Introduction
Hepatocellular carcinoma (HCC) is the second leading
cause of cancer-related mortality worldwide, accounting for
~700,000 deaths/year (1). Although
the application of surgical resection and liver transplantation
techniques improves the outcomes and decreases the mortality of HCC
patients, the 5-year survival rate is still less than 30%. Hence,
the molecular mechanisms involved in HCC progression require
further investigation.
Iroquois homeobox 3 (IRX3) is a member of the
Iroquois family of homeobox (IRX) genes, which are highly conserved
among species and play crucial roles in embryonic development ().
Recently, members of this family were shown to closely correlate
with tumor progression (). The relevance of IRX3 throughout cancer
progression is contradictory. It has been reported that IRX3 is
overexpressed in tumor tissues and cell lines of glioblastoma,
astrocytoma and cholangiocarcinoma (8,9). In
pheochromocytomas and paragangliomas, IRX3 is overexpressed in
malignant tumors compared with its expression in benign tumors
(10). In a study on
transcriptional profiles of human colorectal adenoma tissues, IRX3
was identified as one of the most upregulated transcription factors
compared to healthy tissues (11).
In breast cancer, IRX3 was identified as a target of WHSC1L1, an
oncogene which promotes tumor cell proliferation (12). Thus, these studies suggest that IRX3
is a possible oncogene. In contrast, another study showed that
hypermethylation of the 5′-CpG island region of IRX3 contributed to
its downregulation in a mouse model of prostate cancer (13). A low level of IRX3 is associated
with high-stage disease in Wilms' tumor, suggesting IRX3 as a
tumor-suppressor (14). However,
the potential role and the regulatory mechanism of IRX3 in HCC have
not previously been studied.
MicroRNAs (miRNAs) are a class of small noncoding
endogenous RNAs that suppress gene expression at the
post-transcriptional levels through complementary base pairing with
mRNAs. Accumulating evidence suggests that miRNAs play crucial
roles in tumor development and progression, where they function as
oncogenes or tumor suppressors (15). miRNAs are emerging as potential
biomarkers or therapeutic targets in a variety of cancers including
HCC (16,17). miRNAs have become a research focus
due to their important roles in gene regulation.
In the present study, we found that IRX3 was
upregulated in HCC cell lines, suggesting the tumor-promoting
potential of IRX3. We further investigated the regulatory mechanism
of IRX3, and demonstrated that IRX3 is a direct target of miR-377.
Downregulation of miR-377 may contribute to the upregulation of
IRX3.
Materials and methods
Cell culture
The human HCC cell lines HepG2 and SMMC7721, and
normal liver cell line LO2 were obtained from the Cell Bank of the
Chinese Academy of Sciences (Shanghai, China) and maintained in
Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal bovine serum (Gibco, Carlsbad, CA, USA) at 37°C with 5%
CO2.
Construction of stable cell lines with
overexpression of IRX3
Recombinant lentiviruses containing human IRX3
(Lv-IRX3) or control were purchased from Biowit (Shenzhen, China).
To obtain cell lines stably expressing IRX3, SMMC7721 cells were
infected with Lv-IRX3 at a multiplicity of infection (MOI) of 30.
Forty-eight hours after infection, SMMC7721 cells were selected for
two weeks using puromycin (2.5 µg/ml). The stable cell line
was identified by qRT-PCR.
Quantitative real-time PCR
Total RNA was extracted using TRIzol reagent
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer's
manual. Reverse transcription was performed using PrimeScript™ RT
reagent kit (Takara, Dalian, Japan) with random primers or specific
primers. Then, real-time PCR was performed using SYBR-Green PCR
Master Mixture (Takara). All reactions were run on StepOne™
Real-Time PCR system (Applied Biosystems, Foster City, CA, USA)
under the conditions as follows: 95°C for 1 min, followed by 95°C
for 5 sec and 60°C for 1 min for 40 cycles. The relative expression
levels of miR-377 and mRNAs were calculated by the
2−ΔΔCt method using U6 or GAPDH as an internal control.
All reactions were performed in triplicate. The primer for miR-377
was purchased from RiboBio (Guangzhou, China). The specific primers
used for real-time PCR were: IRX3-F, 5′-ATCGATTTGGAGAACTTAGACG-3′
and IRX3-R, 5′-TTTGGAGTCCGAAATGGGT-3′; GAPDH-F,
5′-GAAGGTGAAGGTCGGAGTC-3′ and GAPDH-R, 5′-GAAGATGGTGATGGGATTTC-3′;
U6-F, 5′-CTCGCTTCGGCAGCACA-3′ and U6-R,
5′-AACGCTTCACGAATTTGCGT-3′.
