Introduction
Colorectal cancer (CRC) is one of the three leading
causes of cancer-related deaths worldwide. Despite rapidly
developing novel targeted therapies, 5-fluorouracil (5-Fu) is still
recognized as the mainstay in colorectal cancer chemotherapy.
However, the response rate to 5-Fu in colorectal cancer adjuvant
chemotherapy is less than 50%, and recurrence of colon cancer
occurs in almost 50% of patients treated by standard therapeutics,
according to NCCN (National Comprehensive Cancer Network) (1). Resistance to 5-Fu has been attributed,
at least in part, to enhance DNA repair.
The B7 family is an immunoglobulin superfamily,
which consists of B7-1, B7-2, B7-H1, B7-H2, B7-DC, B7-H3, B7-H4,
B7-H5 and B7-H6 (2). As key
immunity modulating molecules, the B7 family is important for the
treatment of cancer, transplantation and autoimmune diseases
(3). B7-H3 was first identified in
monocytes and dendritic cells (4),
which regulate T cell functions, including proliferation, apoptosis
and adhesion. B7-H3 is also overexpressed in tumor tissue, where it
is associated with cancer metastasis and poor prognosis (5–9),
suggesting a role for B7-H3 in tumor progression (8). Our previous results indicated that
overexpressed B7-H3 significantly increased anti-apoptosis and the
metastasis ability of CRC cells.
According to our mRNA microarray data, increasing
the expression of B7-H3 induced a subsequent increase in BRCC3
expression. BRCC3 and BRCA1/BRCA2 are known to form a complex that
repairs DNA damage (10).
Downregulation of BRCC3 causes glioma to become more sensitive to
chemotherapy (11). The roles of
BRCA1 and BRCA2 in breast, ovarian, and other cancer have been
reported in many clinical and experimental studies (12,13).
Results indicate that the BRCA1/BRCA2/BRCC3 DNA damage repair
complex is closely related to tumor recurrence and metastasis. In
the present study, we focused on the relationship between B7-H3 and
BRCC3 to investigate the mechanism of B7-H3-induced 5-Fu resistance
due to enhanced DNA repair.
Materials and methods
Cells and reagents
Two human CRC cell lines, SW480 and HCT-8, exhibit
different expression levels of B7-H3. We constructed SW480 cells
that expressed high levels of B7-H3 (SW480-B7-H3) and HCT-8 cells
that were stably transfected with B7-H3 shRNA (HCT-8-shB7-H3).
Cells transfected with a mock vector were used as negative controls
(SW480-NC and HCT-8-NC). Cells were maintained in Dulbecco's
modified Eagle's medium or RPMI-1640 medium, supplemented with 10%
fetal bovine serum (FBS), at 37°C in a humidified atmosphere with
5% CO2.
5-fluorouracil (5-Fu) was obtained from
Sigma-Aldrich (St. Louis, MO, USA). Anti-human B7-H3, β-actin and
BRCC3 antibodies were purchased from Abcam (Cambridge, MA, USA);
the α-tubulin antibody was purchased from Biotime (Shanghai,
China); the horseradish peroxidase-conjugated secondary anti-rabbit
antibodies were purchased from Beyotime Institute of Biotechnology
(Nantong, China); the secondary antibodies fluorophore-labeled
donkey anti-rabbit IgG (A11374) were purchased from Invitrogen
(Carlsbad, CA, USA).
CCK-8 assay
SW480-NC/SW480-B7-H3 or HCT-8-NC/HCT-8-shB7-H3 cells
were seeded at 3,000 cells/well in 96-well plates in triplicate.
The next day, the medium was replaced with medium containing 5-Fu
at a 2-fold concentration gradient. After 48 h, 10 µl CCK-8
was added to each well, and the absorbance was recorded on a BioTek
microplate reader at the wavelength of 450 nm. Cell viability was
assessed in percent wherein vehicle-treated cells were taken as
100%. Half maximal inhibitory concentration (IC50) was
calculated using GraphPad Prism 5 (GraphPad Software, Inc., La
Jolla, CA, USA).
Real-time quantitative polymerase chain
reaction
Total RNA was isolated from 1.5×106 cells
using TRIzol, following the manufacturer's instructions, and then
treated with RNase-free DNase to remove residual genomic DNA. The
first strand cDNA was synthesized from 1 µg RNA using a
SuperScript First-Strand Synthesis system (Invitrogen).
First-strand cDNA was amplified in a 20 µl PCR reaction
mixture: 10 µl 2X SYBR-Green PCR Master Mix, 0.4 µl
50X ROX, 0.4 µl of each specific primer set and
ddH2O added to 20 µl. The sequences of primers
were as follows: β-actin 5′-AGCGAGCATCCCCCAAAGTT-3′ (sense) and
5′-GGGCACGAAGGCTCATCATT-3′ (antisense); BRCC3
5′-AGAGTTCAGAGTATGAGAGAATCG-3′ (sense) and
5′-TCTTGGTTACTGAGTCCAGATGT-3′ (antisense); The PCR cycling
consisted of 40 cycles of amplification of the cDNA with annealing
at 60°C.
Western blot assay
Western blot assays were performed on whole-cell
extracts prepared by lysing 1×106 cells containing 50 mM
Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate,
0.1% SDS and PMSF protease inhibitor. Equal protein loading of the
lysates was achieved by standardization with the BCA protein assay
kit from Biotime. Samples were separated on SDS-PAGE gels and
transferred to nitrocellulose transfer membranes. After blocking
with 5% skim milk in TBST, the membranes were incubated with the
appropriate primary antibody (α-tubulin or BRCC3) overnight at 4°C,
followed by incubation with HRP-conjugated secondary antibody for 2
h at room temperature. The protein bands were visualized with an
enhanced chemiluminescence.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 20 min
and permeabilized with 0.1% Triton X-100 for 10 min at room
temperature (RT). Cells were washed three times in PBS after each
treatment. After blocking with 2% BSA in PBS for 30 min at room
temperature, the cells were incubated with primary antibody in 2%
BSA/PBS overnight at 4°C and then incubated with fluorescent
secondary antibody for 1 h at room temperature. Cell nuclei were
stained with DAPI for 10 min at RT. Images were acquired with the
TSC SP8 confocal laser scanning microscope (Leica Microsystems,
Wetzlar, Germany).
