Introduction
Liver cancer, predominantly hepatocellular carcinoma
(HCC), is the fifth most deadly cancer worldwide, and considered as
the third most common cause of cancer mortality. Hepatitis C virus
(HCV) infection is a major etiological factor for developing HCC.
Studies showed that HCC patients with HCV infection usually had a
poorer prognosis associated with higher rate of intrahepatic
recurrence and extrahepatic metastasis, and shorter patient
survival than those without HCV infection (1,2).
Increasing evidence indicates that HCV encoded viral proteins play
a critical role in the modulation of HCV-induced HCC malignancy.
HCV core protein has attracted special attention, numerous studies
demonstrated the ability of HCV core protein in regulating gene
transcription, cell proliferation, apoptosis, transformation and
also immortalization of human hepatocytes, which contributes
largely to the pathogenesis of HCC (3,4).
Furthermore, it has been reported that in transgenic mice, HCV core
protein directly induces hepatic steatosis and finally HCC,
indicating a direct involvement of HCV core protein in
tumorigenesis of HCC (5). Although
there is a strong correlation between HCV infection and HCC
development, the molecular mechanism of HCV core protein in
hepatocarcinogenesis remains to be elucidated.
Octamer-binding protein 4 (OCT4, also known as
OCT3/4 or POU5F1), is a nuclear transcription factor belonging to
the POU-homeodomain family of DNA binding proteins. OCT4, together
with the transcription factor NANOG and SOX2, form a core
transcriptional network to modulate the maintenance of pluripotency
and self-renewal in undifferentiated embryonic stem cells (ESCs)
(6,7). OCT4 also plays a key role in the
induced pluripotent stem cell (iPSC) process, which could reprogram
human somatic fibroblasts and hepatocytes into embryonic stem
cell-like pluripotent cells (8,9). At
first, OCT4 was only found to be expressed in germline and
pregastrulation embryos and in embryonal carcinomas, but not
expressed in mature somatic cells (10). Recently, accumulating evidence shows
that OCT4 is abnormally expressed in numerous types of human solid
tumors and cell lines, such as breast (11), glioma (12) and lung cancer (13), suggesting that OCT4 may play a
potential role in tumorigenesis. Moreover, recent studies
demonstrated that hepatitis B virus X protein, a potential
oncoprotein of hepatitis B virus, was associated with the
modulation of OCT4 expression in HCC (14), indicating that aberrant OCT4
expression and function may be involved in hepatic tumorigenesis of
virus-related HCC. However, any correlation of HCV core protein and
OCT4 expression in HCC remains unclear.
In the present study, we investigated the
relationship between HCV core protein and OCT4 expression and found
that HCV core protein upregulated OCT4 expression and promoted cell
cycle progression in hepatocellular carcinoma cells. Our findings
provide new insight into the mechanism of hepatocarcinogenesis by
HCV infection.
Materials and methods
Cell culture and plasmids
Human hepatoma cell line HepG2 was purchased from
the American Type Culture Collection (ATCC; Manassas, VA, USA) and
stored in our laboratory. Cells were cultured in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% fetal
bovine serum (FBS; (HyClone Laboratories, Inc., Logan, UT, USA).
The full-length HCV core sequence (genotype 1b) was cloned into
pcDNA3.1 to construct the HCV core-expressing plasmid pcDNA-core
(pCore), and pcDNA-vector (pVector) was used as control. HepG2
stable cell lines were established by transfection with either
pCore or pVector, followed by selection with 500 µg/ml G418
(Gibco), as previously described (15).
RNA interference
HCV core- and OCT4-specific small interfering RNAs
(siRNAs) were employed to knock down expression of HCV core and
OCT4. The target mRNA sequences for siRNAs were as follows: HCV
core, 5′-AAG GCG ACA ACC TAT CCC CAA-3′ and OCT4, 5′-CCC TCA CTT
CAC TGC ACT G-3′. Non-targeting scrambled siRNAs was used to
control for non-specific effects. siRNAs at 100 picomolar/ml medium
were transfected into cells using Lipofectamine™ 2000 (Invitrogen),
according to the manufacturer's protocols. The Mock transfected
cells were transfected with Lipofectamine™ 2000 without siRNAs.
Real-time PCR assay
Total RNA isolation, reverse transcription and PCR
were as previously described (16).
All gene-specific mRNA expression values were normalized to GAPDH
expression levels. Primer sequence used in the present study were
as follows: OCT4: forward, 5′-TCT CGC CCC CTC CAG GT-3′ and
reverse, 5′-GCC CCA CTC CAA CCT GG-3′; CCNDl: forward, 5′-CCG CCT
CAC ACG CTT CCT CTC-3′ and reverse, 5′-CGG CCT TGG GGT CCA TGT
TCT-3′; GAPDH: forward, 5′-GCA CCG TCA AGG CTG AGA AC-3′ and
reverse, 5′-TGG TGA AGA CGC CAG TGG A-3′.