Western blotting
Cells were lysed using RIPA buffer reagent (Beyotime
Institute of Biotechnology, Jiangsu, China), and the cellular
proteins were prepared in a sample buffer. Then, identical
quantities of proteins were separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto a polyvinylidene difluoride (PVDF) membrane
(Milipore, Billerica, MA, USA). After blocking with 5% skim milk
powder in Tris-buffered saline and Tween-20 (TBST) for 1 h at room
temperature, the membrane was incubated with rabbit anti-human
primary antibodies specific for IRX3 (ab25703; Abcam, Cambridge,
MA, USA) and GAPDH (#2118; Cell Signaling Technology, Danvers, MA,
USA) at 4°C overnight. Then, the membranes were washed with
phosphate-buffered saline (PBS) 3 times, and incubated with
HRP-conjugated goat anti-rabbit secondary antibody (1:5,000; #7074;
Cell Signaling Technology) for 1 h at room temperature. The blots
were detected using BeyoECL Plus kit (Beyotime) and the intensity
of proteins was quantified. GAPDH was used as loading control.
Cell proliferation assay
Cells were seeded in a 96-well plate at a density of
2×103/well and incubated for 72 h. Ten micro-liters of
Cell Counting Kit-8 (CCK-8) reagent (Beyotime) was added into each
well at different time points. After incubating the mixture at 37°C
for 1 h, the optical density (OD) at 450 nm was measured by a
microplate reader. All experiments were repeated at least 3
times.
Colony formation assay
Cells were seeded in 6-well plates (500 cells/well)
and maintained in complete medium for 2 weeks. Then, the cells were
fixed and stained with 1% crystal violet, and the number of
colonies were counted using a microscope (IX83; Olympus
Corporation, Tokyo, Japan).
Wound healing assay
Cells were plated in 6-well plates and grown until
90% confluency. The cells were then starved in 0.5% serum medium
overnight. A line was drawn on the bottom of the well by a sterile
200-µl pipette. After rinsing with PBS, the cells were
cultured in 0.5% serum medium for 48 h before imaging. The wound
healing ability of the tested cells was evaluated by measuring the
wound width.
Transwell invasion assays
The invasive ability of the HCC cells was measured
using a 24-well Transwell plate (8-µm pores; Corning,
Corning, NY, USA). Cells (5×105) were plated in the top
chamber with a Matrigel-coated membrane. The bottom chamber was
filled with complete medium. After culture for 24 h, the cells that
migrated to the underside of the membrane were stained with Giemsa
and counted under a microscope. All experiments were performed in
triplicate.
Construction of plasmids
To construct the IRX'UTR-WT plasmid, a 404-bp DNA
fragment of IRX3 3′UTR containing the predicted binding site of
miR-377 was amplified by PCR and cloned into a reporter vector as
previously described (18). The
primers were: GTAGAATTCGCTCTCTCCTCATCCTAGTTC
(forward) and GCAAAGCTTCGGGTTATAGTCAAAGTG
(reverse). The underlined bases represent the restriction enzyme
cutting sites. The predicted miR-377 binding site was mutated by
DpnI-mediated site-directed mutagenesis using the primers as
follows: AGAGAAATGTACATACTCGAGAACCAAATTGTACGAG (forward) and
TCGTACAATTTGGTTCTCGAGTATGTACATTTCTCTG (reverse).
Cell transfection and luciferase reporter
assay
The control and miR-377 mimics were purchased from
RiboBio. For transfection, the cells were seeded in culture plates
until 70% confluency, and then transfected with control or miR-377
mimics at a final concentration of 100 nM, using Lipofectamine 2000
reagent (Invitrogen) according to the manufacturer's instructions.
For the luciferase reporter assay, the cells were seeded into
96-well plates. Then, 50 ng of the constructed IRX3 3′UTR reporter
plasmids were co-transfected with either the miR-377 mimics or
control mimics, and 5 ng of pRL-TK plasmid (Promega, Madison, WI,
USA). Forty-eight hours after transfection, the luciferase activity
was measured using the Dual-Luciferase Reporter Assay System
(Promega).
Statistical analysis
Data are expressed as the mean ± standard deviation
(SD). Differences between two groups were evaluated using the
two-tailed Student's t-test. One-way ANOVA was used for comparisons
between more than two groups, followed by Tukey's post hoc test. A
value of p<0.05 was considered significant. All statistical
analyses were performed using GraphPad Prism 6.0 software.
