Introduction
Lung cancer is the world's most common cause of
cancer fatality, accounting for 1/3 of tumor causes (1,2). In
recent years, lung cancer cases in China showed a rapid growth
trend. A total of 80% of lung cancer fatalities are due to
non-small cell lung cancer (NSCLC) (3–5).
However, traditional chemotherapy treatment programs are not
optimal for NSCLC. Currently, molecular targeted drugs are widely
used in the treatment of advanced NSCLC because of their high
specificity, significant therapeutic effects and minimal side
effects.
With the in-depth studies of the molecular
mechanisms of lung cancer in recent years, tremendous progress has
been made regarding the treatment of advanced NSCLC by tyrosine
kinase inhibitors (TKI) which target the epidermal growth factor
receptor (EGFR) (6–9). As the main drug within EGFR-TKIs,
gefitinib is one of the first drugs used in the treatment of NSCLC
(10–12). However, many patients taking
gefitinib will develop drug resistance. Currently, the T790M
mutation and c-MET gene amplification are the main accepted reasons
for this resistance (13). It is
also reported that epithelial-mesenchymal transition (EMT) of tumor
cells (14), which can cause drug
resistance in lung cancer, is thought to be inhibited by histone
deacetylase inhibitors, but the effect of histone deacetylase
inhibitors is still uncertain (15). In our study, we investigated the
resistance mechanism of EMT and provided a basis for the study of
NSCLC resistance to gefitinib.
It is known that peptidylarginine deiminase IV
(PAD4) downregulation induces EMT (16). Therefore, we hypothesized that
overexpression of PAD4 could inhibit EMT and thus suppress drug
resistance. Peptidylarginine deiminase IV is a nuclear enzyme that
converts histone arginine residues to citrulline at
apoptosis-related gene promoters and represses gene expression,
amongst other functions (16,17).
In recent years, studies have indicated that PAD4 is related to
many diseases, such as rheumatoid arthritis, cancer, ankylosing
spondylitis, and Alzheimer's disease (16,18).
Studies have shown that PAD4 is increased in cancer patients and
possibly contributes to tumorigenesis (18). However, it is not clear if PAD4
plays a role in the resistance mechanism of NSCLC to gefitinib.
ETS-domain containing protein (Elk1), a
transcription factor belonging to the ETS oncogene family, can be
phosphorylated by the MAPK cascade (19), and is a regulated factor of oncogene
c-fos (20). Additionally, Elk1 is
relevant to cell differentiation, proliferation, apoptosis and
tumorigenesis (21). It may be an
important part of the development of malignant tumors and cell
invasion. It has been reported to be involved in the regulation of
EMT in osteosarcoma (22). Studies
have also revealed that PAD4 deiminates Elk1, and this
post-translational modification has a bearing on the
phosphorylation of Elk1 (23).
However, the possibility that PAD4 can regulate Elk1 has not yet
been confirmed. For this reason, we hypothesized that PAD4
regulates EMT through controlling the expression of Elk1.
Epithelial-mesenchymal transition is a process where
epithelial cells become mesenchymal cells (24). In this process, epithelial markers
such as E-cadherin and α-catenin are reduced, and mesenchymal
markers such as N-cadherin and vimentin are upregulated (25,26).
Epithelial-mesenchymal transition is not only involved in embryonic
development and normal physiology, but is also involved in many
pathologic processes. Additionally, increasing evidence has
demonstrated that EMT plays a role in tumorigenesis, and the
development and migration of tumor cells (27). The resistance to gefitinib in NSCLC
cell lines has been reported be related to EMT. Non-small cell lung
cancer lines that had undergone EMT obtained resistance to the
growth inhibitory effects of EGFR-TKIs such as gefitinib, both
in vitro and in xenografts (14).
In this study, we investigated the molecular
mechanism of PAD4 in the resistance of NSCLC to gefitinib. We found
that overexpression of PAD4 could inhibit EMT and the resistance to
gefitinib. Further analysis showed that PAD4 could suppress
resistance by inhibiting Elk1 expression. Peptidylarginine
deiminase IV may thus be a promising molecular target for treatment
of the resistance of NSCLC to gefitinib.
Materials and methods
Establishment of the NSCLC cell lines
resistant to gefitinib
The curative effect of gefitinib is closely related
to EGFR mutations such as the deletion mutation of exon-19. The
NSCLC cell lines HCC827 and H1650, which contain the deletion
mutation of exon-19, were purchased from the American Type Culture
Collection (ATCC; Manassas, VA, USA), and the established method
was in accordance with Engelman et al (28). The cell lines HCC827/G, H1650/G
resistant to gefitinib and NSCLC cell lines HCC827 and H1650 were
cultured in RPMI-1640 medium (Hyclone, Salt Lake City, UT, USA)
with 10% fetal bovine serum (FBS; Gibco, Rockville, MD, USA),
penicillin and streptomycin (100 U/ml and 100 mg/ml, respectively;
Gibco) in an atmosphere of 5% CO2 at 37°C.
Cell growth and viability
The cell growth and viability were measured by
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay based on the manufacturer's instructions. In brief, the cells
were plated in the 96-well plate and the density was
1×104 cells/well. Then, MTT was diluted in phosphate
buffered saline to 5 g/l, and 20 µl was added to each well
to replace the culture medium. Thereafter, the 96-well plate was
stored at 37°C for 4 h. Subsequently, dimethyl sulfoxide (DMSO) was
added (150 µl/well) in the plate to dissolve the crystals.
Ultimately, absorbance was detected by the microplate reader at 490
nm (Thermo Fisher Scientific, Waltham, MA, USA).
Annexin V-FITC/PI
Annexin V-fluorescein isothiocyanate
(FITC)/propidium iodide (PI) apoptosis detection kit (Invitrogen,
Carlsbad, CA, USA) was used according to the manufacturer's
instructions to identify cell apoptosis. Cells were suspended in
binding buffer, 10 µl of Annexin V-FITC solution was added,
and they were incubated at 4°C for 25 min. Next, cells were washed
with PBS, 10 µl of PI was added, and they were incubated for
5 min. Cellular apoptosis was measured by a FACS analyzer (Thermo
Fisher Scientific, Waltham, MA, USA).
Construction of pCMV-2a/2b-PAD4 and
pCMV-2a/2b-Elk1
The full-length cDNA of PAD4 (accession no.
NM_012387) and Elk1 (accession no. AB016194.1) were both amplified
by reverse transcription PCR, and then cloned into the pCMV-2a/2b
vector (Novagen, Madison, WI, USA). The restriction sites included
EcoRI and BamHI in the amplified cDNA and pCMV-2a/2b
vector. Next, the two different recombinant plasmids were
separately transformed into DH5α competent cells (Takara
Biotechnology, Dalian, China), and the transformed DH5α was grown
overnight at 37°C. Plasmids were extracted and then sequenced to
identify the correct ones. The correct plasmids were named as
pCMV-2a/2b-PAD4 and pCMV-2a/2b-Elk1.
Transfection of the recombinant plasmids
and siRNA
HCC827/G and H1650/G (5×104/well) were
separately plated in the 24-well plate and incubated in a
humidified atmosphere containing 5% CO2 at 37°C for 24 h
followed by the transfection strictly according to the
manufacturer's instruction. A total of 3 µl TurboFect
(Thermo Fisher Scientific) was separately added to the recombinant
plasmids (pCMV-2a/2b-PAD4, pCMV-2a/2b-Elk1or pCMV-2a/2b), PAD4
siRNA (5′-GCGAAGACCTGCAGGACAT-3′) or non-specific siRNA with 100
µl serum-free RPMI-1640 medium, and the mixtures were each
added to the wells. Finally, the plate was placed in the incubator
(Thermo Fisher Scientific) with 5% CO2 at 37°C for
another 24 h.