Comet assay
The comet assay was performed under alkaline
conditions according to Singth et al (14). Cells were seeded in 12-well
tissue-culture plates and incubated for 24 h for cell attachment.
Subsequently, cells were treated with increasing concentrations of
5-Fu (positive control) for 24 h. Cells were harvested, washed and
resuspended in ice-cold PBS. Approximately 25 µl of the
resuspended cells were mixed with 75 µl of low melting point
agarose at 37°C; the suspension was spread over the well with the
pipette tip. The slides were placed at 4°C in the dark until
gelling occurred and then were immersed in pre-chilled lysis buffer
at 4°C. After incubating for 2 h, the buffer was aspirated and
replaced with pre-chilled alkaline solution for 30 min at 4°C.
After lysis and unwinding, the slides were placed in a horizontal
electrophoresis tank filled with freshly prepared alkaline
electrophoresis buffer. The electrophoresis was run at 20 V and 200
mA for 30 min. After electrophoresis, the slides were transferred
to pre-chilled distilled water for 2 min, then aspirated and
repeated twice. The final rinse was aspirated and replaced with
cold 70% ethanol for 5 min. Thereafter, the slides were allowed to
air dry, and 100 µl/well of PI was added to each slide for
20 min in the dark at room temperature for DNA staining. DNA
migration was observed using a Nikon 90i fluorescence microscope.
For each concentration, 6–10 randomly selected cells were analyzed.
All of the comet images were analyzed using CASP software (CASPlab,
Wroclaw, Poland) (15), and the
percentage of DNA in the comet tail (TDNA%), tail length (TL), tail
moment (TM) and olive tail moment (OTM) were recorded to
characterize the cell DNA damage. All the experiments were repeated
once, and the variation between experiments was analyzed.
Colony formation assay
Colony formation assays were performed by replating
cells at a density of 300 cells/well onto a 6-well culture plate in
medium containing 10% FBS. After 3 weeks, the cells were fixed with
ice-cold methanol for 10 min and then stained by using 0.05%
