Introduction
Angiogenesis, which is the formation of new
capillaries from pre-existing blood vessels, is crucial for normal
embryonic development and also responsible for many pathological
states including tumor growth and the development of metastases
(1,2). When solid tumors grow beyond the
diameter of ~1–2 mm, new blood vessels are needed to provide oxygen
and nutrients and take metabolic waste away (3). Tumor angiogenesis is a complex process
dependent on angiogenic factors and also involve in multiple
molecular events in tumor microenvironment (4,5).
The tumor microenvironment is composed of a variety
of signaling molecules that influence angiogenesis. Previous
studies illustrate that tumor cells create the microenvironment by
secreting cytokines and growth factors to activate normal,
quiescent cells around them and initiate a cascade of events. For
example, tumor cell can release vascular endothelial growth factor
(VEGF) to stimulate the sprouting and proliferation of endothelial
cells (6,7). Furthermore, it has been proved as a
credible method to simulate the tumor microenvironment by the
cancer cell supernatant (8,9).
There are many ways for tumor angiogenesis such as
sprouting angiogenesis, intussusceptive angiogenesis, vasculogenic
mimicry and lymphangiogenesis (10). Moreover, tumor microenvironment
secrete numerous cytokines and growth factors to recruit normal
endothelial cells differentiate into tumor endothelial cells for
tumor angiogenesis, which was considered as an important way for
tumor neovascularization (6,7).
Esophageal carcinoma is one of the most common
cancers in China. The main type of esophageal cancer is esophageal
squamous cell carcinoma which is characterized by poor prognosis as
well as strong invasiveness (11).
Currently, there are no other good methods to manage it except
operation, chemo-or radiotherapy, from which only people who are
diagnosed during the early period can benefit. In the 1970s,
Folkman hypothesized a link between angiogenesis, tumor growth and
metastasis, thus angiogenesis became a putative target for
anticancer therapy (12). Recent
studies revealed that blocking tumor vascularization is a crucial
way to curtail tumor growth for tumor therapy (13). Moreover, controlling of pathological
angiogenesis is known as a potential therapeutic strategy for the
prevention of tumor progression and the treatment of vascular
diseases (14).
The purpose of the present study is to analyze the
effects of tumor microenvironment on gene expression of endothelial
cells and clear the link between tumor microenvironment and
angiogenesis, which would be clinically important as the potential
anti-angiogenesis target for esophageal carcinoma.
Materials and methods
Cell preparation
KYSE70, poorly differentiated esophageal squamous
cancer cells, were cultured in RPMI-1640 (Biological Industries,
Kibbutz Beit Haemek, Israel) medium with 10% FBS, then replenished
with fresh medium after reaching 60–80% confluence, the supernatant
was collected and centrifuged after 24 h incubation and stored at
−20°C. Human umbilical vein endothelial cells (HUVECs) were
cultured in endothelial cell medium (ECM) (ScienCell, Carlsbad, CA,
USA). After reaching 60–80% confluence the HUVECs were induced with
condition medium (60% KYSE70 supernatant and 40% ECM) for 48 h as
induced HUVECs (I-1, I-2, I-3), while normal HUVECs (N-1, N-2, N-3)
were used as control. The preparation of tissue homogenate
supernatant of human esophageal carcinoma and pericarcinoma is
according to our previous study (15). Briefly, tumor specimens of ESCC
tissue and pericarcinoma tissue (>5 cm) were collected, to
prepare to the tissues homogenate supernatant by grinding and
centrifuging. The HUVECs were induced by 40% tissue homogenate
supernatant of human esophageal carcinoma or pericarcinoma for
further verified by qRT-PCR.
Complementary RNA preparation and
microarray hybridization
RNA extraction and microarray hybridization were
performed at CapitalBio Technology Corp. (Beijing, China). Total
RNA isolation was performed using the TRIzol reagent and was
further purified using Qiagen RNeasy Mini kit according to the
manufacturer's instructions. RNA samples with RIN values >8,
260/280 absorbance ratios >1.8 and 260/230 absorbance ratios
>1.5 were considered suitable for microarray analysis. The
processes of labeling, hybridization and scanning were performed at
a platform of CapitalBio Technology Corp. Aliquots (100 ng) of
total RNA from induced HUVECs (I-1, I-2, I-3) and normal HUVECs
(N-1, N-2, and N-3) were used for synthesis and amplification of
first-strand cDNAs, double stranded cDNAs, and biotin-labeled
antisense RNAs, with a Message Amp™ Premier RNA Amplification kit
(Ambion) on a PCR apparatus (MJ, PTC-225).