Western blotting assay
Cells were lysed with Laemmli's sample solution. The
lysates were electrophoresed, transferred onto nitrocellulose
membranes, and immunoblotted with antibodies against core, OCT4
(Abcam), CCND1 (Cell Signal Technology) and GAPDH (Sigma). After
membranes were incubated with HRP-conjugated secondary antibody
(Bio-Rad Laboratories), the SuperSignal West Pico Chemiluminescent
Substrate (Thermo Fisher Scientific, Waltham, MA, USA) was used to
visualize protein bands on X-ray film.
Cell proliferation assay
Cell proliferation assay was performed by using the
CCK-8 method. Cells were seeded in 96-well plates (Corning Costar,
Corning, NY, USA) and incubated under the indicated conditions.
CCK-8 assay was used to detect the viability of cells following the
manufacturer's instructions. The absorbance at 450 nm was
measured using a microplate reader.
Cell cycle analysis
Cell cycle profile was analyzed using flow
cytometry. Briefly, 1×106 cells were harvested and fixed
with ice-cold 75% ethanol overnight, and incubated with propidium
iodide (PI) (50 µg/ml; Sigma) in the dark at room
temperature for 30 min. Flow cytometric analysis was performed on a
Beckman Coulter EPICS analyzer.
Chromatin immunoprecipitation assay
The ChIP kit (Millipore, Billerica, MA, USA) was
used according to the manufacturer's instructions. Cells
were bathed in 1% formaldehyde for cross-linking of proteins and
DNA, and then the chromatin samples were prepared. The prepared
