SK: Cell cycle
control as a basis for cancer chemoprevention through dietary
agents. Front Biosci. 13:2191–2202. 2008. View Article : Google Scholar :
|
|
23
|
León-Galicia I, Díaz-Chávez J,
García-Villa E, Uribe-Figueroa L, Hidalgo-Miranda A, Herrera LA,
Alvarez-Rios E, Garcia-Mena J and Gariglio P: Resveratrol induces
downregulation of DNA repair genes in MCF-7 human breast cancer
cells. Eur J Cancer Prev. 22:11–20. 2013. View Article : Google Scholar
|
|
24
|
Lee SY, Jang C and Lee KA: Polo-like
kinases (plks), a key regulator of cell cycle and new potential
target for cancer therapy. Dev Reprod. 18:65–71. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Zhang DY, Ren Q, Ye F, Zhao X,
Daniels G, Wu X, Dynlacht B and Lee P: Regulation of a novel
androgen receptor target gene, the cyclin B1 gene, through
androgen-dependent E2F family member switching. Mol Cell Biol.
32:2454–2466. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fustier P, Le Corre L, Chalabi N,
Vissac-Sabatier C, Communal Y, Bignon YJ and Bernard-Gallon DJ:
Resveratrol increases BRCA1 and BRCA2 mRNA expression in breast
tumour cell lines. Br J Cancer. 89:168–172. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zou J, Rezvani K, Wang H, Lee KS and Zhang
D: BRCA1 downregulates the kinase activity of Polo-like kinase 1 in
response to replication stress. Cell Cycle. 12:2255–2265. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Joe AK, Liu H, Suzui M, Vural ME, Xiao D
and Weinstein IB: Resveratrol induces growth inhibition, S-phase
arrest, apoptosis, and changes in biomarker expression in several
human cancer cell lines. Clin Cancer Res. 8:893–903.
2002.PubMed/NCBI
|