Quantitative real-time polymerase chain
reaction (qRT-PCR)
TRIzol reagent (Takara Biotechnology) was used to
extract the total RNA from the cells with the different treatments.
A total of 6 µg of the extracted RNA was synthesized into
the cDNA that would be used as the template in the qRT-PCR.
Additionally, the synthesis process of the cDNA was in terms of the
reverse transcription kit (Invitrogen). A volume of 10 µl
SsoFast™ EvaGreen Supermix (Bio-Rad, Hercules, CA, USA) was added
into 20-µl reactive volume in the qRT-PCR. The qRT-PCR
protocol was as follows: 95°C for 20 sec; 35 cycles of denaturation
at 95°C for 30 sec, annealing at 52°C (Elk1), 58°C [PAD4,
glyceraldehyde-3-phosphate dehydrogenase (GAPDH)] or 60°C
(E-cadherin, α-catenin, vimentin and N-cadherin) for 20 sec and
extension at 72°C for 30 sec. The data obtained were calculated by
2−ΔΔCt and the target gene relative expression levels
were normalized to GAPDH. The primers are presented in Table I.
 | Table IThe primers of qRT-PCR. |
Table I
The primers of qRT-PCR.
| Gene names | Primer
sequences |
|---|
| PAD4 | Sense:
5′-GGACTGCGAGGATGATG-3′ |
| Anti-sense:
5′-GCTGTCTTGGAACACCAC-3′ |
| Elk1 | Sense:
5′-CCTTGCGGTACTACTATGAC-3′ |
| Anti-sense:
5′-GGCTGCGGCTGCAGAGACTGG-3′ |
| E-cadherin | Sense:
5′-TACACTGCCCAGGAGCCAGA-3′ |
| Anti-sense:
5′-TGGCACCAGTGTCCGGATTA-3′ |
| α-catenin | Sense:
5′-CTCTACTGCCACCAGCTGAACATC-3′ |
| Anti-sense:
5′-ATGCCTTCACTGTCTGCACCAC-3′ |
| N-cadherin | Sense:
5′-CGAATGGATGAAAGACCCATCC-3′ |
| Anti-sense:
5′-TAGCAGCTTCAACGGCAAAGTTC-3′ |
| Vimentin | Sense:
5′-TGAGTACCGGAGACAGGTGCAG-3′ |
| Anti-sense:
5′-GGAGCCACTGCCTTCATAGTCAA-3′ |
| GAPDH | Sense:
5′-CGTCTTCACCACCATGGAGA-3′ |
| Anti-sense:
5′-CGGCCATCACGCCACAGTTT-3′ |
Western blot analysis
The proteins were extracted from the cells using
lysis buffer with phenylmethanesulfonyl fluoride (PMSF) for 30 min
on ice, and the concentrations of the proteins were measured by the
BCA kit (Pierce, Rockford, IL, USA). Twenty-five micrograms of
protein was loaded in 12% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred by a semi-dry blotting
apparatus (Bio-Rad). Blots were washed with Tris buffered saline
with Tween (TBST), and then blocked using 5% skim milk in Tris
buffered saline for 2 h. Next, blots were incubated with the
primary antibodies (Cell Signaling Technology, Danvers, MA, USA),
diluted (1:500) in the blocking buffer at 4°C overnight, incubated
in the horseradish peroxidase-conjugated secondary antibody (Abcam,
Cambridge, UK) and diluted (1:1,000) in the blocking buffer at room
temperature for 1 h. Blots were analyzed in the Bio-Rad ChemiDoc
apparatus.
Statistical analysis
The differences were determined by Student's t-test
for two groups or by one-way ANOVA followed by Bonferroni's test
for multiple groups. A P<0.05 was recognized as statistically
significant. Data were processed as mean ± standard deviation
(SD).
Results
Demonstration of the drug resistance of
cell lines to gefitinib and the protein expression of PAD4
Drug resistance was demonstrated using the Annexin
V-FITC/PI apoptosis detection kit and MTT assay. The detections
were performed following the cell treatments. The treated cells
were cultured in 180 µl RPMI-1640 medium containing 10% FBS
and 20 µl of various concentrations of gefitinib for 72 h at
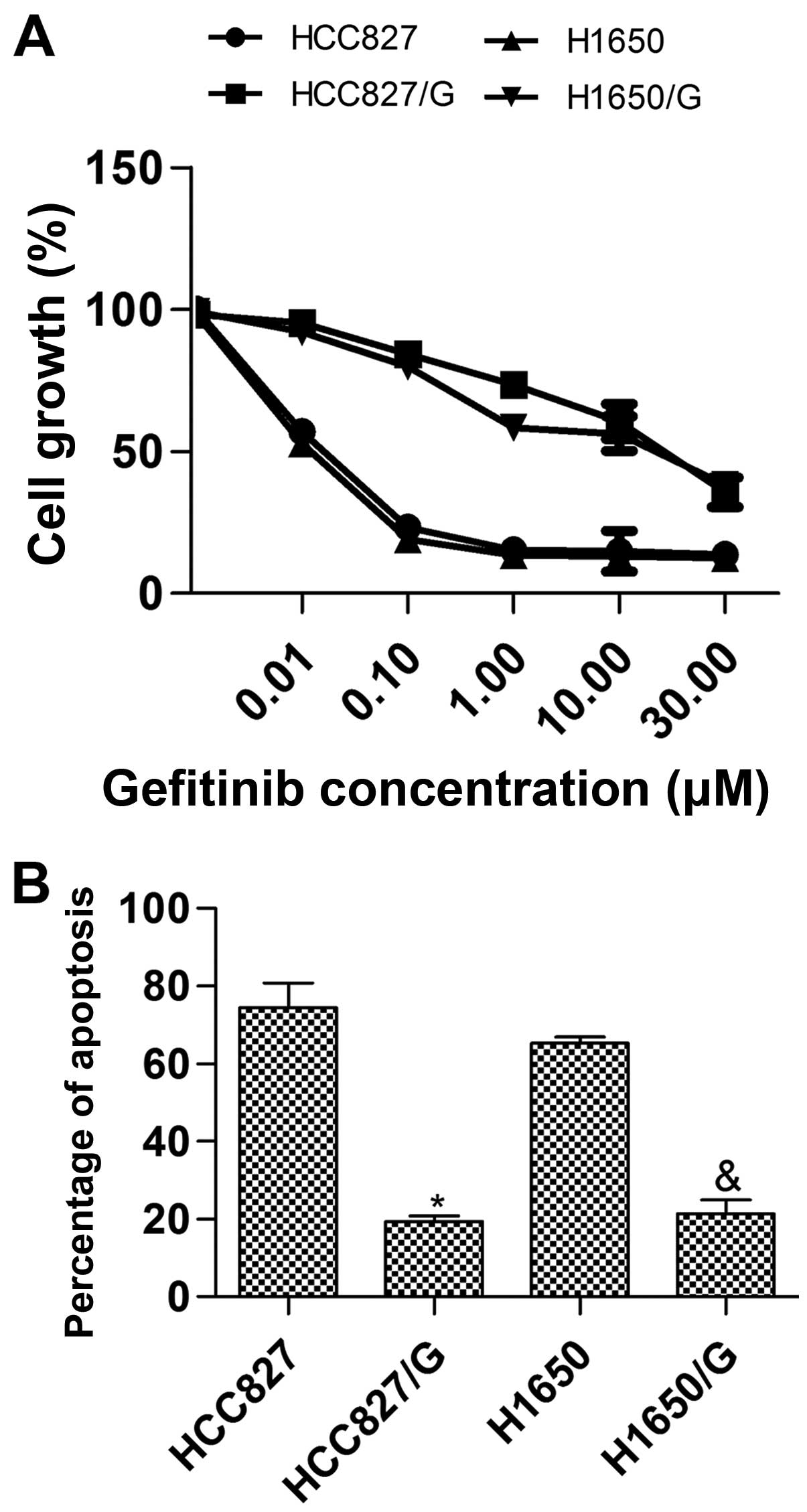
37°C. In the MTT assay, the percent survival (Fig. 1A) was calculated as: (mean
absorbance of six replicate wells containing drugs - mean
absorbance of six replicate background wells)/(mean absorbance of
six replicate drug-free wells - mean absorbance of six replicate
background wells) ×100. The results showed that the half maximal
inhibitory concentration (IC50) of HCC827/G and H1650/G
cell lines were 16.5±6.3 and 14.1±3.7 µmol/l, respectively,
and the IC50 of HCC827 and H1650 cell lines were
0.03±0.02 and 0.02±0.01 µmol/l, respectively. The drug
resistance of HCC827/G or H1650/G was significantly higher than
that of HCC827 or H1650 (P<0.01). In the Annexin V-FITC/PI
apoptosis detection assay, the cells cultured with 1 µmol/l
gefitinib were measured, and the results indicated that the
apoptosis induced by gefitinib in HCC827/G or H1650/G was lower
than that in HCC827 or H1650, respectively (Fig. 1B). In conclusion, the establishment
of drug resistant cell lines to gefitinib was successful.