samples were immunoprecipitated with anti-Oct4 antibody (Abcam) or
negative control rabbit IgG (Cell Signal Technology). DNA released
from precipitated complexes was amplified by PCR using the CCND1
primers (forward, 5′- AGA TTC TTT GGC CGT CTG TC-3′ and reverse,
5′-GCA GCG AGG GGC AGA GCC CA-3′). The PCR reaction conditions were
as follows: denaturation at 50°C for 2 min, followed by 40 cycles
of 95°C (15 sec), 60°C (15 sec) and 72°C (32 sec), and a final
extension at 72°C for 2 min.
Statistical analysis
Results are presented as means of three independent
experiments (± SD). Statistical analyses were performed with the
two-tailed Student's t-test or ANOVA using the SPSS 13.0 software.
P<0.05 was considered statistically significant.
Results
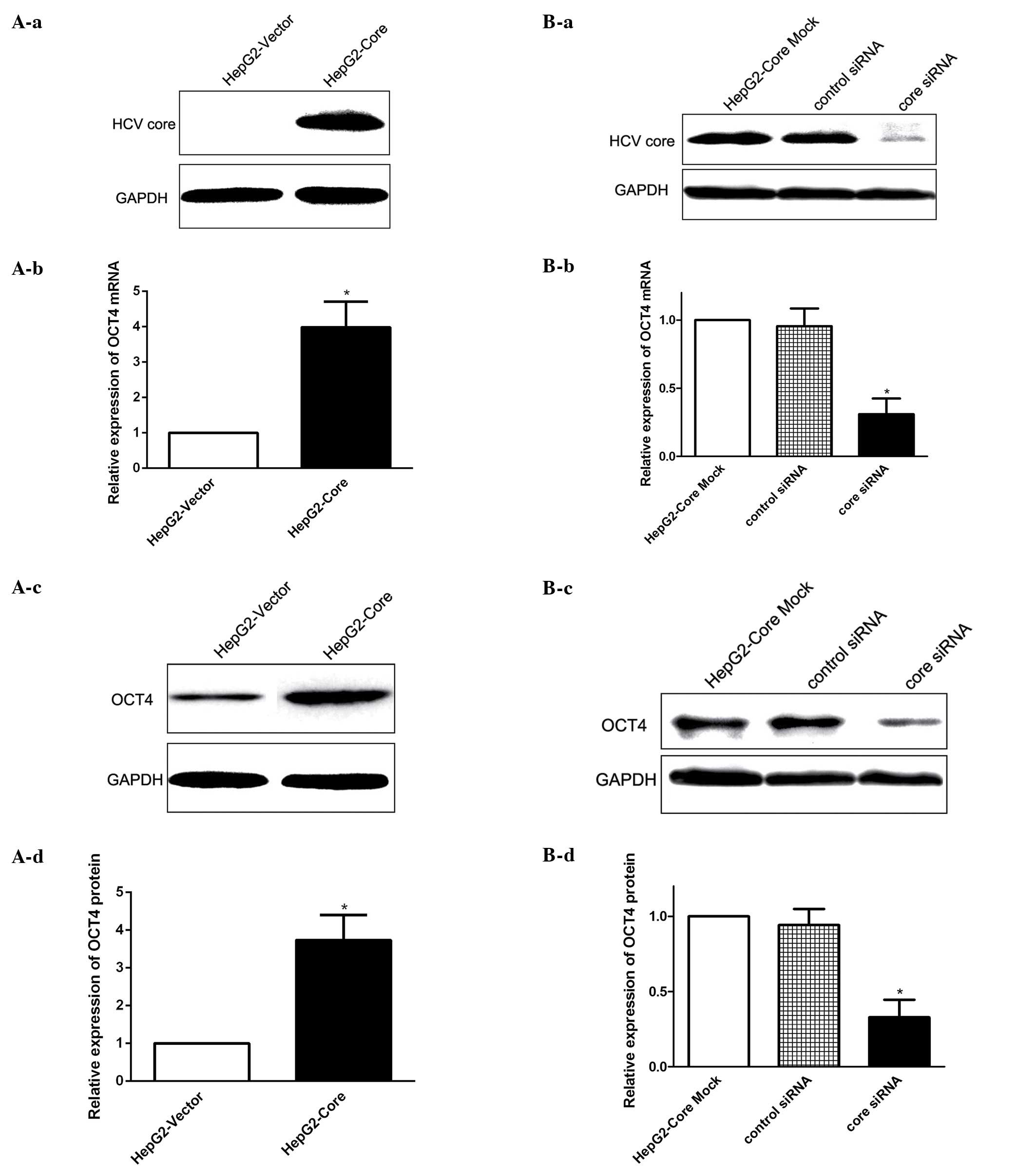
HCV core regulates OCT4 expression
To explore whether HCV core has a potential role in
modulating OCT4 expression in HCC, we first established human HepG2
cells stably expressing HCV core protein (HepG2-core) to
investigate the functional effect of HCV core on OCT4 expression
regulation. We found that stable expression of HCV core increased
the expression level of OCT4 mRNA (Fig.
1A-a and -b), as determined by real-time PCR, and western blot
results showed that the expression level of OCT4 protein in
HepG2-Core cells was similarly enhanced by HCV core expression
(Fig. 1A-c and -d). To further
confirm the correlation between HCV core and OCT4 expression, we
measured OCT4 expression following knockdown of HCV core by using