After measuring the concentrations of the labeled
RNAs by ultraviolet spectrophotometer, 1 µg of each
preparation was fragmented and verified onto 1.2% formaldehyde
denatured agarose electrophoresis. The biotinylated cRNAs were
hybridized to a commercial gene chip, Affymetrix GeneChip Human
Genome U133 (HG-U133) Array. Microarray hybridization was performed
at 45°C for 16 h with constant rotation at the speed of 60 rpm
using an Affymetrix GeneChip Hybridization Oven 640. After washing
and staining automatically on an Affymetrix fluidics station 450,
and using the hybridization, Wash and Stain kit (Affymetrix), the
chips were scanned on Affymetrix GeneChip® scanner 3000.
Overall, six microarray chips were analyzed in this study.
Data processing and microarray data
analysis
The obtained scanned images were first assessed by
visual inspection and then analyzed using Affymetrix GeneChip
Operating software (GCOS 1.4). An invariant set normalization
procedure was performed to normalize the different arrays using
DNA-chip analyzer (dChip). In a comparison analysis, we applied a
two-class unpaired method of the Significance Analysis of
Microarrays software (SAM version 3.02, Stanford University,
Stanford, CA, USA) to compare significantly differentially
expressed genes (DEGs) in the induced HUVECs and normal HUVECs. The
algorithm used to sort the statistically significant DEGs was a
modified t-test, and the criteria for DEGs were FDR <0.05 and
fold change >2.0 or <0.5. FDR was the corrected p-value by