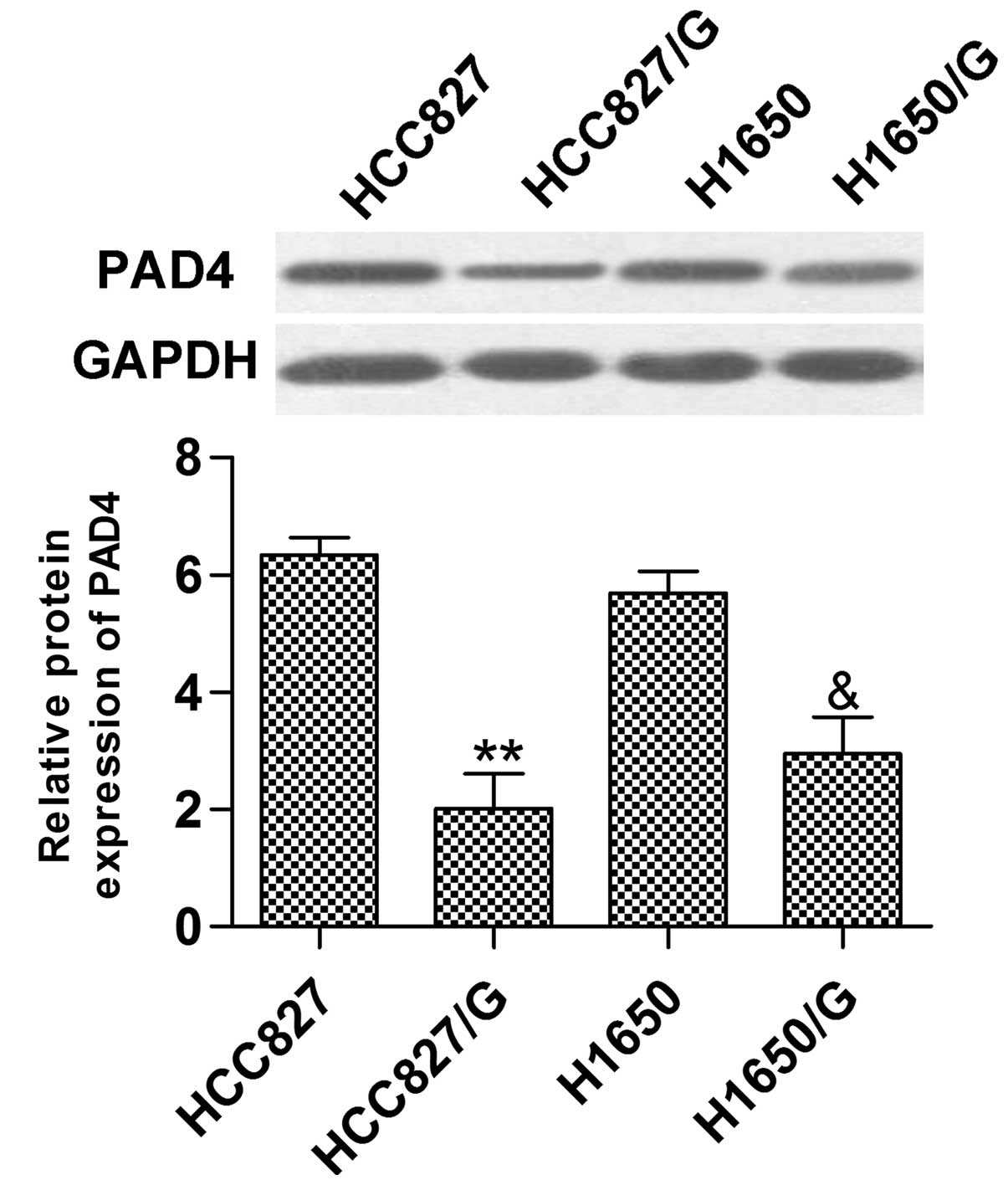
Additionally, the relative protein expression of PAD4 in HCC827/G
or H1650/G was distinctly reduced compared with HCC827 or H1650
(Fig. 2).