specific siRNAs (Fig. 1B-a). The
loss-of-function analysis revealed that OCT4 expression at both the
mRNA and protein level was decreased following the inhibition of
HCV core (Fig. 1B-b-d). Taken
together, these results suggested that HCV core upregulated OCT4
expression at both the mRNA and protein level.
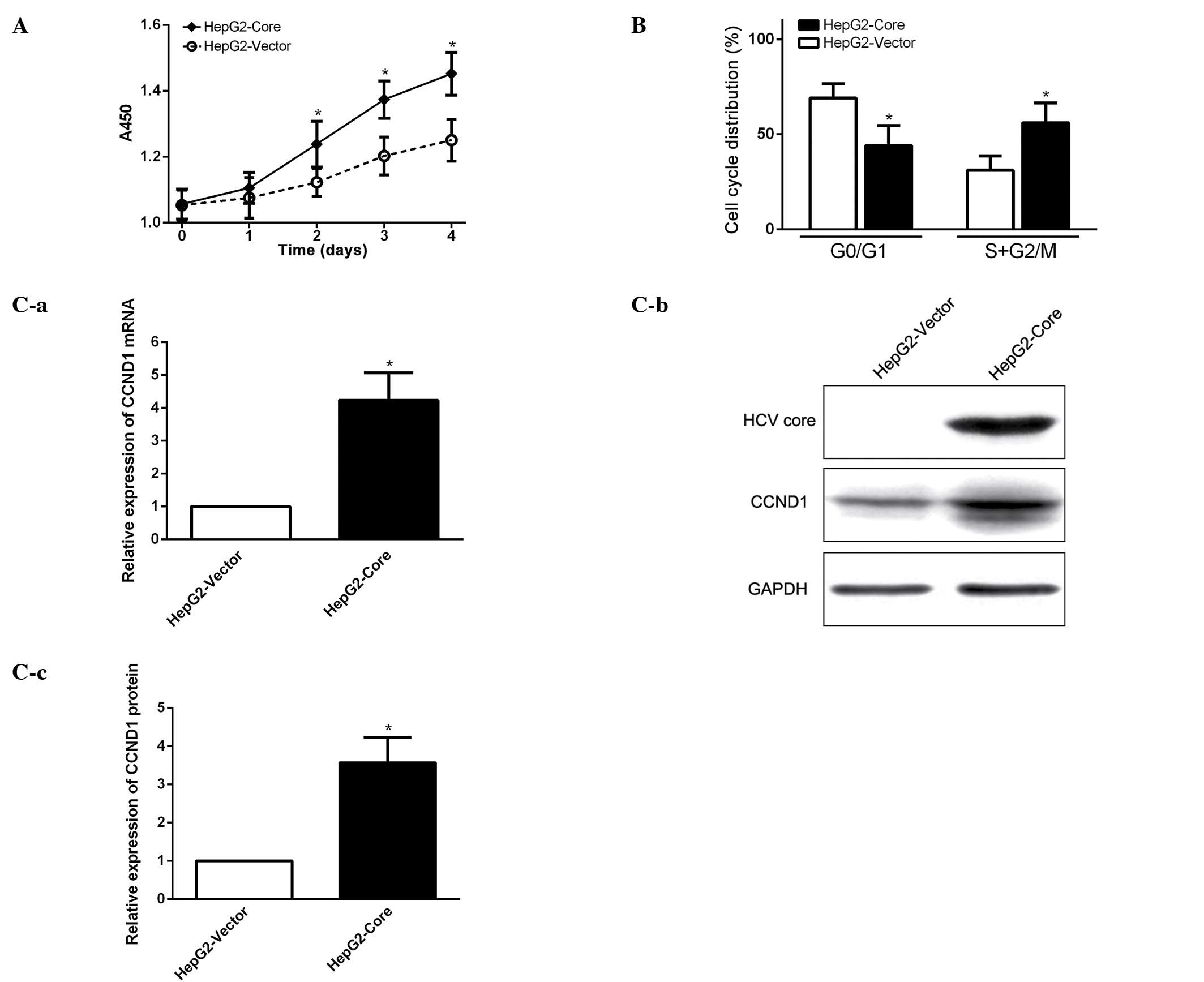
HCV core promotes cell cycle progression
and elevates CCND1 expression
In order to investigate the biological function of
enhanced expression of OCT4 in HCV core-expressing HepG2 cells, we
first examined whether HCV core promotes cancer cell growth and
proliferation. Cell proliferation assay showed that cell growth
capacity of HCV core-expressing cells (HepG2-core) was much
stronger than that of the parental HepG2 cells without HCV core
expression (Fig. 2A). FCM analysis
revealed that HCV core effectively stimulated cell cycle
progression from G1 to S phase (Fig.
2B). The experimental data suggest that HCV core promotes cell
growth by enhancing cell cycle progression from G1 to S phase.
Western blot analysis demonstrated that cell cycle related protein
CCND1 (cyclin D1) expression was significantly elevated in HCV
core-expression cells (Fig. 2C),
indicating that CCND1 might be a downstream target gene, regulated
by the HCV core, to promote cell cycle progression in
HCV-associated HCC.
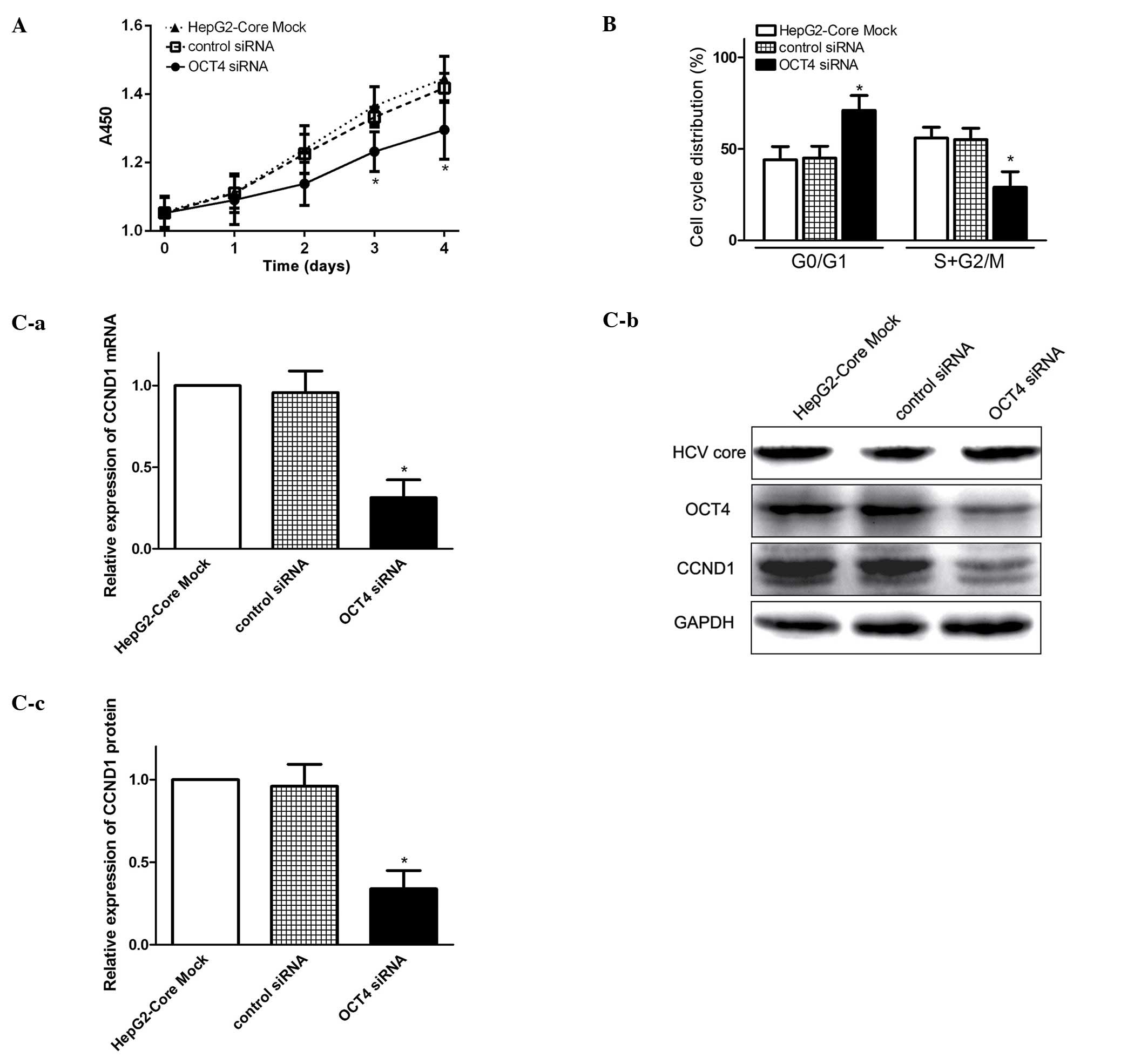
Knockdown of OCT4 blocks cell cycle
progression and inhibits CCND1 expression
OCT4 is a pivotal transcription factor involved in