Results
IRX3 is upregulated in HCC cell
lines
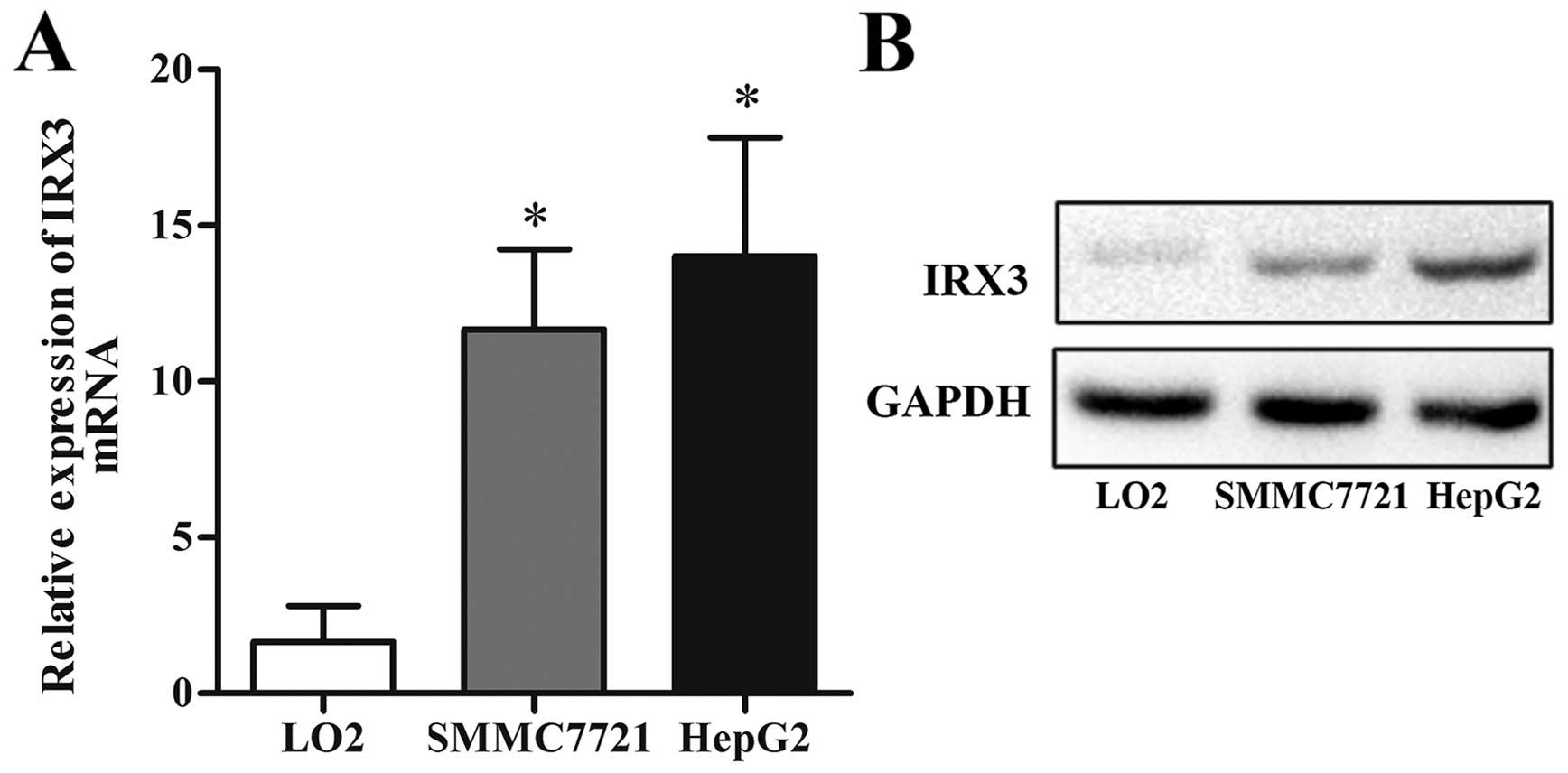
To explore the potential role of IRX3 in HCC, we
measured the expression level of IRX3 in normal live cell line
(LO2) and HCC cell lines (HepG2 and SMMC7721). As shown in Fig. 1, both mRNA and protein expression
levels of IRX3 were markedly higher in the HepG2 and SMMC7721 cells
than that in the LO2 cells.