Overexpression of PAD4 suppresses drug
resistance to gefitinib
To investigate the influence of PAD4 expression
changes on the drug resistance of NSCLC cell lines to gefitinib, we
promoted and inhibited the expression of PAD4, and cultured the
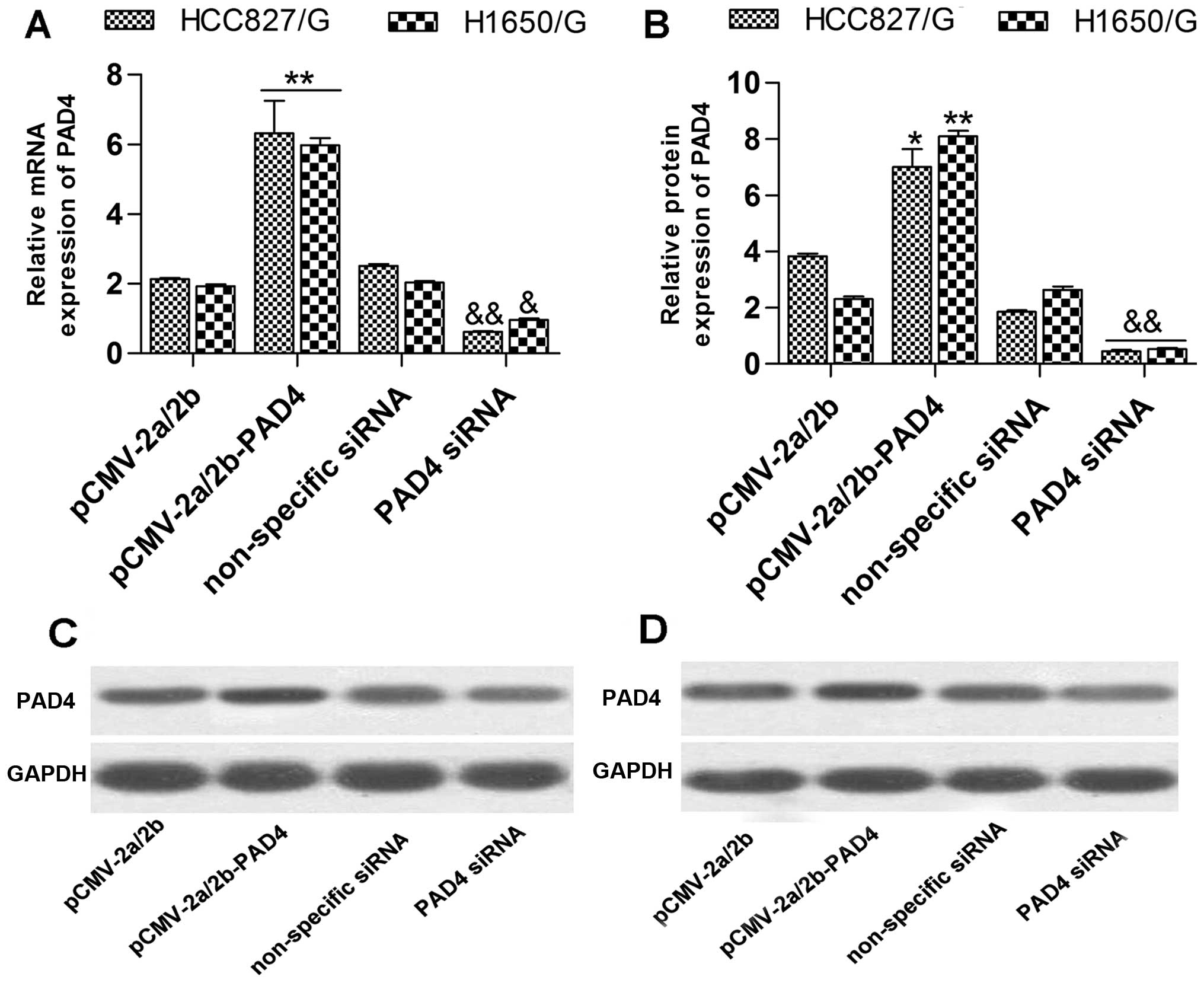
cells with 1 µmol/l of gefitinib. The detection showed that
the mRNA (Fig. 3A) and protein
(Fig. 3B–D) expression levels of
PAD4 in the PAD4 siRNA transfected group were significantly lower
than in the non-specific siRNA transfected group. The mRNA
(Fig. 3A) and protein (Fig. 3B–D) expression levels of PAD4 in the
pCMV-2a/2b-PAD4 transfected group were significantly upregulated
contrasting with the pCMV-2a/2b transfected group. Subsequently,
the effect of PAD4 overexpression or inhibition on drug resistance
was measured by MTT assay and Annexin V-FITC/PI apoptosis
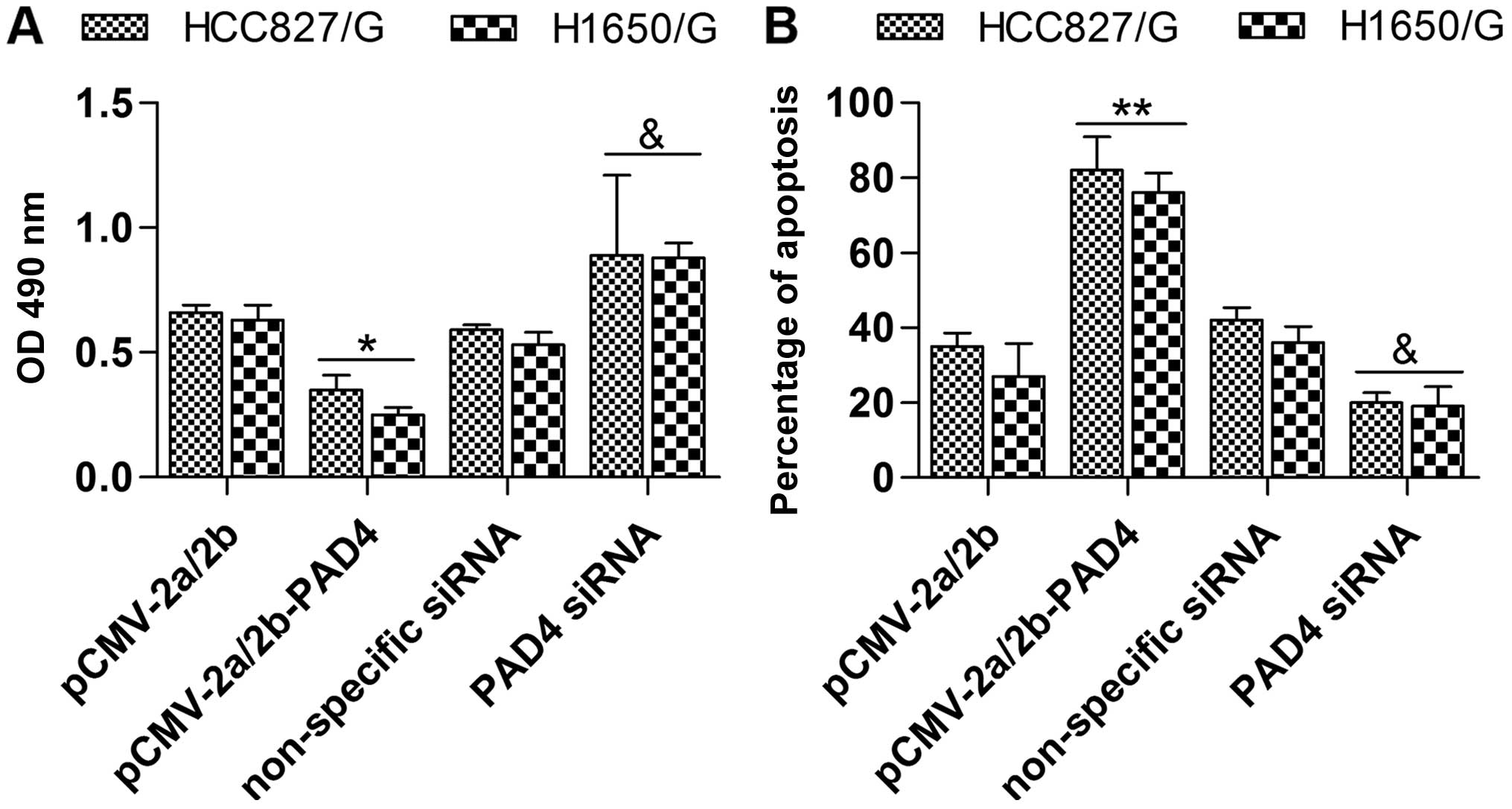
detection. In the MTT assay, the cell growth and viability of the
pCMV-2a/2b-PAD4 transfected group was obviously decreased in
contrast with the pCMV-2a/2b transfected group (Fig. 4A). Conversely, inhibition of PAD4 by
transfection of PAD4 siRNA exhibited the opposite effect (Fig. 4A). Moreover, in the Annexin
V-FITC/PI apoptosis detection, pCMV-2a/2b-PAD4 transfection induced
apoptosis compared with the pCMV-2a/2b transfection (Fig. 4B). Additionally, PAD4 siRNA
transfection had the opposite effect of pCMV-2a/2b-PAD4
transfection (Fig. 4B). To
summarize, upregulation of PAD4 suppressed drug resistance of NSCLC
cell lines to gefitinib.