maintaining pluripotency and self-renewal in ESCs (7). However, the biological function of
OCT4 in HCV associated HCC remains unclear. Using RNA interference
technology to silence OCT4 mRNA, we examined the effect of OCT4 on
cell cycle progression and proliferation in HepG2-core cells. We
found that knockdown of OCT4 inhibited cell growth of HCV
core-expressing cells (Fig. 3A).
FCM analysis revealed that cell cycle was mainly blocked in the
G0/G1 phases in the OCT4-inhibited cells (Fig. 3B), therefore, the G0/G1 ratio was
much higher and the proportion of S + G2/M was much lower than that
in HCV core-expressing cells (Fig.
3B). Q-PCR and western blot analysis demonstrated that CCND1
expression was significantly decreased in OCT4-inhibited cells as
compared to parental cells (Fig.
3C). These results indicated that enhanced OCT4 expression by
HCV core, may modulate cell growth and promote cell proliferation
at least in part via the OCT4/CCND1 pathway.
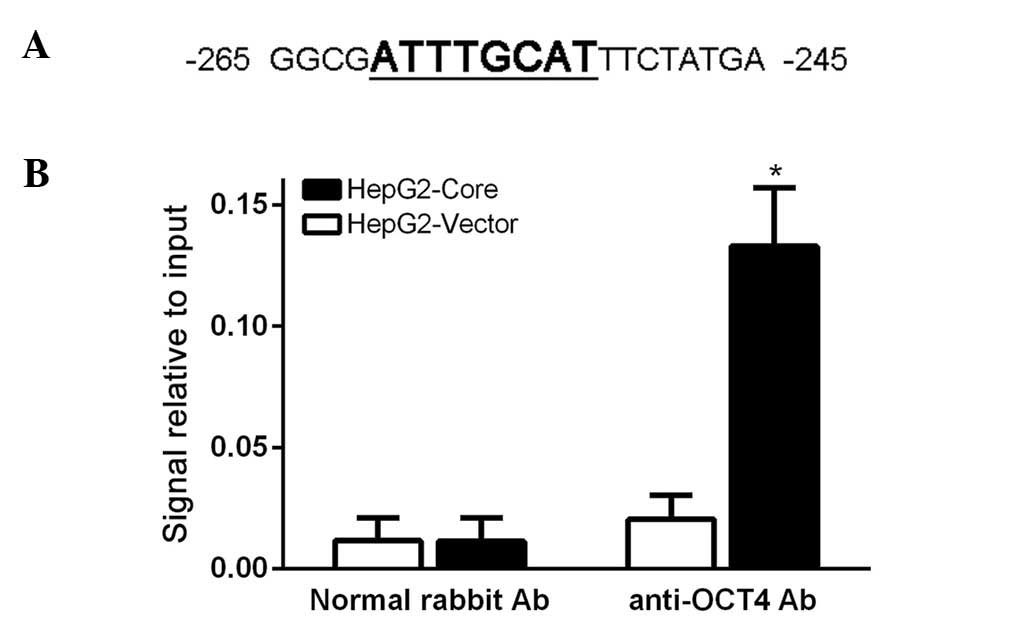
OCT4 directly binds to the promoter of
CCND1 and regulates its expression
Since OCT4 is known as a DNA binding protein to
directly activate its target gene transcription and expression, and
we also demonstrated that CCND1 expression was significantly
decreased via OCT4 inhibition, we hypothesized OCT4 might regulate
CCND1 expression directly. To test the hypothesis, we first
analyzed the 5′-UTR of CCND1 gene and found an octamer motif for
OCT4 in the CCND1 promoter region (Fig.