post-hoc test. Each set of DEGs was further subjected to the
CapitalBio® Molecule Annotation System V3.0 (MAS3.0) for
gene ontology (GO) and KEGG pathway analyses. As for GO and KEGG
pathways, we calculated the p-value and Q-value for the
significantly affected biological processes and pathways and ranked
them by the p-values.
Quantitative RT-PCR
qRT-PCR was conducted on candidate genes that were
differentially expressed in the microarrays, and whose functions
were found upon a biological function analysis, to be closely
related to cell differentiation and angiogenesis. qRT-PCR was
performed according to the manufacturer's guideline using
Quantifast SYBR Green PCR kit, and qRT-PCR were run using the
following program: 95°C/10 min + 40 × (95°C/10 sec + 60°C/30 sec).
Each sample was run in triplicate. The qRT-PCR data analysis was
performed on 7500 software v2.0.5. The comparative threshold cycle
(Ct) method, i.e., 2−ΔΔCt was used to calculate fold
amplification.
Statistical analysis
Data were expressed as means ± standard deviation
(SD) with at least three separate experiments and analyzed by
One-way ANOVA and t-test. Significance was defined as
p<0.05.
Results
Microarray analysis
We used Affymetrix GeneChip 4.0 Array to analysis
differentially expressed genes in induced HUVECs and normal HUVECs.
Of the 47000 probe sets on the arrays, 19536 had a present call,
except the similar expression between the induced HUVECs and normal
HUVECs, there were 3769 genes that had differential expression