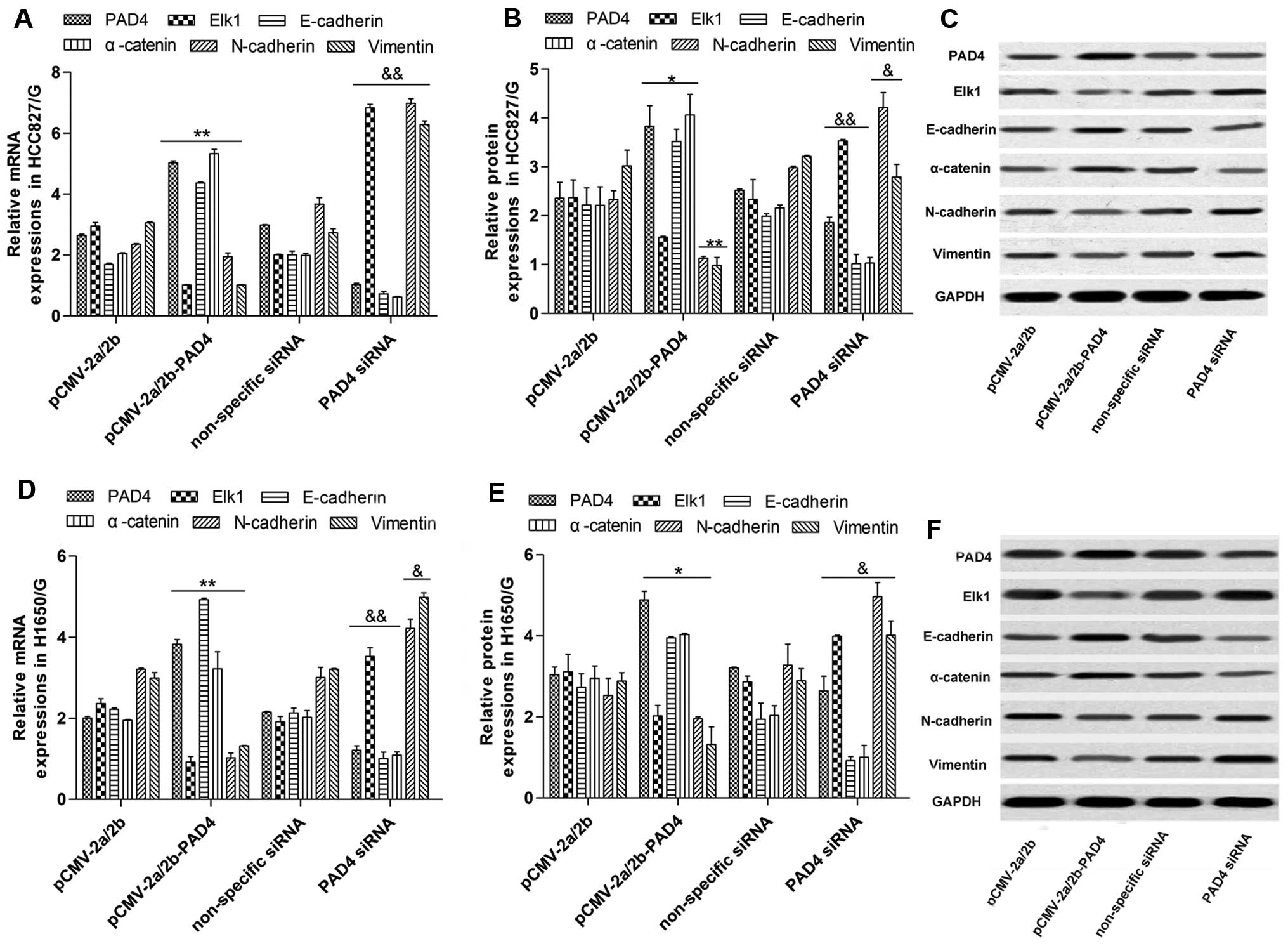
Overexpression of PAD4 constrains Elk1
and EMT
We identified that Elk1 expression and the EMT
activity were both inhibited by PAD4 overexpression in the cells
cultured with 1 µmol/l of gefitinib. The mRNA (Fig. 5A and D) and protein (Fig. 5B, C, E and F) of Elk1, N-cadherin
and vimentin were significantly downregulated by pCMV-2a/2b-PAD4
transfection while E-cadherin and α-catenin were upregulated. In
addition, PAD4 siRNA transfection had a reverse impact compared
with pCMV-2a/2b-PAD4 transfection (Fig.
5).
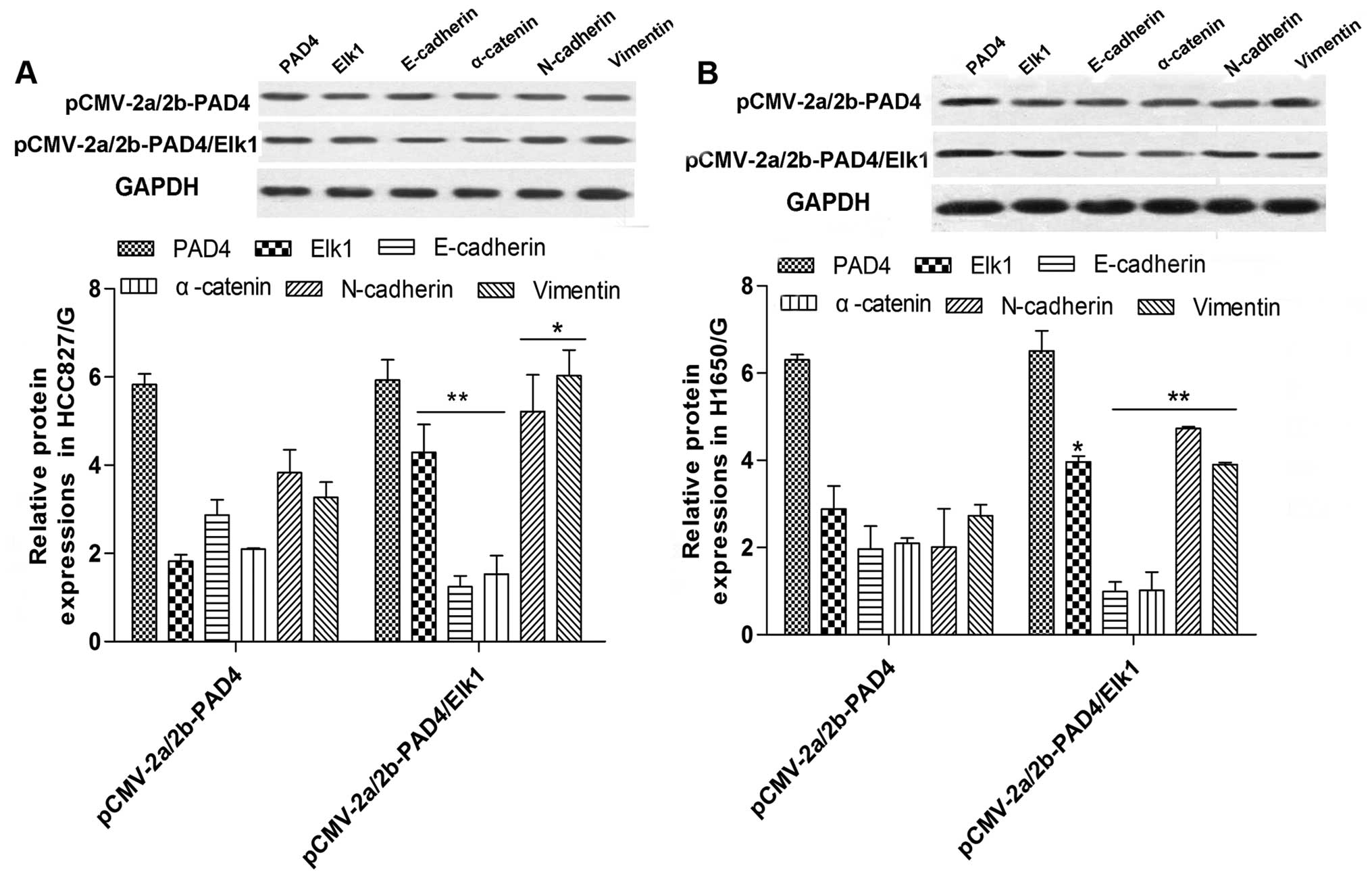
Overexpression of PAD4 inhibits EMT
through suppression of Elk1
We demonstrated that PAD4 overexpression inhibits
both EMT and Elk1. Moreover, it has been reported that Elk1 can
regulate the process of EMT (22).