IRX3 is a target of miR-377 in HCC
cells
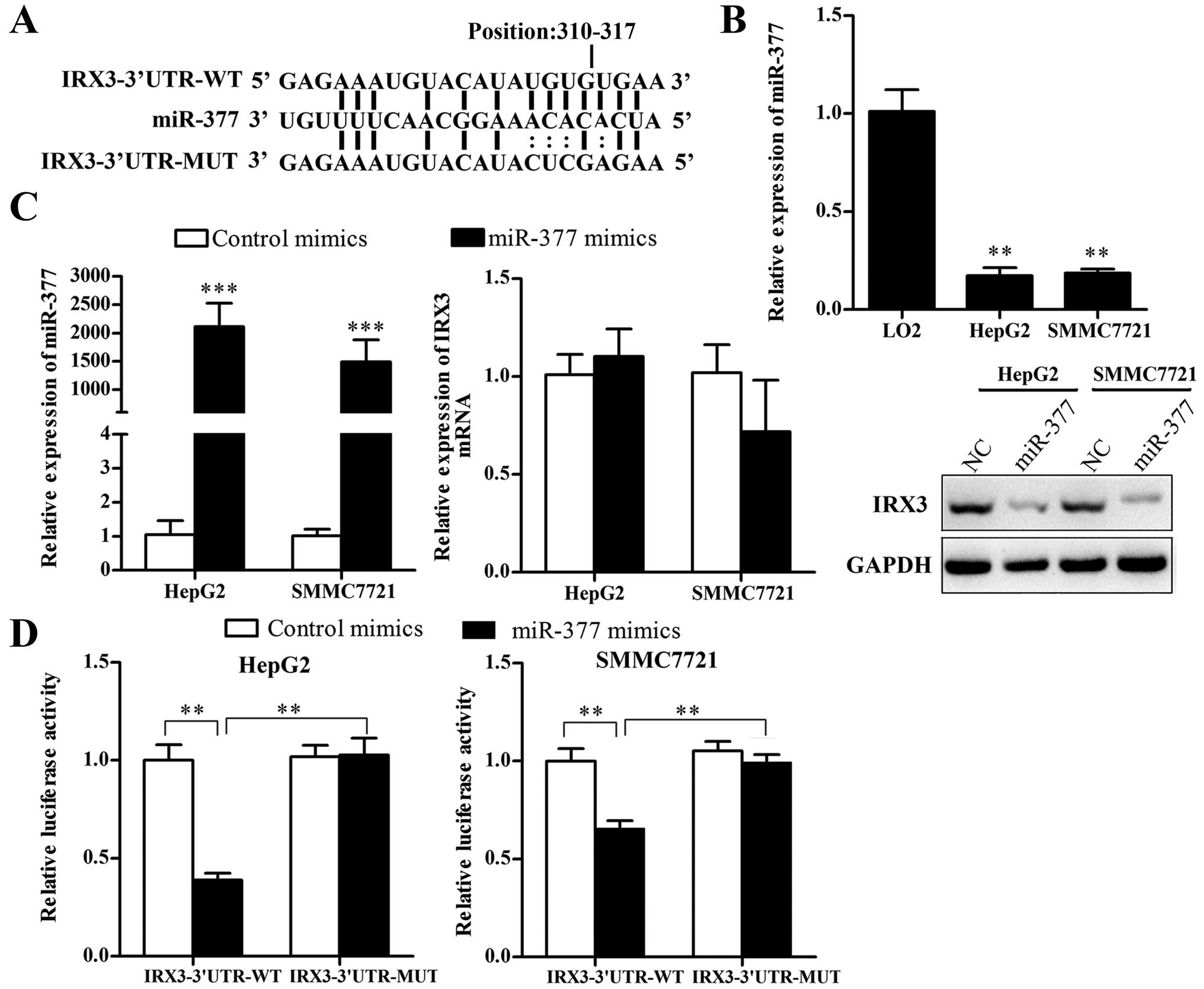
To explore the regulatory mechanism of IRX3
overexpression in HCC cell lines, we searched the putative miRNAs
that target IRX3 in the websites TargetScan human 7.0 (www.targetscan.org) and miRanda (www.microrna.org), and noted that miR-377 has a
potential binding site at the 3′UTR of IRX3 mRNA (Fig. 2A). We investigated the expression of
miR-377 in LO2, SMMC7721 and HepG2 cell lines, and found that
miR-377 expression was markedly lower in the SMMC7721 and HepG2
cells than that in the LO2 cells (Fig.
2B). To verify whether miR-377 directly targets IRX3, we
performed luciferase reporter gene assays. The results showed that
ectopic expression of miR-377 significantly reduced the luciferase
activity of IRX3 3′UTR wild-type reporter gene but had no effect on
mutant IRX3 3′UTR (Fig. 2D),
indicating that miR-377 directly targets IRX3 3′UTR. Then, we
transfected the HCC cells with miR-377 mimics and further
investigated the regulation by qRT-PCR and western blotting. As
shown in Fig. 2C, the protein level
but not the mRNA level of IRX3 was decreased in the miR-377
mimic-transfected HCC cells, compared with the negative
controls.