(p<0.001), of which 1609 were upregulated and 2160 were
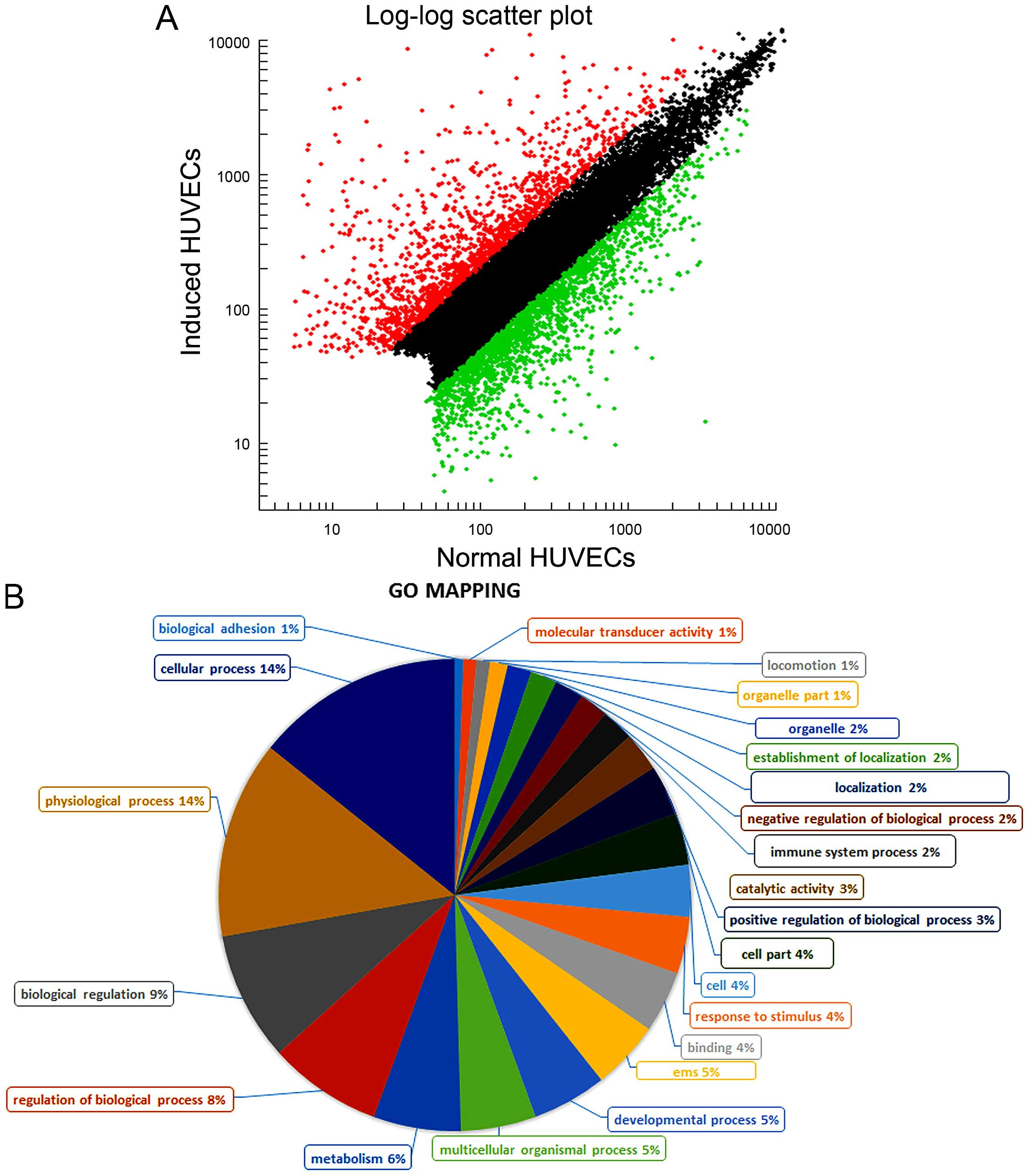
downregulated. Scatter plot for all the detected genes are shown
(Fig. 1A). Red spots represent
upregulated genes, green spots represent downregulated genes, black
spots represent no change genes between the induced HUVECs and
normal HUVECs. The distance of red or green spots from the black
spots represents markedly up- or downregulated (p<0.001). To
compare the functions of differential genes, the functional
annotations of all identified genes were collected and displayed
(Fig. 1B). The following will focus
on the genes involved in cell differentiation and angiogenesis, for
their potential roles in normal endothelial cells differentiation
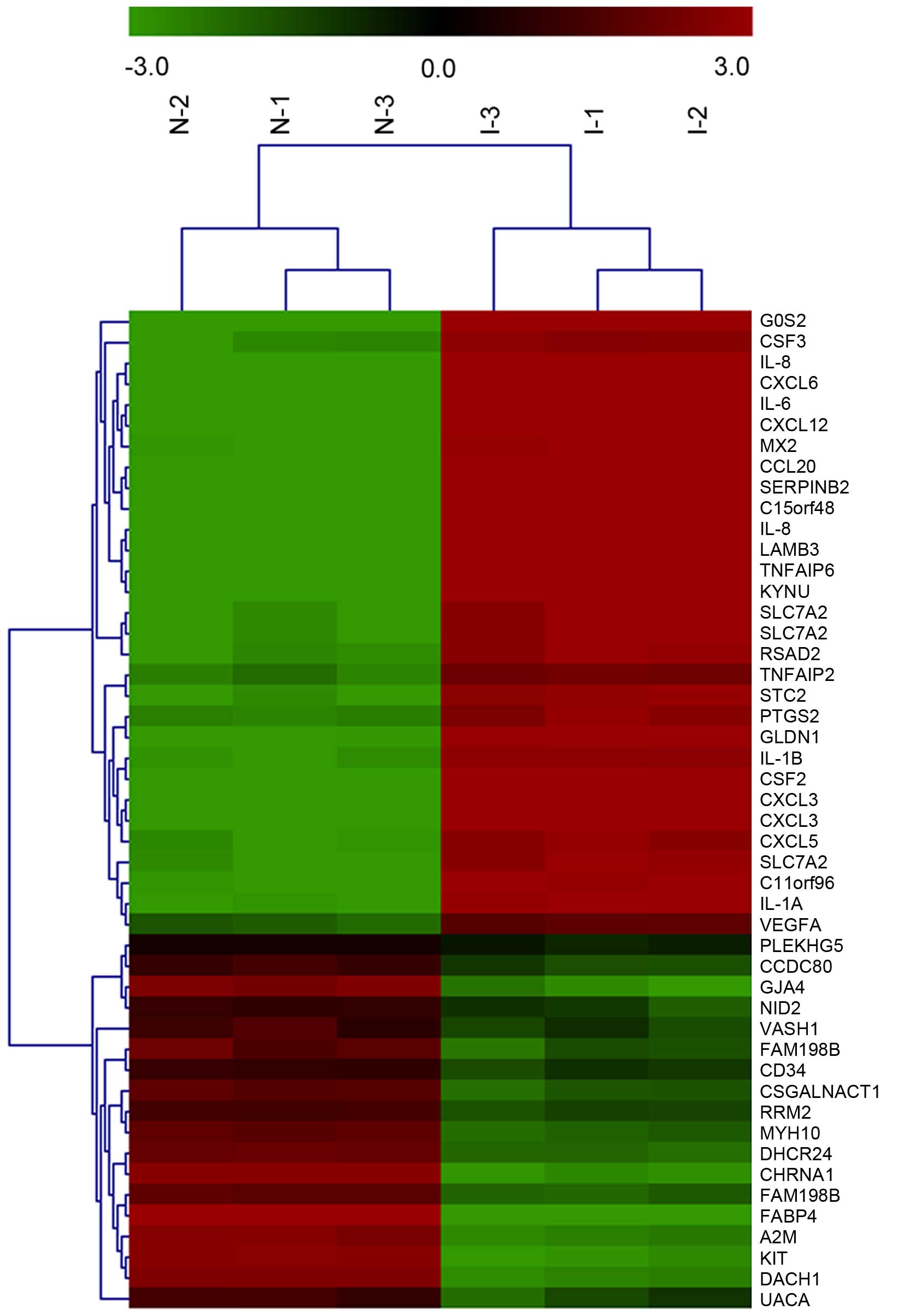
into tumor vascular endothelial cells. Clustering analysis of the
arrays showed the 48 genes differentially expressed in induced
HUVECs compared with normal HUVECs, of which 30 were upregulated
and 18 were downregulated. Among them, the upregulated genes
included IL-6, VEGF-A, S1PR1, and TYMP (Fig. 2).
All the differential genes were
categorized based on their cellular component, molecular function,
biological process and pathway
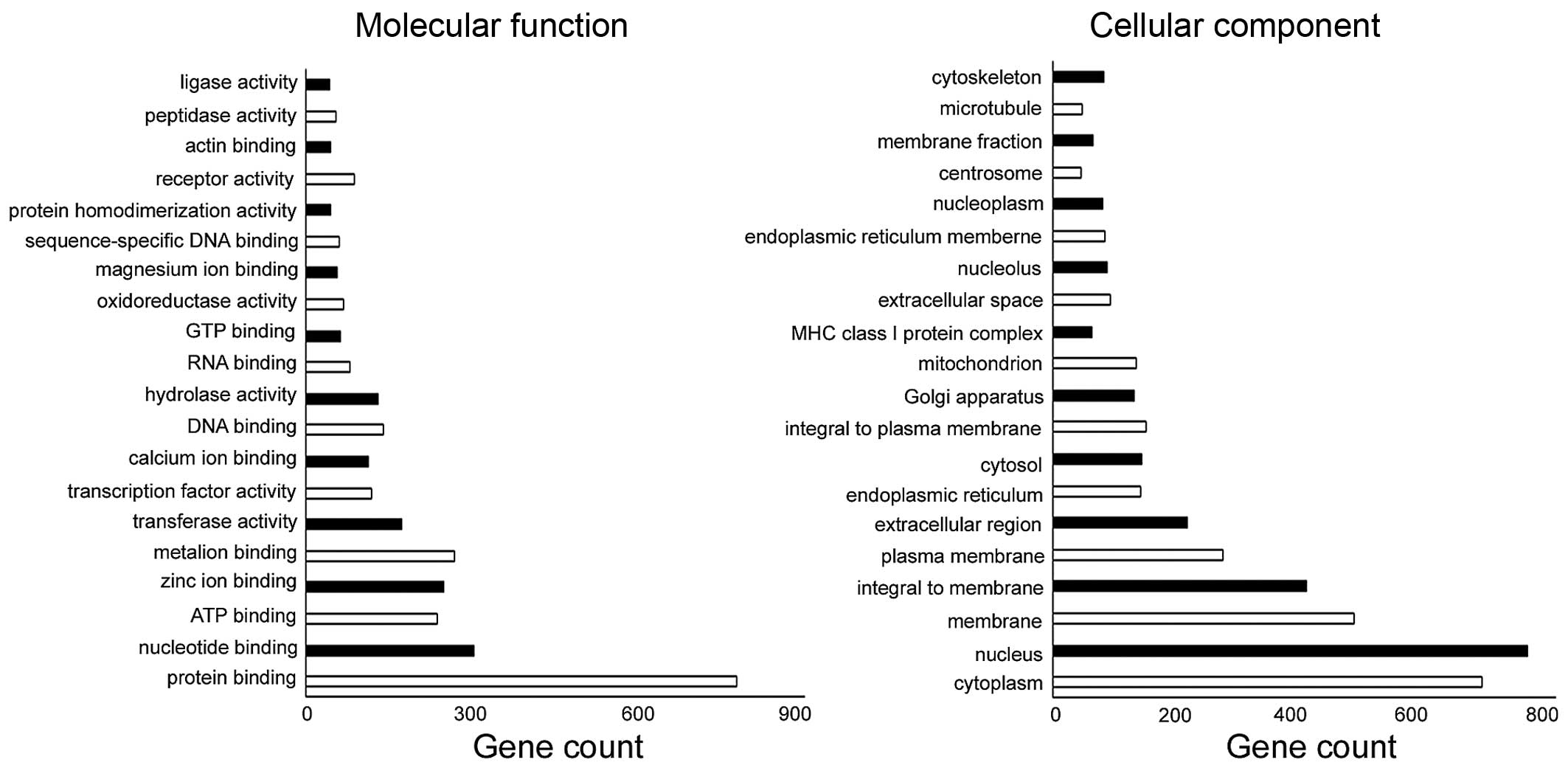
The number of changed genes enriched in the cellular
component and molecular function is shown (Fig. 3). Among the differentially genes, 28
were categorized based on their involvement in one or more of the
biological processes. Of these processes, angiogenesis, cell