4A). Then, we employed chromatin immunoprecipitation (ChIP) to
investigate whether OCT4 protein can bind to the CCND1 promoter
region. After gathering the crosslink product, we used ultrasound
to shear the chromatin DNA into an average size of 100–1000 bp, and
then ChIP assay was carried out according to the manufacturer's
instructions. Our PCR amplification experiment revealed that the
OCT4 protein can directly bind to the CCND1 promoter region
(Fig. 4B). These results indicated
that OCT4 regulates CCND1 transcription and expression directly.
Together with the results described above, our findings demonstrate
a novel positive link between the HCV core expression, the OCT4
activation and the CCND1 transcription.
Discussion
Accumulating evidence has demonstrated that OCT4 may
be a type of human oncogene involved in the tumor initiation and
development (17). Aberrant
expression of OCT4 was associated with proliferation, invasiveness,
recurrence, metastasis and unsatisfactory clinical outcome in a
variety of human solid tumors, including HCC (18). Ectopical expression of OCT4
positively regulated survivin expression and promoted cancer cell
proliferation in esophageal squamous cell carcinoma (19), knockdown of OCT4 inhibits colorectal
cancer cell migration and invasion (20), and reduced liver cancer cell
resistance to chemotherapeutic drugs (21). These findings indicated that OCT4
may play a particularly important role in HCC carcinogenesis.
It is well accepted that HCV infection may lead to
the development of HCC and the core protein of HCV induces HCC in
transgenic mice, however, whether HCV infection, especially HCV
core protein modulates OCT4 expression and activity was not well
understand. It is therefore imperative to investigate whether HCV
core regulate OCT4 expression in liver cancer cells and to better
understand the biological function of OCT4 in HCV-associated HCC.
In the present study, we found that HCV core induced upregulation
of OCT4 expression in HepG2 cells. The loss-of-function analysis
confirmed that knockdown of HCV core downregulates the expression
of OCT4 at both the mRNA and protein level. Furthermore, inhibition
of OCT4 expression in HCV core-expressing cells attenuated the HCV
core-induced cell proliferation and migration. These results
established a novel link between HCV core and OCT4 in liver cancer
cells, and indicated that OCT4 may play a role in HCV core
associated hepatocellular carcinogenesis. Moreover, recent studies
showed that OCT4 was highly expressed in cancer stem cells (CSCs)
(21,22), a sub-population of heterogeneous
cancer cells with the capacity of self-renewal and differentiation
into non-tumorigenic cells, contribute to cancer development,
metastasis, recurrence and multidrug resistance. The CSCs
properties of OV6+ HCC cells such as self-renewal,
tumorigenicity and chemoresistance were modulated partly by
elevated OCT4 expression (23). In
addition, exogenous expression of OCT4 was sufficient to directly
reprogram adult neural stem cells to pluripotency (24), and exogenous expression of OCT4 in
cancer cells could promote expansion of CSCs and mediate tumor
initiating properties (25,26). Therefore, our finding of HCV core
induced OCT4 expression in HCC cells suggested that HCV core might
play a particularly potential role in promoting the appearance of
CSCs and consequently promoting HCC development and
progression.
Uncontrolled cell proliferation is one of the
essential hallmarks of tumor malignancy, which may be associated
with cell cycle dysregulation. The CCND1 gene, encoding cyclin D1
protein, plays a critical role in controlling G1/S phase transition
of cell cycle (27). Overexpression
of CCND1 is commonly found in several human cancers, including HCC
(28). A variety of carcinogenic
factors including HCV infection contribute to aberrant expression
of CCND1 (29). In the present
study, we found that HCV core-stimulated cell proliferation of
HepG2 cells by increasing CCND1 expression, and downregulation of
OCT4 inhibited HCV core-induced cell proliferation, indicating
enhanced cell growth capacity and proliferative ability of
core-expressing cells may be partially associated with upregulation
of OCT4 expression. OCT4 can activate the transcription of its
target gene by binding to the octamer motif (5′-ATGCAAAT-3′) within
the promoter regions. We found that there is an octamer motif for
OCT4 in the CCND1 promoter, indicating that OCT4 may directly
participate in the regulation of CCND1 transcription and
expression. Our ChIP assay confirmed that OCT4 protein can directly
bind to the promoter of CCND1. Therefore, this study presents, for
the first time, evidence that HCV core protein induces CCND1
expression in an OCT4-dependent manner, raising the possibility
that OCT4 could serve as a new target for inhibiting HCV induced
proliferation. Furthermore, a previous study demonstrated that
silencing of NANOG, another core regulator for stem cell
self-renewal and stemness, reduces cell proliferation and induces
G0/G1 cell cycle arrest by inhibiting CCND1 expression in breast
cancer cells, and CCND1 promoter also contain the ATTA binding
motifs for NANOG (30), suggesting
CCND1 as one of the co-occupied target genes for both OCT4 and
NANOG. In addition, it has been reported that OCT4 and NANOG,
together with SOX2, collaborate to form regulatory circuitry
consisting of autoregulatory and feed-forward loops and co-occupy a
substantial portion of their target genes (7,31),
indicating a complex regulatory network in regulating the CCND1
expression and consequently modulating cell proliferation of cancer
cells or CSCs.