miR-377 restoration inhibits IRX3-induced
HCC cell proliferation, migration and invasion
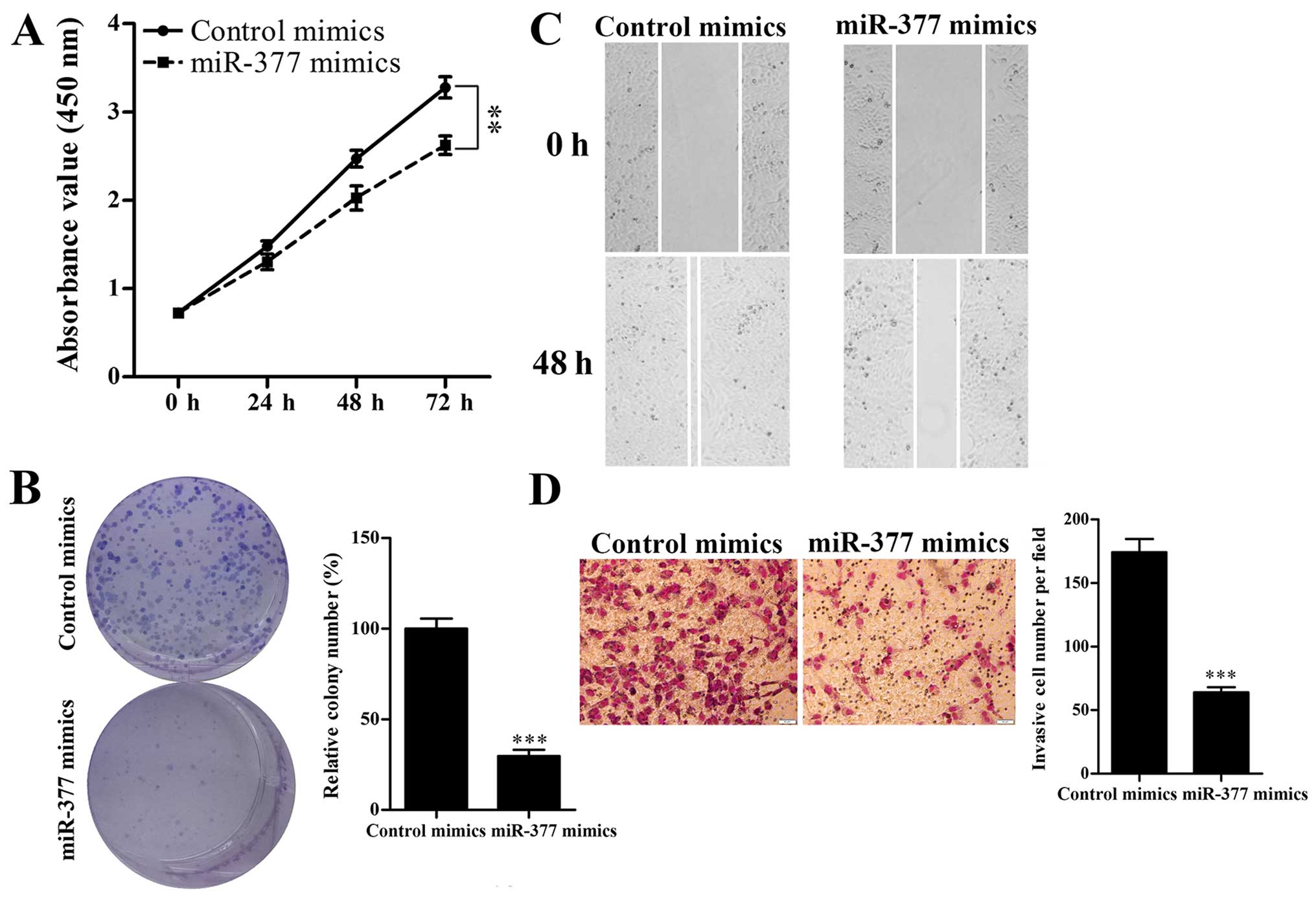
To investigate the effect of miR-377 on HCC cell
proliferation, migration and invasion, HepG2 cells were transiently
transfected with miR-377 mimics, and then CCK-8, colony formation,
Transwell and wound healing assays were performed. The CCK-8 assay
showed that overexpression of miR-377 significantly reduced the
growth rate of HepG2 cells compared with the control cells
(Fig. 3A). Colony formation assay
revealed that miR-377-transfected cells formed few and smaller
colonies compared with the negative control cells (Fig. 3B). Moreover, wound healing and
Transwell assays revealed that upregulation of miR-377
significantly decreased the migration and invasion of HepG2 cells
(Fig. 3C and D). Taken together,
these results suggested that miR-377 inhibited HepG2 cell
proliferation, migration and invasion.