crystal violet in ddH2O for 15 min. Alternatively, for
examination of clonogenic ability of SW480 and HCT-8 cells with
5-Fu treatment, a density of 600 cells/well were seeded onto a
6-well culture plate in medium containing 10% FBS and 2
µg/ml 5-Fu or vehicle was added to the cultures at day 3
after seeding. The cultures were continuously maintained for
another 2 weeks and subjected to the colony formation assay.
Colonies produced by each cell-group were counted and measured
using ImageJ software.
Knockdown of BRCC3 by small interfering
RNA (siRNA)
siRNA against BRCC3 were chemically synthesized by
RiboBio Co., Ltd. (Guangzhou, China). Transfection of siRNAs was
performed using the Oligofectamine reagent (Invitrogen). The
sequences of siRNA-BRCC3 are as follows:
siRNA1-GCAGGAATTACAACAAGAA, siRNA2-GAAGGACCGAGTAGAAATT.
Statistical analyses
All experiments were re-performed at least three
times with similar results. Quantitative data were presented as the
average value of replicates ± SD (standard deviation) within the
representative experiment.
Results
Overexpression of B7-H3 weakens the
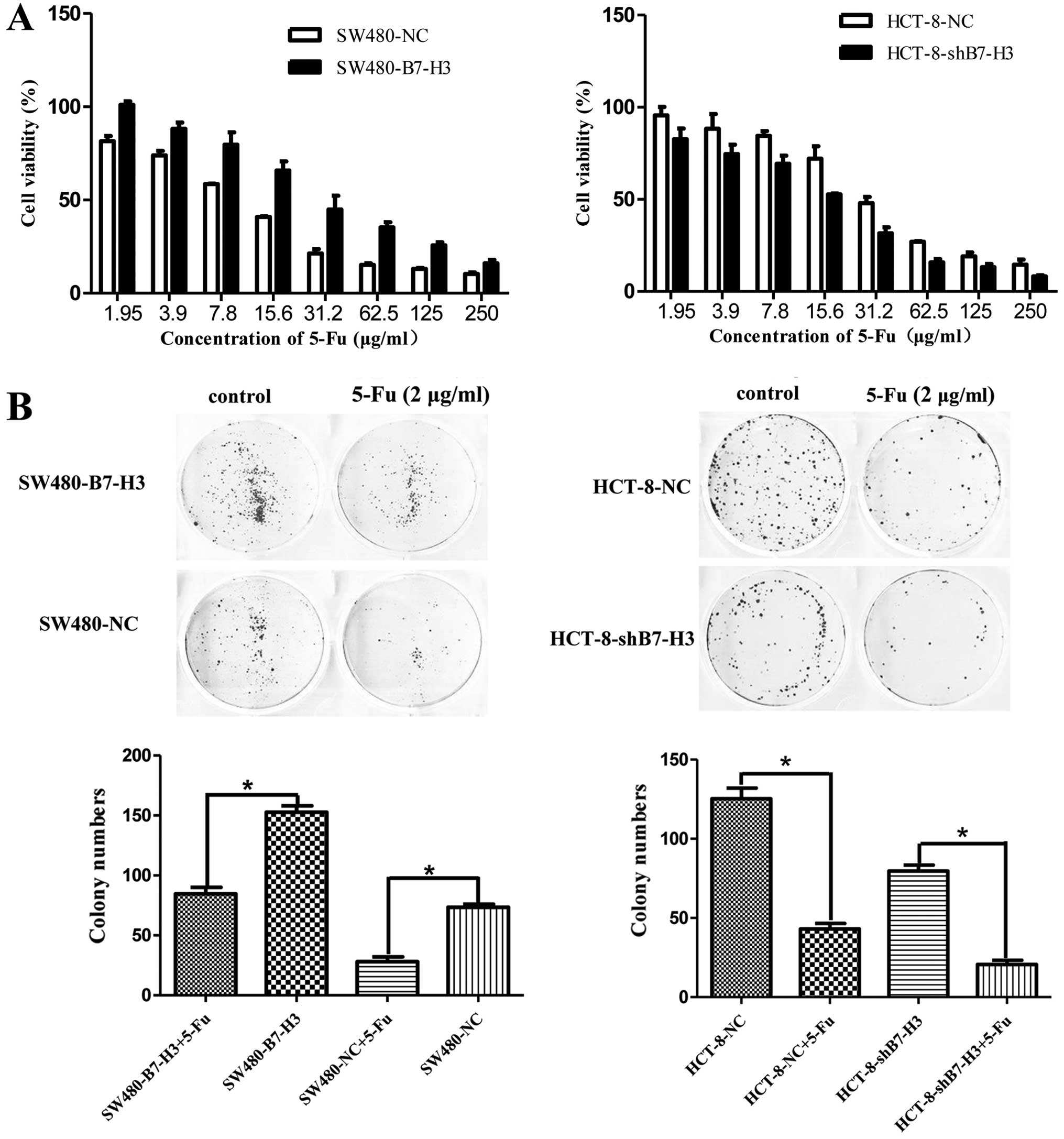
sensitivity of CRC cells to 5-Fu
To inhibit tumor growth, we incubated SW480-NC,
SW480-B7-H3, HCT-8-NC, or HCT-8-shB7-H3 cells with a concentration
gradient of 5-Fu for 48 h (Fig.
1A). CCK-8 assays showed that the inhibition rate of
SW480-B7-H3 was less than SW480-NC at any concentration of 5-Fu
(P<0.05). The IC50 of 5-Fu increased from 10.58 to
32.46 µg/ml after B7-H3 was upregulated. The HCT-8 pairs
showed similar results (P<0.05): the IC50 of 5-Fu
decreased from 31.28 to 14.87 µg/ml after B7-H3 was knocked
down. We next studied the ability of cancer cells to form colonies
on 6-well plates in presence or absence of 5-Fu. Consistent with
the CCK-8 assay results, the colony formation of cells
overexpressing B7-H3 (SW480-B7-H3 and HCT-8-NC) was increased
compared with the cells with low expression of B7-H3 (SW480-NC and
HCT-8-shB7-H3) (Fig. 1B).
Therefore, these observations demonstrate that the overexpression