In summary, we found that HCV core upregulated OCT4
expression and subsequently promoted CCND1 expression and cell
cycle progression in HepG2 cells. In addition, inhibition of OCT4
expression in HCV core-expressing cells led to decreased CCND1
expression and cell cycle arrest at the G0/G1 phases. Collectively,
the present findings provide new insight into the mechanism of
HCV-induced hepatocarcinogenesis, and highlight OCT4 as a novel
potential molecular target for HCV-related HCC.
Abbreviations:
|
HCV
|
hepatitis C virus
|
|
OCT4
|
octamer-binding protein 4
|
|
HCC
|
hepatocellular carcinoma
|
|
ESCs
|
embryonic stem cells
|
|
iPSC
|
induced pluripotent stem cell
|
|
siRNAs
|
small interfering RNAs
|
|
CCND1
|
cyclin D1
|
|
ChIP
|
chromatin immunoprecipitation, CSCs,
cancer stem cells
|
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (no. 81000889), the Key Program
of Natural Science Foundation of Guangdong Province, China (no.
2014A030311047), the Science and Technology Planning Project of
Guangdong Province, China (no. 2014A020212094). The study was also
supported by the Grant [2013]163 by the Key Laboratory of Malignant
Tumor Molecular Mechanism and Translational Medicine of Guangzhou
Bureau of Science and Information Technology; the grant KLB09001 by
the Key Laboratory of Malignant Tumor Gene Regulation and Target
Therapy of Guangdong Higher Education Institutes.
References
|
1
|
Caselmann WH and Alt M: Hepatitis C virus
infection as a major risk factor for hepatocellular carcinoma. J
Hepatol. 24(Suppl): 61–66. 1996.PubMed/NCBI
|
|
2
|
Bartosch B, Thimme R, Blum HE and Zoulim
F: Hepatitis C virus-induced hepatocarcinogenesis. J Hepatol.
51:810–820. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamashita T, Honda M and Kaneko S:
Molecular mechanisms of hepatocarcinogenesis in chronic hepatitis C
virus infection. J Gastroenterol Hepatol. 26:960–964. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arzumanyan A, Reis HM and Feitelson MA:
Pathogenic mechanisms in HBV- and HCV-associated hepatocellular
carcinoma. Nat Rev Cancer. 13:123–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moriya K, Fujie H, Shintani Y, Yotsuyanagi
H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T and
Koike K: The core protein of hepatitis C virus induces
hepatocellular carcinoma in transgenic mice. Nat Med. 4:1065–1067.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pan GJ, Chang ZY, Schöler HR and Pei D:
Stem cell pluripotency and transcription factor Oct4. Cell Res.
12:321–329. 2002. View Article : Google Scholar
|
|
7
|
Boyer LA, Lee TI, Cole MF, Johnstone SE,
Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG,
et al: Core transcriptional regulatory circuitry in human embryonic
stem cells. Cell. 122:947–956. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takahashi K, Tanabe K, Ohnuki M, Narita M,
Ichisaka T, Tomoda K and Yamanaka S: Induction of pluripotent stem
cells from adult human fibroblasts by defined factors. Cell.
131:861–872. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aoi T, Yae K, Nakagawa M, Ichisaka T,
Okita K, Takahashi K, Chiba T and Yamanaka S: Generation of
pluripotent stem cells from adult mouse liver and stomach cells.
Science. 321:699–702. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Freberg CT, Dahl JA, Timoskainen S and
Collas P: Epigenetic reprogramming of OCT4 and NANOG regulatory
regions by embryonal carcinoma cell extract. Mol Biol Cell.
18:1543–1553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Oskarsson T, Acharyya S, Zhang XH,
Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K,
Brogi E and Massagué J: Breast cancer cells produce tenascin C as a
metastatic niche component to colonize the lungs. Nat Med.
17:867–874. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y,
Cao X, Ling EA and Hao A: Oct4 is expressed in human gliomas and
promotes colony formation in glioma cells. Glia. 57:724–733. 2009.
View Article : Google Scholar
|
|
13
|
Chiou SH, Wang ML, Chou YT, Chen CJ, Hong
CF, Hsieh WJ, Chang HT, Chen YS, Lin TW, Hsu HS, et al:
Coexpression of Oct4 and Nanog enhances malignancy in lung
adenocarcinoma by inducing cancer stem cell-like properties and
epithelial-mesenchymal transdifferentiation. Cancer Res.
70:10433–10444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arzumanyan A, Friedman T, Ng IO, Clayton
MM, Lian Z and Feitelson MA: Does the hepatitis B antigen HBx
promote the appearance of liver cancer stem cells? Cancer Res.