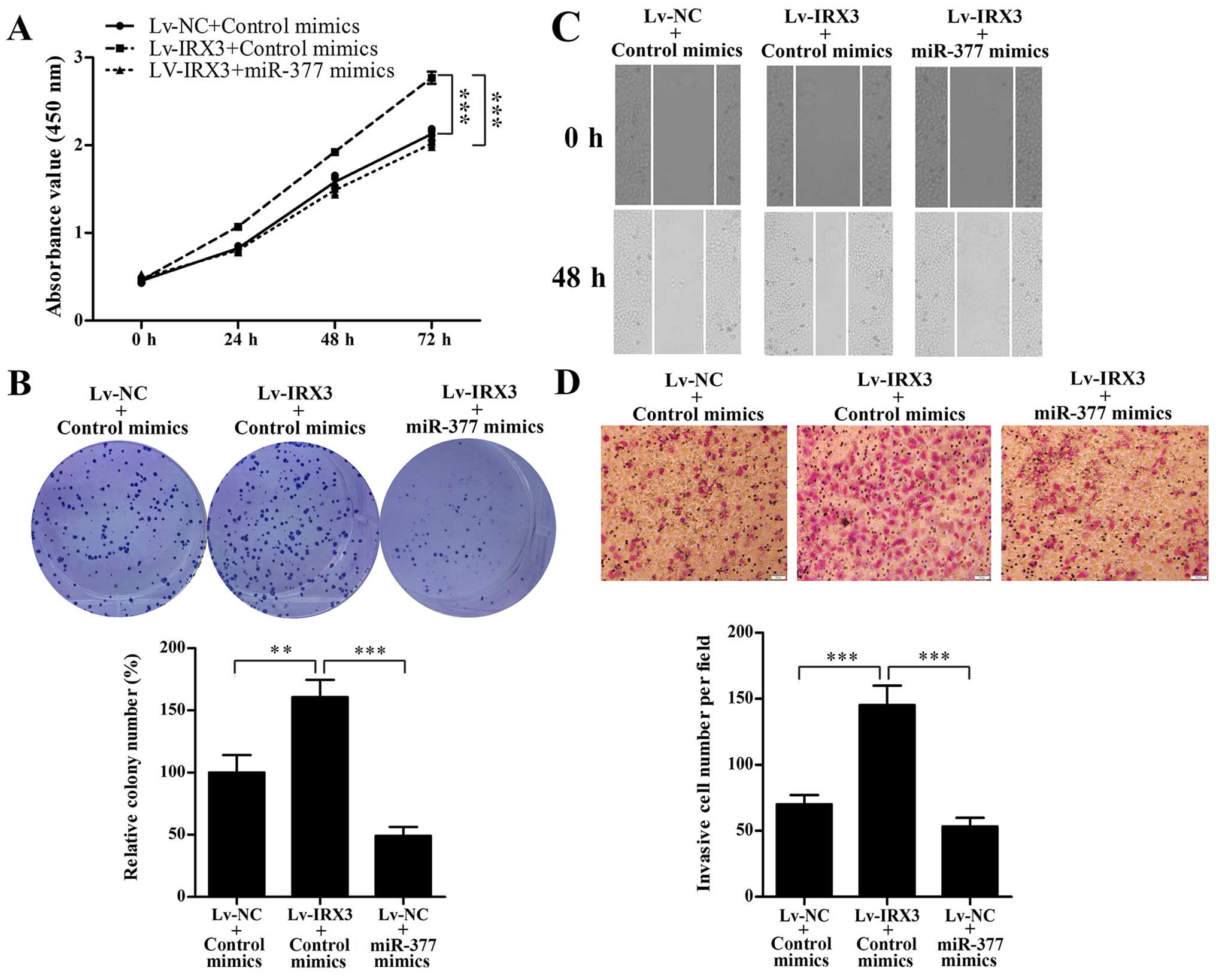
Subsequently, we evaluated whether restoration of
miR-377 could counteract the effect of IRX3 on proliferation,
migration and invasion. Two stable cell lines, SMMC7721-IRX3 and
the corresponding negative control (SMMC7721-NC) were established.
SMMC7721-NC and SMMC7721-IRX3 cells were transfected with miR-377
or control mimics, then CCK-8, colony formation, Transwell and
wound healing assays were performed. As shown in Fig. 4, the results showed that miR-377
significantly abrogated IRX3-induced proliferation, migration and
invasion of SMMC7721 cells.
Discussion
Iroquois homeobox (IRX) gene family, which plays
essential roles in embryonic development, has recently been found
to be involved in the development of many types of cancers. For
instance, IRX1 was reported as a potential tumor-suppressor gene in
gastric cancer (5). IRX2 has been
shown to act as a negative regulator of cellular motility in breast
cancer (6). Knockdown of IRX5 led
to cell cycle arrest and increased apoptosis in prostate cancer
(7). However, there is limited data
on the association between IRX3 and cancer. Previous studies showed
different expression trends of IRX3 in different cancers (4), suggesting that IRX3 may play both
tumor-promoting and tumor-suppressing roles depending on the cancer
type. Similarly, IRX genes also show spatial and temporal
restricted expression patterns during the development of many
embryonic tissues (3,19,20).