of B7-H3 weakened the sensitivity of CRC cells to 5-Fu.
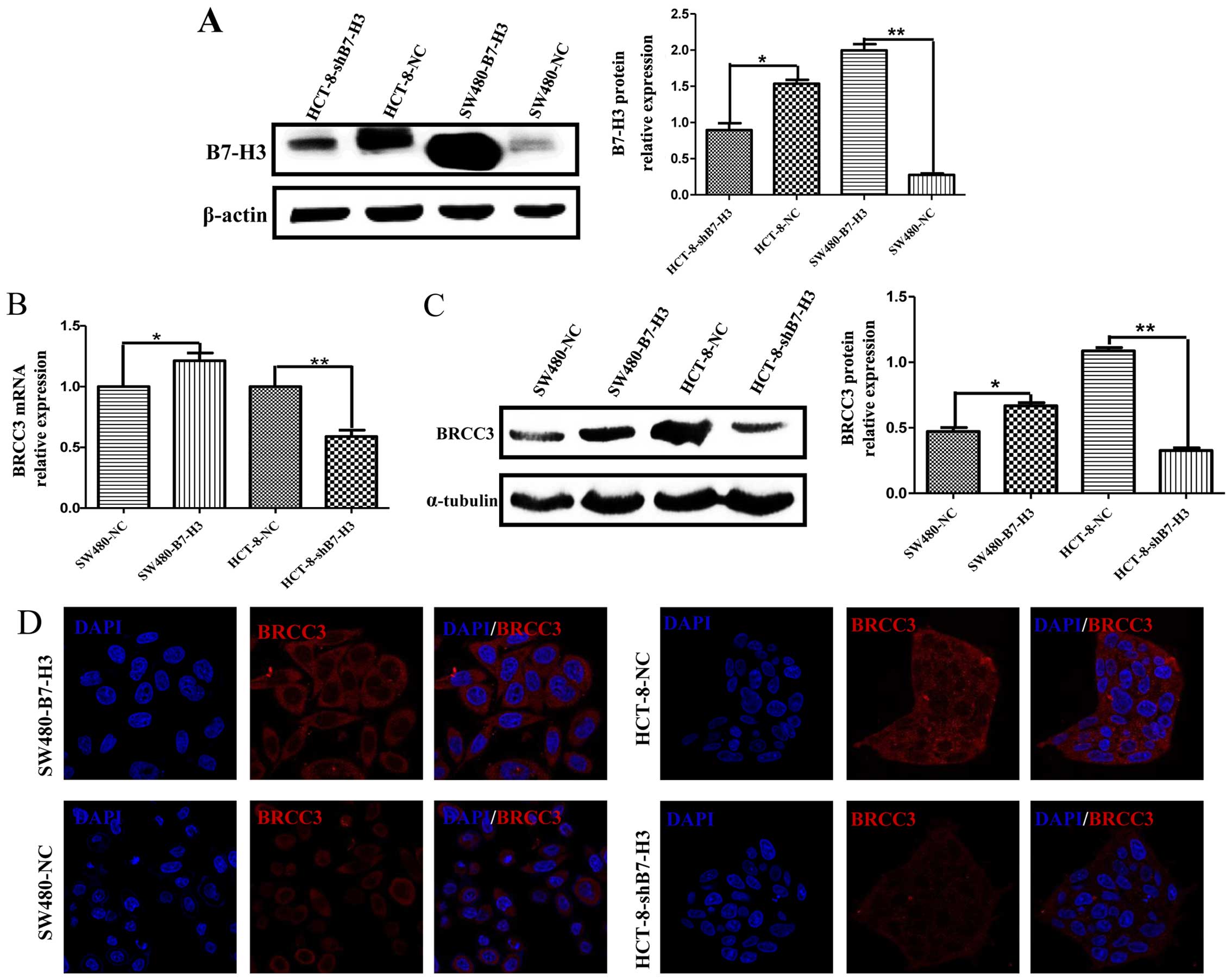
Overexpression of B7-H3 upregulated BRCC3
expression in CRC cells
The mechanism by which B7-H3 induces resis tance to
5-Fu in cancer cells is very complicated. We have previously
reported that B7-H3 could enhance cancer cell resistance to
apoptosis by increasing the expression of anti-apoptotic proteins
Bcl-2 and Bcl-xl. Here, we examined the expression of B7-H3 in
stably transfected CRC cells using western blot analysis (Fig. 2A) and analyzed BRCC3 expression in
response to B7-H3 overexpression or knockdown using real-time PCR
(Fig. 2B), western blot analysis
(Fig. 2C) and immunofluorescence
(Fig. 2D). Both mRNA and protein
levels of BRCC3 in SW480-NC, SW480-B7-H3, HCT-8-NC, and
HCT-8-shB7-H3 cells corresponded with B7-H3 expression, such that
B7-H3-overexpressing cells (SW480-B7-H3 and HCT-8-NC) demonstrated
higher BRCC3 expression compared to cells having low B7-H3
expression (SW480-NC and HCT-8-shB7-H3). Immunofluorescence showed
that BRCC3 expression was predominantly cytosolic.
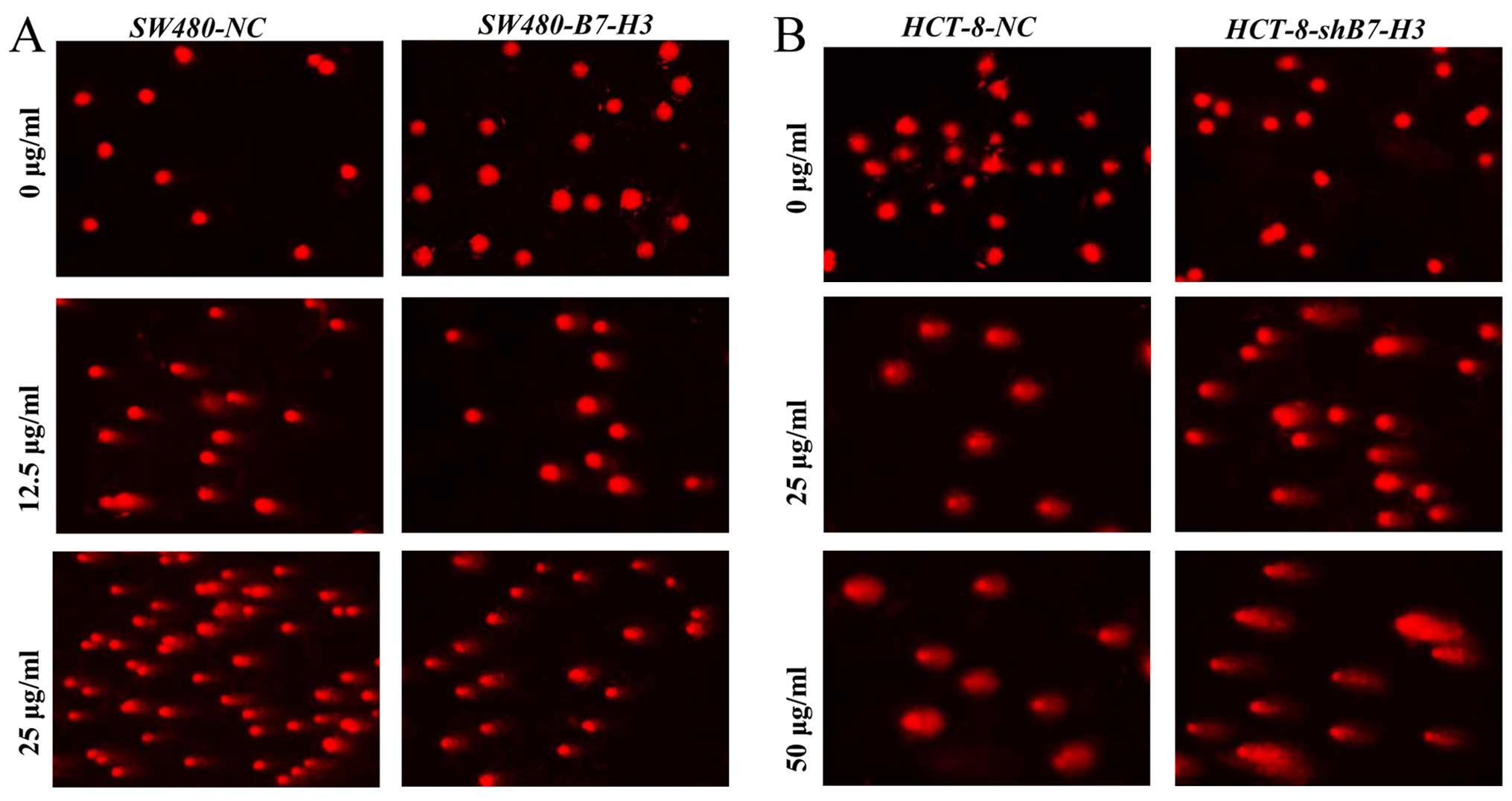
Overexpression of B7-H3 suppresses DNA
damage of CRC cells by 5-Fu
Since BRCC3 has been reported to be involved in
mediating DNA repair (16), we
examined its role in repairing 5-Fu-induced DNA damage in CRC cells
(Fig. 3). SW480-NC and SW480-B7-H3
cells were treated with 5-Fu (0, 12.5 and 25 µg/ml; Fig. 3A), and HCT-8-NC and HCT-8-shB7-H3
cells were treated with 5-Fu (0, 25 and 50 µg/ml; Fig. 3B) for 24 h; subsequent DNA damage
was measured by DNA comet assay. As depicted, treatment with 12.5
or 25 µg/ml 5-Fu induced longer DNA tails and higher
tail-DNA content in SW480-NC versus SW480-B7-H3 cells, indicating
greater DNA damage in cells expressing less B7-H3. Similarly,
treatment with 25 or 50 µg/ml 5-Fu induced longer DNA tails
and higher tail-DNA content in HCT-8-shB7-H3 vs. HCT-8-NC
cells. Percent TDNA, TL, TM and OTM were quantitatively analyzed by