So we hypothesized that PAD4 upregulation limits EMT by decreasing
Elk1 expression. To this end, we overexpressed both Elk1 and PAD4
by transfecting the plasmids pCMV-2a/2b-Elk1 and pCMV-2a/2b-PAD4
and cultured the cells with 1 µmol/l of gefitinib. The
results demonstrated that Elk1 and PAD4 were highly expressed
(Fig. 6) and the overexpression of
Elk1 apparently reversed the downregulated influence of PAD4
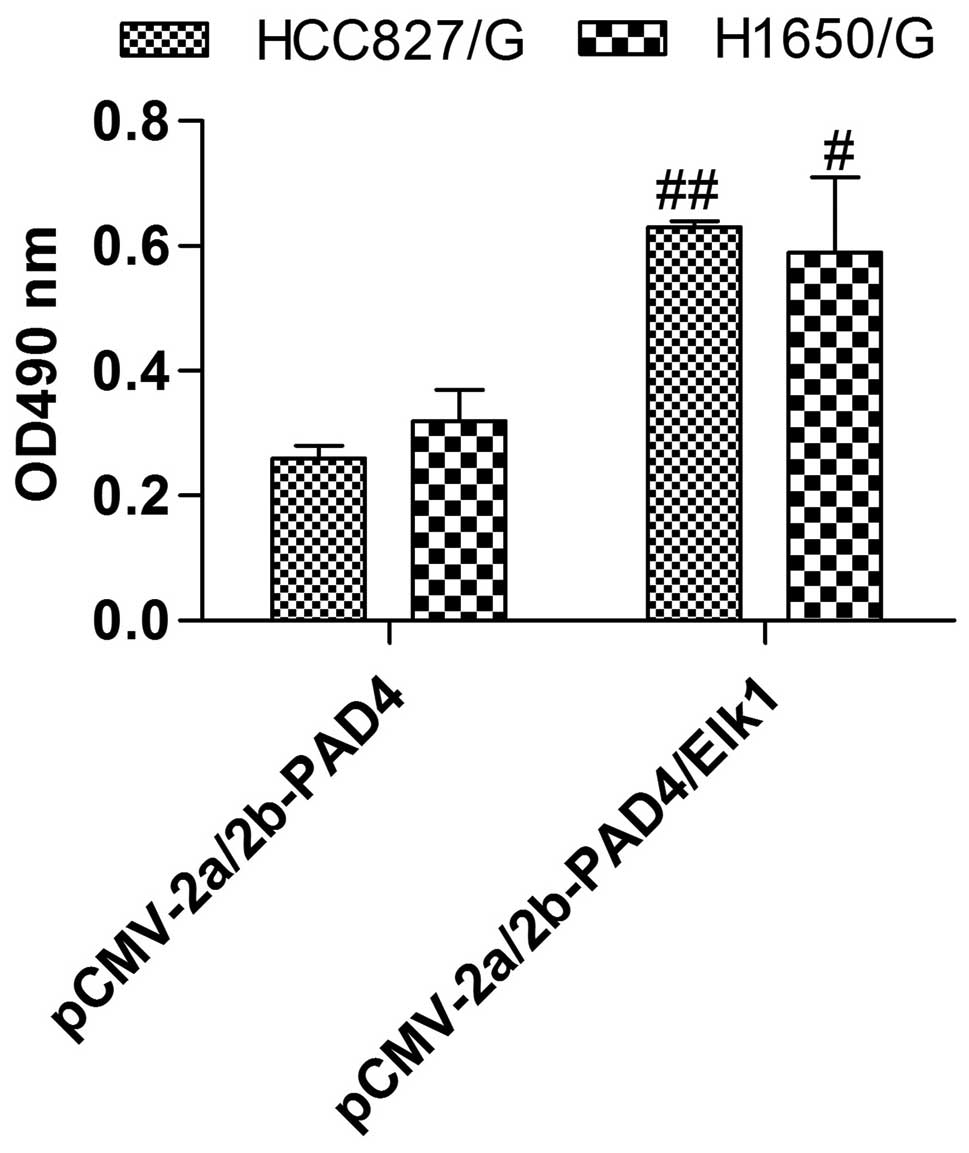
upregulation on EMT (Fig. 6).
Furthermore, Elk1 overexpression evidently abolished the drug
resistance suppression of PAD4 overexpression, so the cell growth
and viability were distinctly increased compared with PAD4
overexpression only (Fig. 7). Above
all, PAD4 overexpression inhibited the drug resistance by
controlling the expression of Elk1.
Discussion
Lung cancer is associated with significant morbidity
and mortality, and seriously threatens human health and life
(29). In the past 50 years, many
countries reported significantly higher lung cancer incidence and
mortality rates. Male lung cancer incidence and mortality rates are
the highest of all malignant tumors, and female lung cancer is in
second place. Non-small cell lung cancer containing glandular
cancer, squamous carcinoma and large cell carcinoma comprises 80%
of lung cancer (30–32). Gefitinib (Iressa), is one of the
effective drugs for advanced NSCLC. Nevertheless, acquired
resistance always appears and puzzles researchers.