However, the reasons for the contradicting functions of IRX3 in
different tissue contexts are largely unknown. In the present
study, we demonstrated that IRX3 is overexpressed in HCC cell
lines, suggesting its tumor-promoting role in HCC.
Then, we sought to investigate how IRX3 is
upregulated in HCC. Bioinformatic analysis revealed that miR-377 is
a candidate that targets IRX3. We then investigated the expression
and functional role of miR-377 in HCC cell lines. The results
showed that miR-377 was downregulated in HCC cells, and that
overexpression of miR-377 inhibited HCC cell proliferation,
migration and invasion. Our result was consistent with a recent
study that reported that miR-377 acts as a tumor suppressor
inhibiting HCC cell proliferation and invasion by targeting TIAM1
(21). We further validated that
miR-377 negatively regulated IRX3 expression by directly targeting
the 3′UTR of IRX3 mRNA. miR-377 restoration inhibited IRX3-induced
SMMC7721 cell proliferation, migration and invasion. These results
suggested that downregulation of miR-377 may contribute to IRX3
upregulation. Notably, our results showed that the mRNA level of
IRX3 was not affected by miR-377 mimics. As both the mRNA and
protein of IRX3 were upregulated in HCC cells, we speculated that
IRX3 could also be transcriptionally regulated by other mechanisms.
In fact, it has been reported that hypermethylation of the CpG
islands within an IRX3 exon was correlated with overexpression of
IRX3 in oligodendroglioma tissues and cell lines relative to normal
brain samples (8). Another study by
Smemo et al showed that IRX3 expression is driven by a
long-range enhancer located in the intron of the FTO gene in
multiple tissues of mice (22).
Whether these mechanisms are involved in the deregulation of IRX3
in HCC needs further investigation. Moreover, IRX3 as a
transcriptional factor has been reported to act as either an
activator or suppressor of gene expression (25). The functional transcription targets
of IRX3 remain to be elucidated in future studies.
In conclusion, our findings indicate that IRX3 may
be a potential oncogene. Moreover, we confirmed that IRX3 is a
target of miR-337, and that downregulation of miR-377 may
contribute to the upregulation of IRX3. The present study provides
a novel potential therapeutic target for HCC treatment.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (nos. 30871153 and
81370525).
References
|
1
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing, and treating hepatocellular carcinoma.
CA Cancer J Clin. 62:39992012. View Article : Google Scholar
|
|
2
|
Gómez-Skarmeta JL and Modolell J: Iroquois
genes: Genomic organization and function in vertebrate neural
development. Curr Opin Genet Dev. 12:40082002. View Article : Google Scholar
|
|
3
|
Kudoh T and Dawid IB: Role of the
iroquois3 homeobox gene in organizer formation. Proc Natl Acad Sci
USA. 98:7858572001. View Article : Google Scholar
|
|
4
|
Kim KH, Rosen A, Bruneau BG, Hui CC and
Backx PH: Iroquois homeodomain transcription factors in heart
development and function. Circ Res. 110:1515242012. View Article : Google Scholar
|
|
5
|
Guo X, Liu W, Pan Y, Ni P, Ji J, Guo L,
Zhang J, Wu J, Jiang J, Chen X, et al: Homeobox gene IRX1 is a
tumor suppressor gene in gastric carcinoma. Oncogene.
29:3909202010. View Article : Google Scholar
|
|
6
|
Werner S, Stamm H, Pandjaitan M, Kemming
D, Brors B, Pantel K and Wikman H: Iroquois homeobox 2 suppresses
cellular motility and chemokine expression in breast cancer cells.
BMC Cancer. 15:8962015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Myrthue A, Rademacher BL, Pittsenbarger J,
Kutyba-Brooks B, Gantner M, Qian DZ and Beer TM: The iroquois
homeobox gene 5 is regulated by 1,25-dihydroxyvitamin D3
in human prostate cancer and regulates apoptosis and the cell cycle
in LNCaP prostate cancer cells. Clin Cancer Res. 14:3565702008.
View Article : Google Scholar
|
|
8
|
Ordway JM, Bedell JA, Citek RW, Nunberg A,
Garrido A, Kendall R, Stevens JR, Cao D, Doerge RW, Korshunova Y,
et al: Comprehensive DNA methylation profiling in a human cancer
genome identifies novel epigenetic targets. Carcinogenesis.