CASP software; results are listed in Tables I (corresponding to Fig. 3A) and II (corresponding to Fig. 3B).
 | Table ISW480-NC and SW480-B7-H3 cells were
treated with 5-Fu for 24 h, DNA damage was measured by DNA comet
assay. |
Table I
SW480-NC and SW480-B7-H3 cells were
treated with 5-Fu for 24 h, DNA damage was measured by DNA comet
assay.
| Group | 0 µg/ml-5-Fu
| 12.5
µg/ml-5-Fu
| 25 µg/ml-5-Fu
|
|---|
| SW480-NC | SW480-B7-H3 | SW480-NC | SW480-B7-H3 | SW480-NC | SW480-B7-H3 |
|---|
| TDNA% | 1.8±1.2 | 0.2±0.4a | 21.3±3.8 | 12.5±4.7b | 33.1±9.8 | 22.9±4.1a |
| TL | 4.4±1.2 | 3.0±0.0a | 24.1±5.6 | 21.1±3.0a | 30.7±7.1 | 26.3±4.0 |
| TM | 0.1±0.1 | 0.0±0.0 | 5.3±1.9 | 2.8±1.4b | 10.7±6.2 | 6.1±1.8 |
| OTM | 0.2±0.2 | 0.0±0.0 | 3.7±1.0 | 3.0±1.3 | 6.4±2.8 | 4.8±1.1 |
 | Table IIHCT-8-NC and HCT-8-shB7-H3 cells were
treated with 5-Fu for 24 h, DNA damage was measured by DNA comet
assay. |
Table II
HCT-8-NC and HCT-8-shB7-H3 cells were
treated with 5-Fu for 24 h, DNA damage was measured by DNA comet
assay.
| Group | 0 µg/ml-5-Fu
| 25 µg/ml-5-Fu
| 50 µg/ml-5-Fu
|
|---|
| HCT-8-NC | HCT-8-shB7-H3 | HCT-8-NC | HCT-8-shB7-H3 | HCT-8-NC | HCT-8-shB7-H3 |
|---|
| TDNA% | 0.9±0.7 | 1.3±1.4 | 16.1±4.4 | 31.1±8.0a | 27.4±10.3 | 60.6±8.9a |
| TL | 3.6±1.1 | 3.6±1.1 | 15.6±2.3 | 36.3±6.2a | 22.7±5.3 | 55.2±7.0a |
| TM | 0.0±0.0 | 0.1±0.1 | 2.6±0.9 | 12.0±4.3a | 6.7±3.6 | 34.0±9.0a |
| OTM | 0.1±0.1 | 0.2±0.2 | 3.1±0.8 | 7.5±2.4a | 6.4±2.2 | 18.0±4.4a |
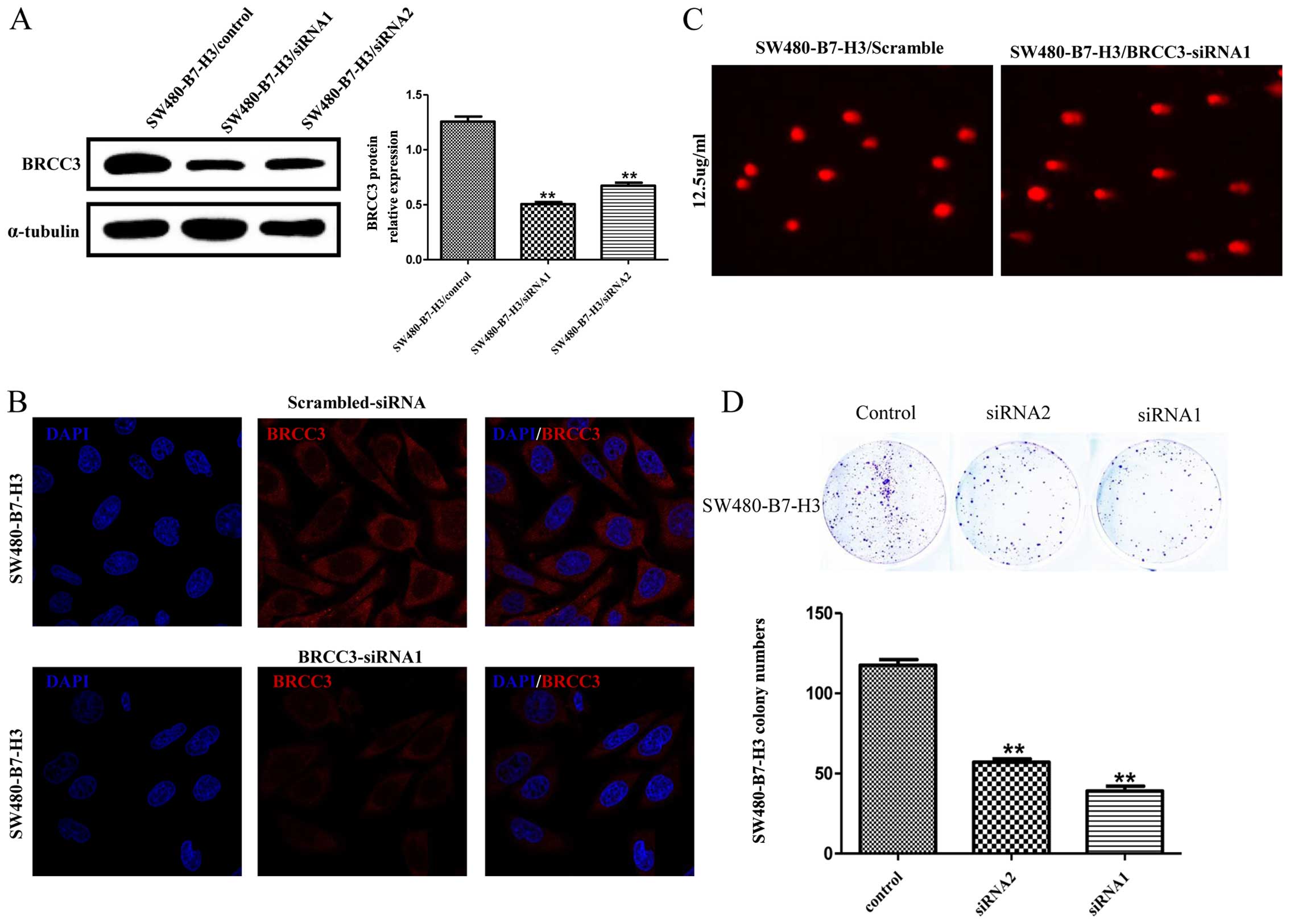
Downregulated expression of BRCC3 blocks
B7-H3
Fig. 4A depicts
western blot analysis demonstrating the effect of knocking down
BRCC3 protein expression with either siRNA1 or siRNA2 (two
different siRNA sequences targeting BRCC3); representative
immunofluorescence images depicting BRCC3 knockdown are shown in
Fig. 4B.
The DNA comet assay shown in Fig. 4C indicates that 5-Fu (25
µg/ml) elicits greater DNA damage in SW480-B7-H3 cells
transfected with BRCC3 siRNA1 vs. scramble control siRNA
(Fig. 4C). The TDNA%, TL, TM and
OTM results are recorded in Table
III. Further results indicated that knocking down BRCC3