71:3701–3708. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou JJ, Chen RF, Deng XG, Zhou Y, Ye X,
Yu M, Tang J, He XY, Cheng D, Zeng B, et al: Hepatitis C virus core
protein regulates NANOG expression via the stat3 pathway. FEBS
Lett. 588:566–573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou JJ, Deng XG, He XY, Zhou Y, Yu M, Gao
WC, Zeng B, Zhou QB, Li ZH and Chen RF: Knockdown of NANOG enhances
chemosensitivity of liver cancer cells to doxorubicin by reducing
MDR1 expression. Int J Oncol. 44:2034–2040. 2014.PubMed/NCBI
|
|
17
|
Hayashi H, Arao T, Togashi Y, Kato H,
Fujita Y, De Velasco MA, Kimura H, Matsumoto K, Tanaka K, Okamoto
I, et al: The OCT4 pseudogene POU5F1B is amplified and promotes an
aggressive phenotype in gastric cancer. Oncogene. 34:199–208. 2015.
View Article : Google Scholar
|
|
18
|
Yuan F, Zhou W, Zou C, Zhang Z, Hu H, Dai
Z and Zhang Y: Expression of Oct4 in HCC and modulation to
wnt/β-catenin and TGF-β signal pathways. Mol Cell Biochem.
343:155–162. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li C, Yan Y, Ji W, Bao L, Qian H, Chen L,
Wu M, Chen H, Li Z and Su C: OCT4 positively regulates Survivin
expression to promote cancer cell proliferation and leads to poor
prognosis in esophageal squamous cell carcinoma. PLoS One.
7:e496932012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dai X, Ge J, Wang X, Qian X, Zhang C and
Li X: OCT4 regulates epithelial-mesenchymal transition and its
knockdown inhibits colorectal cancer cell migration and invasion.
Oncol Rep. 29:155–160. 2013.
|
|
21
|
Wang XQ, Ongkeko WM, Chen L, Yang ZF, Lu
P, Chen KK, Lopez JP, Poon RT and Fan ST: Octamer 4 (Oct4) mediates
chemotherapeutic drug resistance in liver cancer cells through a
potential Oct4-AKT-ATP-binding cassette G2 pathway. Hepatology.
52:528–539. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ikushima H, Todo T, Ino Y, Takahashi M,
Saito N, Miyazawa K and Miyazono K: Glioma-initiating cells retain
their tumorigenicity through integration of the Sox axis and Oct4
protein. J Biol Chem. 286:41434–41441. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qian YW, Chen Y, Yang W, Fu J, Cao J, Ren
YB, Zhu JJ, Su B, Luo T, Zhao XF, et al: p28GANK
prevents degradation of Oct4 and promotes expansion of
tumor-initiating cells in hepatocarcinogenesis. Gastroenterology.
142:1547–58.e14. 2012. View Article : Google Scholar
|
|
24
|
Kim JB, Sebastiano V, Wu G, Araúzo-Bravo
MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, et
al: Oct4-induced pluripotency in adult neural stem cells. Cell.
136:411–419. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Beltran AS, Rivenbark AG, Richardson BT,
Yuan X, Quian H, Hunt JP, Zimmerman E, Graves LM and Blancafort P:
Generation of tumor-initiating cells by exogenous delivery of OCT4
transcription factor. Breast Cancer Res. 13:R942011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsai LL, Hu FW, Lee SS, Yu CH, Yu CC and
Chang YC: Oct4 mediates tumor initiating properties in oral
squamous cell carcinomas through the regulation of
epithelial-mesenchymal transition. PLoS One. 9:e872072014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hui AM, Makuuchi M and Li X: Cell cycle
regulators and human hepatocarcinogenesis. Hepatogastroenterology.
45:1635–1642. 1998.PubMed/NCBI
|
|
28
|
Sundarrajan M, Gupta S and Rao KV:
Overexpression of cyclin D1 is associated with the decondensation
of chromatin during den-induced sequential hepatocarcinogenesis.
Cell Biol Int. 26:699–706. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sato Y, Kato J, Takimoto R, Takada K,
Kawano Y, Miyanishi K, Kobune M, Sato Y, Takayama T, Matunaga T, et
al: Hepatitis C virus core protein promotes proliferation of human
hepatoma cells through enhancement of transforming growth factor
alpha expression via activation of nuclear factor-kappaB. Gut.
55:1801–1808. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han J, Zhang F, Yu M, Zhao P, Ji W, Zhang
H, Wu B, Wang Y and Niu R: RNA interference-mediated silencing of
NANOG reduces cell proliferation and induces G0/G1 cell cycle
arrest in breast cancer cells. Cancer Lett. 321:80–88. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sharov AA, Masui S, Sharova LV, Piao Y,
Aiba K, Matoba R, Xin L, Niwa H and Ko MS: Identification of
Pou5f1, Sox2, and Nanog downstream target genes with statistical
confidence by applying a novel algorithm to time course microarray
and genome-wide chromatin immunoprecipitation data. BMC Genomics.
9:2692008. View Article : Google Scholar : PubMed/NCBI
|