27:2404232006. View Article : Google Scholar
|
|
9
|
Seol MA, Chu IS, Lee MJ, Yu GR, Cui XD,
Cho BH, Ahn EK, Leem SH, Kim IH and Kim DG: Genome-wide expression
patterns associated with oncogenesis and sarcomatous
transdifferentation of cholangiocarcinoma. BMC Cancer. 11:782011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Evenepoel L, Van Nederveen FH, Oudijk L,
Papathomas TG, Restuccia DF, Belt EJ, Franssen GJ, Feelders RA, Van
Eeden S, Timmers H, et al: 9b09: Identification of markers
predictive for malignant behavior of pheochromocytomas and
paragangliomas. J Hypertens. 33(Suppl 1): e1222015. View Article : Google Scholar
|
|
11
|
Sabates-Bellver J, Van der Flier LG, de
Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA,
Bujnicki JM, Menigatti M, et al: Transcriptome profile of human
colorectal adenomas. Mol Cancer Res. 5:1262752007. View Article : Google Scholar
|
|
12
|
Yang ZQ, Liu G, Bollig-Fischer A, Giroux
CN and Ethier SP: Transforming properties of 8p12 amplified genes
in human breast cancer. Cancer Res. 70:8484972010. View Article : Google Scholar
|
|
13
|
Morey SR, Smiraglia DJ, James SR, Yu J,
Moser MT, Foster BA and Karpf AR: DNA methylation pathway
alterations in an autochthonous murine model of prostate cancer.
Cancer Res. 66:116516672006. View Article : Google Scholar
|
|
14
|
Mengelbier LH, Karlsson J, Lindgren D, Øra
I, Isaksson M, Frigyesi I, Frigyesi A, Bras J, Sandstedt B and
Gisselsson D: Deletions of 16q in Wilms tumors localize to
blastemal-anaplastic cells and are associated with reduced
expression of the IRXB renal tubulogenesis gene cluster. Am J
Pathol. 177:2606212010. View Article : Google Scholar
|
|
15
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:21332009. View Article : Google Scholar
|
|
16
|
Szabo G and Bala S: MicroRNAs in liver
disease. Nat Rev Gastroenterol Hepatol. 10:54522013. View Article : Google Scholar
|
|
17
|
He Y, Lin J, Kong D, Huang M, Xu C, Kim
TK, Etheridge A, Luo Y, Ding Y and Wang K: Current state of
circulating microRNAs as cancer biomarkers. Clin Chem.
61:1131552015. View Article : Google Scholar
|
|
18
|
Li Y, Xie J, Xu X, Wang J, Ao F, Wan Y and
Zhu Y: MicroRNA-548 down-regulates host antiviral response via
direct targeting of IFN-λ1. Protein Cell. 4:13412013. View Article : Google Scholar
|
|
19
|
van Tuyl M, Liu J, Groenman F, Ridsdale R,
Han RN, Venkatesh V, Tibboel D and Post M: Iroquois genes influence
proximo-distal morphogenesis during rat lung development. Am J
Physiol Lung Cell Mol Physiol. 290:L777–L789. 2006. View Article : Google Scholar
|
|
20
|
Houweling AC, Dildrop R, Peters T,
Mummenhoff J, Moorman AF, Rüther U and Christoffels VM: Gene and
cluster-specific expression of the Iroquois family members during
mouse development. Mech Dev. 107:16742001. View Article : Google Scholar
|
|
21
|
Chen G, Lu L, Liu C, Shan L and Yuan D:
MicroRNA-377 suppresses cell proliferation and invasion by
inhibiting TIAM1 expression in hepatocellular carcinoma. PLoS One.
10:e01177142015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Smemo S, Tena JJ, Kim KH, Gamazon ER,
Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR,
Wasserman NF, et al: Obesity-associated variants within FTO form
long-range functional connections with IRX3. Nature. 507:37752014.
View Article : Google Scholar
|
|
23
|
Scarlett K, Pattabiraman V, Barnett P, Liu
D and Anderson LM: The proangiogenic effect of iroquois homeobox
transcription factor Irx3 in human microvascular endothelial cells.
J Biol Chem. 290:6303152015. View Article : Google Scholar
|
|
24
|
Martorell Ò, Barriga FM, Merlos-Suárez A,
Stephan-Otto Attolini C, Casanova J, Batlle E, Sancho E and Casali
A: Iro/IRX transcription factors negatively regulate Dpp/TGF-β
pathway activity during intestinal tumorigenesis. EMBO Rep.
15:1212182014. View Article : Google Scholar
|
|
25
|
Zhang SS, Kim KH, Rosen A, Smyth JW,
Sakuma R, Delgado-Olguín P, Davis M, Chi NC, Puviindran V, Gaborit
N, et al: Iroquois homeobox gene 3 establishes fast conduction in
the cardiac His-Purkinje network. Proc Natl Acad Sci USA.
108:135735812011.
|