expression in SW480-B7-H3 cells effectively suppressed colony
formation (Fig. 4D). Collectively,
our data showed that downregulated BRCC3 can induce DNA damage and
reduce colony formation, thereby antagonizing two characteristics
of B7-H3. These results suggest that BRCC3 may play a key role in
5-Fu resistance and DNA repair activation caused by B7-H3.
 | Table IIISW480-B7-H3 cells, transfected with
siRNA targeting BRCC3 or with scramble siRNA, were treated with
5-Fu for 24 h, DNA damage was measured by DNA comet assay. |
Table III
SW480-B7-H3 cells, transfected with
siRNA targeting BRCC3 or with scramble siRNA, were treated with
5-Fu for 24 h, DNA damage was measured by DNA comet assay.
| Group | 12.5
µg/ml-5-Fu
|
|---|
|
SW480-B7-H3/scramble |
SW480-B7-H3/BRCC3-siRNA1 |
|---|
| TDNA% | 7.7±3.9 | 21.2±6.4a |
| TL | 25.0±8.9 | 56.7±15.7a |
| TM | 2.2±1.4 | 12.9±6.0a |
| OTM | 3.7±1.8 | 12.0±3.4a |
Discussion
The present study first demonstrated that B7-H3
could upregulate BRCC3 expression to increase the ability to repair
DNA and to antagonize DNA damage caused by 5-Fu. We and other
researchers have discovered that co-stimulatory molecule B7-H3
could cause cancer cells resistance to 5-Fu or other agents, and
reported few probable mechanisms to explain this phenomenon; for
example, B7-H3 has been shown to induce Bcl-2 and Bcl-xl
overexpression via the Jak2-STAT3 signaling pathway to inhibit
cancer cell apoptosis (17). Here,
we propose that B7-H3 may enhance DNA repair to attenuate DNA
damage induced by chemotherapy medications.
5-Fu and its derivatives are recognized as the
cornerstone pharmaceuticals in gastrointestinal cancer
chemotherapy. The classic chemotherapy regimens FOLFOX and FOLFIRI
were both developed from 5-Fu. 5-Fu, a uracil analog, blocks
synthesis of the pyrimidine thymidine, interrupting DNA
replication. 5-Fu also antagonizes homologous recombination repair,
leading to persistent DNA damage (18). Therefore, we analyzed 5-Fu
resistance in colorectal cancer cells after upregulating or
downregulating B7-H3.