Peptidylarginine deiminase IV is a
Ca2+-dependent enzyme that converts arginine and
methylarginine residues to citrulline (16). It is said that PAD4 helps to
regulate immune cell differentiation and cell death (33). Additionally, it plays an important
role in many diseases such as rheumatoid arthritis (RA), and it is
the main treatment of RA. For breast cancer, inhibition of PAD4 can
induce activity of EMT, and EMT is related to the resistance of
NSCLC to gefitinib (14,16). In this study, we established the
NSCLC cell lines that are resistant to gefitinib according to the
literature, and we found that the expression of PAD4 was
significantly downregulated in those cells. Next, we constructed
the recombinant plasmid pCMV-2a/2b-PAD4, and then transfected the
plasmid into the cells, obtaining resistance to gefitinib. The
results indicated that the resistance of gefitinib was obviously
inhibited following the successful overexpression of PAD4.
Moreover, PAD4 siRNA was transfected and the reduction of PAD4
expression had the distinct reverse effect compared with
upregulation of PAD4. Other studies demonstrated that PAD4 could
act on Elk-1, and Elk1 could influence the process of EMT. However,
studies have not determined if PAD4 can regulate Elk1
expression.
Elk1 encodes a related ternary complex factor (TCF)
subfamily protein of ETS-domain transcription factors (34). Also, Elk-1 controls the expression
of other genes such as oncogene c-fos, smooth muscle-specific
genes, and immediate early genes (IEGs) and plays a role in cell
apoptosis, proliferation and cancer development (35,36).
It was demonstrated that Elk1 is upregulated in NSCLC and is nearly
undetectable in normal tissue. Therefore, it was speculated that
Elk1 probably plays a role in tumorigenesis (37). In our investigation, we found that
Elk1 could be regulated by PAD4 in the NSCLC cell lines resistant
to gefitinib. Elk1 was distinctly reduced by overexpression of
PAD4, whereas it was increased with PAD4 downregulation.
Epithelial-to-mesenchymal transition (EMT) is a
basic physiologic and pathologic phenomena involved in embryonic
formation and development and tumor invasion and metastasis
(38,39). Research findings related to EMT
originated from embryonic development studies, whereas in recent
years, more are concerned with the study of tumors. Studies have
shown that EMT promotes acquired resistance to various apoptotic
stimuli (40). In NSCLC treatment,
acquired resistance to gefitinib always appears. However, research
demonstrated that EMT is induced in gefitinib-acquired resistance
in lung cancer cells (14). In our
study, we found that overexpression of PAD4 obviously inhibited
Elk1 and EMT activity, inducing the suppression of resistance to
gefitinib. Nevertheless, the co-overexpression of PAD4 and Elk1
promoted EMT, reversing the effect on resistance. Therefore, we
confirmed that PAD4 overexpression could suppress the resistance of
NSCLC cells to gefitinib.
In summary, this study revealed that upregulation of
PAD4 inhibited the resistance of NSCLC to gefitinib through
reducing the expression of Elk1. Our study identified the role of
PAD4 in gefitinib-acquired resistance in NSCLC, providing a novel
target and potential therapeutic strategy to prevent treatment
resistance.
Abbreviations:
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
PAD4
|
peptidylarginine deiminase IV
|
|
NSCLC
|
non-small cell lung cancer
|
|
TKI
|
tyrosine kinase inhibitors
|
|
EGFR
|
epidermal growth factor receptor
|
|
RA
|
rheumatoid arthritis
|
|
MTT
|
3-(4,5-dimethyl
thiazol-2-yl)-2,5-diphenyltetrazolium bromide
|
|
DMSO
|
dimethyl sulfoxide
|
|
Annexin V-FITC/PI
|
Annexin V-fluorescein
isothiocyanate/propidium iodide
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
PMSF
|
phenylmethanesulfonyl fluoride
|
|
SDS-PAGE
|
sodium dodecyl sulfate-polyacrylamide
gel electrophoresis
|
Acknowledgments
This study was supported by grants from the Natural
Science Foundation of China (nos. 81371891 and 31170880).
References
|
1
|
Church TR, Black WC, Aberle DR, Berg CD,
Clingan KL, Duan F, Fagerstrom RM, Gareen IF, Gierada DS, Jones GC,
et al National Lung Screening Trial Research Team: Results of
initial low-dose computed tomographic screening for lung cancer. N
Engl J Med. 368:1980–1991. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saghir Z, Dirksen A, Ashraf H, Bach KS,
Brodersen J, Clementsen PF, Døssing M, Hansen H, Kofoed KF, Larsen
KR, et al: CT screening for lung cancer brings forward early
disease. The randomised Danish Lung Cancer Screening Trial: Status
after five annual screening rounds with low-dose CT. Thorax.
67:296–301. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Das M, Riess JW, Frankel P, Schwartz E,
Bennis R, Hsieh HB, Liu X, Ly JC, Zhou L, Nieva JJ, et al: ERCC1
expression in circulating tumor cells (CTCs) using a novel
detection platform correlates with progression-free survival (PFS)
in patients with metastatic non-small-cell lung cancer (NSCLC)
receiving platinum chemotherapy. Lung Cancer. 77:421–426. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maemondo M, Minegishi Y, Inoue A,
Kobayashi K, Harada M, Okinaga S, Morikawa N, Oizumi S, Tanaka T,
Isobe H, et al: First-line gefitinib in patients aged 75 or older
with advanced non-small cell lung cancer harboring epidermal growth
factor receptor mutations: NEJ 003 study. J Thorac Oncol.
7:1417–1422. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Warth A, Muley T, Meister M, Stenzinger A,
Thomas M, Schirmacher P, Schnabel PA, Budczies J, Hoffmann H and
Weichert W: The novel histologic International Association for the
Study of Lung Cancer/American Thoracic Society/European Respiratory
Society classification system of lung adenocarcinoma is a
stage-independent predictor of survival. J Clin Oncol.
30:1438–1446. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cross DA, Ashton SE, Ghiorghiu S, Eberlein
C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MR, Ward RA, Mellor MJ,
et al: AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated
resistance to EGFR inhibitors in lung cancer. Cancer Discov.
4:1046–1061. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wynes MW, Hinz TK, Gao D, Martini M, Marek
LA, Ware KE, Edwards MG, Böhm D, Perner S, Helfrich BA, et al:
FGFR1 mRNA and protein expression, not gene copy number, predict
FGFR TKI sensitivity across all lung cancer histologies. Clin
Cancer Res. 20:3299–3309. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Laurila N and Koivunen JP: EGFR inhibitor
and chemotherapy combinations for acquired TKI resistance in
EGFR-mutant NSCLC models. Med Oncol. 32:2052015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li L, Han R, Xiao H, Lin C, Wang Y, Liu H,
Li K, Chen H, Sun F, Yang Z, et al: Metformin sensitizes
EGFR-TKI-resistant human lung cancer cells in vitro and in vivo
through inhibition of IL-6 signaling and EMT reversal. Clin Cancer
Res. 20:2714–2726. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su KY, Chen HY, Li KC, Kuo ML, Yang JC,
Chan WK, Ho BC, Chang GC, Shih JY, Yu SL, et al: Pretreatment
epidermal growth factor receptor (EGFR) T790M mutation predicts
shorter EGFR tyrosine kinase inhibitor response duration in
patients with non-small-cell lung cancer. J Clin Oncol. 30:433–440.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miyauchi E, Inoue A, Kobayashi K, Maemondo
M, Sugawara S, Oizumi S, Isobe H, Gemma A, Saijo Y, Yoshizawa H, et
al North-East Japan Study Group: Efficacy of chemotherapy after
first-line gefitinib therapy in EGFR mutation-positive advanced
non-small cell lung cancer-data from a randomized Phase III study
comparing gefitinib with carboplatin plus paclitaxel (NEJ002). Jpn
J Clin Oncol. 45:670–676. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kosaka T, Yamaki E, Mogi A and Kuwano H:
Mechanisms of resistance to EGFR TKIs and development of a new
generation of drugs in non-small-cell lung cancer. J Biomed
Biotechnol. 2011:1652142011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang JJ, Chen HJ, Yan HH, Zhang XC, Zhou
Q, Su J, Wang Z, Xu CR, Huang YS, Wang BC, et al: Clinical modes of
EGFR tyrosine kinase inhibitor failure and subsequent management in
advanced non-small cell lung cancer. Lung Cancer. 79:33–39. 2013.