Our experiments showed that overexpressing B7-H3 in
SW480 cells lead to increased resistance to 5-Fu; similarly, HCT-8
cells became more sensitive to 5-Fu after B7-H3 had been knocked
down. The relationship between B7-H3 expression and an increased
5-Fu IC50 may be due to the B7-H3 induction of the Bcl-2
and Bcl-xl anti-apoptotic proteins, as previously presented, in
addition to its induction of the DNA repair complex protein BRCC3,
as presented here.
According to our previous mRNA microarray results,
we discovered that the expression of BRCC3 fluctuated following
changes in B7-H3 expression. Confirmation of these results using
both real-time PCR to measure mRNA levels and western blot analysis
to determine protein levels underscored a regulatory relationship
between B7-H3 and BRCC3. However, little has been reported
regarding the regulation of BRCC3 expression and B7-H3 signal
transduction (17). Thus, the
detail mechanism between B7-H3 and BRCC3 need more projects to be
elucidated.
Comet analysis is a classic method to measure DNA
damage: The length and densities of the comet tails have been used
to quantify the extent of DNA damage. Comet analysis of SW480-B7-H3
vs. SW480-NC cells after 24-h treatment with 12.5 or 25
µg/ml 5-Fu indicated less DNA damage in cells expressing
higher levels of B7-H3, consistent with the hypothesis that B7-H3
induces BRCC3 expression, which enhances DNA repair. In this way,
B7-H3 and BRCC3 expression can be thought to antagonize the effects
of 5-Fu. In order to test this hypothesis, we transiently
transfected BRCC3 siRNA into SW480-B7-H3 cells. Results showed that
knocking down BRCC3 expression attenuated DNA damage repair.
Altogether, comet analyses indicated that downregulated BRCC3
(SW480-B7-H3 cells) or downregulated B7-H3 expression (SW480-NC vs.
SW480-B7-H3 cells or HCT-8-shB7-H3 vs. HCT-8-NC cells)
resulted in increased DNA damage, presumably due to attenuated DNA
repair.
Although B7-H3 was first identified as an immunity
checkpoint molecule, recently studies, including ours, have focused
on it due to its abnormal expression in malignant tumors and its
association with poor prognosis. A false subcellular location of
B7-H3 is also observed in colorectal cancer patients. Previous
in vivo experiments implied that B7-H3 could activate
Jak2-STAT3, PI3K-Akt pathway to subsequently induce the expression
of Bcl-2, Bcl-xl and MMP9 (17,19).
In the present study, we have shown that B7-H3 can upregulate the
DNA repair process in cancer cells through inducing BRCC3
expression.
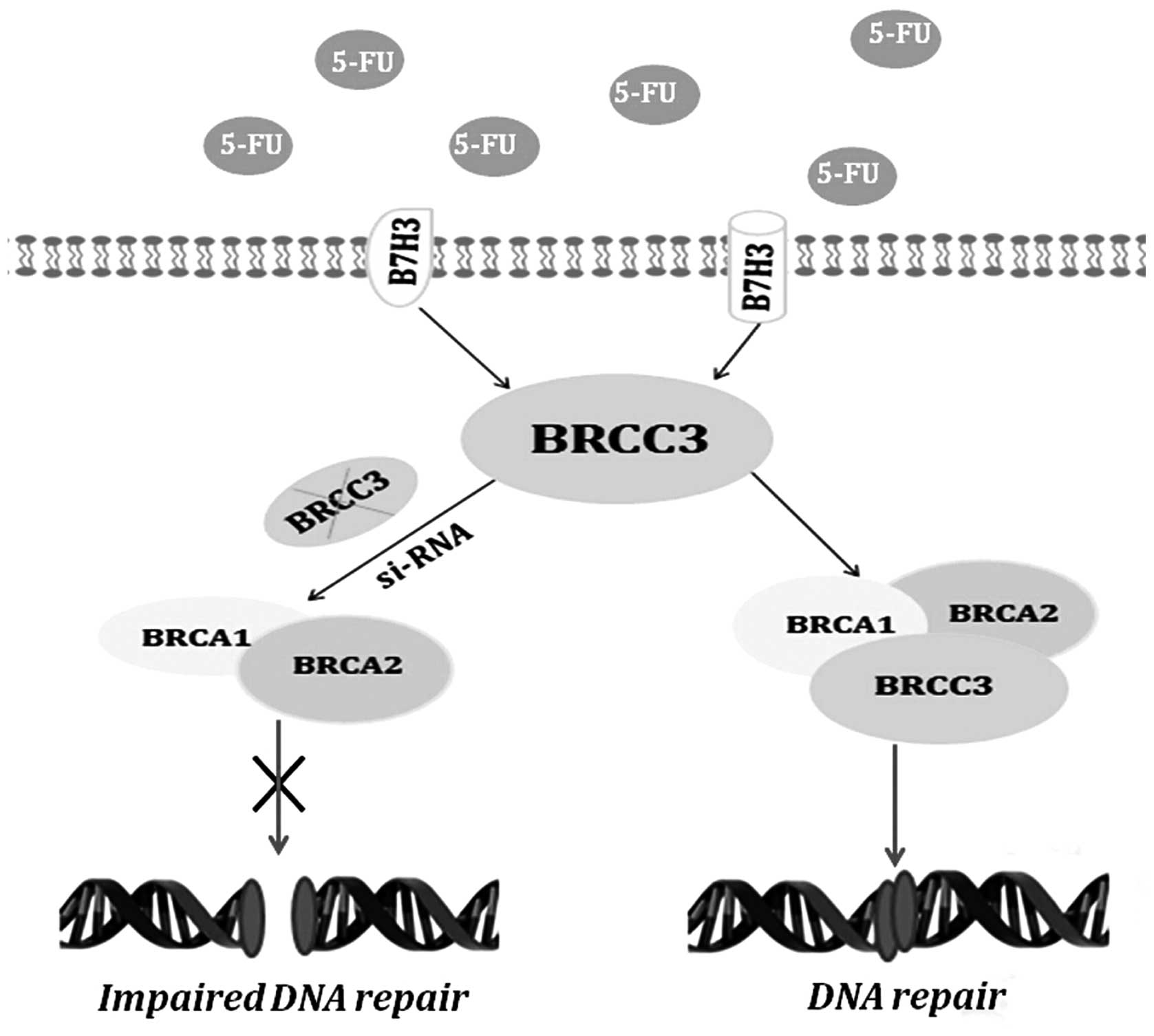
In summary, this study demonstrated that B7-H3 can
reduce the resistance to 5-Fu by mediating DNA damage repair via
BRCC3 (Fig. 5). These results
suggest that new CRC treatments could target B7-H3 overexpression
or associated signaling pathways in tumors as a novel approach to
antagonize drug resistance.
Acknowledgments
The present study was supported by the following:
the National Natural Science Foundation of China (nos. 81372375 and
81502758); the Wuxi Administration of Science and Technology
Project (no. CSE31N1509); the Clinical Medical Science and
Technology Project of Jiangsu Province (no. BL2014019); the Science
and Technology Foundation of Suzhou (no. SYS201417); and the
Priority Academic Program Development of Jiangsu Higher Education
Institutions (PAPD).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dong H, Strome SE, Salomao DR, Tamura H,
Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, et al:
Tumor-associated B7-H1 promotes T-cell apoptosis: A potential
mechanism of immune evasion. Nat Med. 8:793–800. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hashiguchi M, Kobori H, Ritprajak P,
Kamimura Y, Kozono H and Azuma M: Triggering receptor expressed on
myeloid cell-like transcript 2 (TLT-2) is a counter-receptor for
B7-H3 and enhances T cell responses. Proc Natl Acad Sci USA.
105:10495–10500. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang G, Hou J, Shi J, Yu G, Lu B and
Zhang X: Soluble CD276 (B7-H3) is released from monocytes,
dendritic cells and activated T cells and is detectable in normal
human serum. Immunology. 123:538–546. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Streit M and Detmar M: Angiogenesis,
lymphangiogenesis, and melanoma metastasis. Oncogene. 22:3172–3179.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Merimsky O, Shoenfeld Y, Chaitchik S,
Yecheskel G and Fishman P: Antigens and antibodies in malignant
melanoma. Tumour Biol. 15:188–202. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun J, Chen LJ, Zhang GB, Jiang JT, Zhu M,
Tan Y, Wang HT, Lu BF and Zhang XG: Clinical significance and
regulation of the costimulatory molecule B7-H3 in human colorectal
carcinoma. Cancer Immunol Immunother. 59:1163–1171. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen YW, Tekle C and Fodstad O: The
immunoregulatory protein human B7H3 is a tumor-associated antigen
that regulates tumor cell migration and invasion. Curr Cancer Drug
Targets. 8:404–413. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miller AJ and Mihm MC Jr: Melanoma. N Engl
J Med. 355:51–65. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Harper JW and Elledge SJ: The DNA damage
response: Ten years after. Mol Cell. 28:739–745. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chai KM, Wang CY, Liaw HJ, Fang KM, Yang
CS and Tzeng SF: Downregulation of BRCA1-BRCA2-containing complex
subunit 3 sensitizes glioma cells to temozolomide. Oncotarget.
5:10901–10915. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rebbeck TR, Mitra N, Wan F, Sinilnikova
OM, Healey S, McGuffog L, Mazoyer S, Chenevix-Trench G, Easton DF,
Antoniou AC, et al CIMBA Consortium: Association of type and
location of BRCA1 and BRCA2 mutations with risk of breast and
ovarian cancer. JAMA. 313:1347–1361. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hedau S, Batra M, Singh UR, Bharti AC, Ray
A and Das BC: Expression of BRCA1 and BRCA2 proteins and their
correlation with clinical staging in breast cancer. J Cancer Res
Ther. 11:158–163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Singh NP, McCoy MT, Tice RR and Schneider
EL: A simple technique for quantitation of low levels of DNA damage
in individual cells. Exp Cell Res. 175:184–191. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Końca K, Lankoff A, Banasik A, Lisowska H,
Kuszewski T, Góźdź S, Koza Z and Wojcik A: A cross-platform public
domain PC image-analysis program for the comet assay. Mutat Res.
534:15–20. 2003. View Article : Google Scholar
|
|
16
|
Chen X, Arciero CA, Wang C, Broccoli D and
Godwin AK: BRCC36 is essential for ionizing radiation-induced BRCA1
phosphorylation and nuclear foci formation. Cancer Res.
66:5039–5046. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang T, Jiang B, Zou ST, Liu F and Hua D:
Overexpression of B7-H3 augments anti-apoptosis of colorectal
cancer cells by Jak2-STAT3. World J Gastroenterol. 21:1804–1813.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Srinivas US, Dyczkowski J, Beißbarth T,
Gaedcke J, Mansour WY, Borgmann K and Dobbelstein M: 5-Fluorouracil
sensitizes colorectal tumor cells towards double stranded DNA
breaks by interfering with homologous recombination repair.
Oncotarget. 6:12574–12586. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Bai Y, Cui W, Wang Z, Zhang G, Xu
Y, Zhu X, Li Y and Wang JH: Effects of B7-H3 on the inflammatory
response and expression of MMP-9 in mice with pneumococcal
meningitis. J Mol Neurosci. 50:146–153. 2013. View Article : Google Scholar
|