View Article : Google Scholar
|
|
14
|
Nurwidya F, Takahashi F, Murakami A and
Takahashi K: Epithelial mesenchymal transition in drug resistance
and metastasis of lung cancer. Cancer Res Treat. 44:151–156. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Suda K, Tomizawa K, Fujii M, Murakami H,
Osada H, Maehara Y, Yatabe Y, Sekido Y and Mitsudomi T: Epithelial
to mesenchymal transition in an epidermal growth factor
receptor-mutant lung cancer cell line with acquired resistance to
erlotinib. J Thorac Oncol. 6:1152–1161. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stadler SC, Vincent CT, Fedorov VD,
Patsialou A, Cherrington BD, Wakshlag JJ, Mohanan S, Zee BM, Zhang
X, Garcia BA, et al: Dysregulation of PAD4-mediated citrullination
of nuclear GSK3β activates TGF-β signaling and induces
epithelial-to-mesenchymal transition in breast cancer cells. Proc
Natl Acad Sci USA. 110:11851–11856. 2013. View Article : Google Scholar
|
|
17
|
Ham A, Rabadi M, Kim M, Brown KM, Ma Z,
D'Agati V and Lee HT: Peptidyl arginine deiminase-4 activation
exacerbates kidney ischemia-reperfusion injury. Am J Physiol Renal
Physiol. 307:F1052–F1062. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Baka Z, Barta P, Losonczy G, Krenács T,
Pápay J, Szarka E, Sármay G, Babos F, Magyar A, Géher P, et al:
Specific expression of PAD4 and citrullinated proteins in lung
cancer is not associated with anti-CCP antibody production. Int
Immunol. 23:–414. 4052011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Patki M, Chari V, Sivakumaran S, Gonit M,
Trumbly R and Ratnam M: The ETS domain transcription factor ELK1
directs a critical component of growth signaling by the androgen
receptor in prostate cancer cells. J Biol Chem. 288:11047–11065.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Senecal A, Munsky B, Proux F, Ly N, Braye
FE, Zimmer C, Mueller F and Darzacq X: Transcription factors
modulate c-Fos transcriptional bursts. Cell Rep. 8:75–83. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang R, Wang J, Ma S, Huang Z and Zhang
G: Requirement of Osteopontin in the migration and protection
against Taxol-induced apoptosis via the ATX-LPA axis in SGC7901
cells. BMC Cell Biol. 12:112011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Obacz J, Takacova M, Brychtova V, Dobes P,
Pastorekova S, Vojtesek B and Hrstka R: The role of AGR2 and AGR3
in cancer: Similar but not identical. Eur J Cell Biol. 94:139–147.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bicker KL and Thompson PR: The protein
arginine deiminases: Structure, function, inhibition, and disease.
Biopolymers. 99:155–163. 2013. View Article : Google Scholar
|
|
24
|
Eades G, Yao Y, Yang M, Zhang Y, Chumsri S
and Zhou Q: miR-200a regulates SIRT1 expression and epithelial to
mesenchymal transition (EMT)-like transformation in mammary
epithelial cells. J Biol Chem. 286:25992–26002. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Saitoh M, Shirakihara T and Miyazono K:
Regulation of the stability of cell surface E-cadherin by the
proteasome. Biochem Biophys Res Commun. 381:560–565. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cavallaro U, Schaffhauser B and
Christofori G: Cadherins and the tumour progression: Is it all in a
switch? Cancer Lett. 176:123–128. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu Q, Hou X, Xia J, Qian X, Miele L,
Sarkar FH and Wang Z: Emerging roles of PDGF-D in EMT progression
during tumorigenesis. Cancer Treat Rev. 39:640–646. 2013.
View Article : Google Scholar :
|
|
28
|
Engelman JA, Zejnullahu K, Mitsudomi T,
Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen
J, et al: MET amplification leads to gefitinib resistance in lung
cancer by activating ERBB3 signaling. Science. 316:1039–1043. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Field JK, Oudkerk M, Pedersen JH and Duffy
SW: Prospects for population screening and diagnosis of lung
cancer. Lancet. 382:732–741. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu YH, Mei JS and Zhou J: Randomized study
of gefitinib versus pemetrexed as maintenance treatment in patients
with advanced glandular non-small cell lung cancer. Int J Clin Exp
Med. 8:6242–6246. 2015.PubMed/NCBI
|
|
31
|
Chaft JE, Rekhtman N, Ladanyi M and Riely
GJ: ALK-rearranged lung cancer: Adenosquamous lung cancer
masquerading as pure squamous carcinoma. J Thorac Oncol. 7:768–769.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pao W and Girard N: New driver mutations
in non-small-cell lung cancer. Lancet Oncol. 12:175–180. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wagner S, Stuttmann J, Rietz S, Guerois R,
Brunstein E, Bautor J, Niefind K and Parker JE: Structural basis
for signaling by exclusive EDS1 heteromeric complexes with SAG101
or PAD4 in plant innate immunity. Cell Host Microbe. 14:619–630.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Horn S, Figl A, Rachakonda PS, Fischer C,
Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al:
TERT promoter mutations in familial and sporadic melanoma. Science.
339:959–961. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chevigny M, Guérin-Montpetit K, Vargas A,
Lefebvre-Lavoie J and Lavoie JP: Contribution of SRF, Elk-1, and
myocardin to airway smooth muscle remodeling in heaves, an
asthma-like disease of horses. Am J Physiol Lung Cell Mol Physiol.
309:L37–L45. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Neeb A, Wallbaum S, Novac N,
Dukovic-Schulze S, Scholl I, Schreiber C, Schlag P, Moll J, Stein U
and Sleeman JP: The immediate early gene Ier2 promotes tumor cell
motility and metastasis, and predicts poor survival of colorectal
cancer patients. Oncogene. 31:3796–3806. 2012. View Article : Google Scholar
|
|
37
|
Kawahara T, Shareef HK, Aljarah AK, Ide H,
Li Y, Kashiwagi E, Netto GJ, Zheng Y and Miyamoto H: ELK1 is
up-regulated by androgen in bladder cancer cells and promotes tumor
progression. Oncotarget. 6:29860–29876. 2015.PubMed/NCBI
|
|
38
|
Nieto MA: Epithelial plasticity: A common
theme in embryonic and cancer cells. Science. 342:12348502013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rokavec M, Öner MG, Li H, Jackstadt R,
Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et
al: IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated
colorectal cancer invasion and metastasis. J Clin Invest.
124:1853–1867. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kong D, Li Y, Wang Z and Sarkar FH: Cancer
stem cells and epithelial-to-mesenchymal transition
(EMT)-phenotypic cells: Are they cousins or twins? Cancers (Basel).
3:716–729. 2011. View Article : Google Scholar
|