differentiation, positive regulation of angiogenesis, positive
regulation of cell migration, endothelial cell differentiation,
signal transduction, cell adhesion, positive regulation of cell
proliferation, cell-cell signaling, were significantly represented
(Table I). Differential genes
(3769) were submitted to online DAVID for KEGG analysis. Compared
with the normal HUVECs, the induced HUVECs had 23 differential
gene-involved significant pathways (p<0.001, Table II). These pathways included
Cytokine-cytokine receptor interaction, JAK-STAT signaling pathway,
MAPK signaling pathway, TGF-β signaling pathway, Wnt signaling
pathway, VEGF signaling pathway, mTOR signaling pathway, and Cell
adhesion molecules (CAMs). Most of these pathways are associated
with angiogenesis and cell differentiation.
 | Table IGenes differentially expressed in
induced HUVECs vs. normal HUVECs categorized based on biological
processes. |
Table I
Genes differentially expressed in
induced HUVECs vs. normal HUVECs categorized based on biological
processes.
| Gene ID | Gene description | Probe set | Fold |
|---|
| Positive regulation
of cell migration (n=5)a |
| S1PR1 | Sphingosine
1-phosphate receptor 1 | 11743816_s_at | 3.16 |
| RRAS2 | Ras-related protein
R-Ras2 | 11747262_s_at | 3.2541 |
| EGFR | Epidermal growth
factor receptor | 11725104_a_at | 2.8712 |
| CDH13 | Cadherin 13,
H-cadherin | 11744837_a_at | 2.7747 |
| PDGFA | Platelet derived
growth factor A/B | 11729062_a_at | 2.1934 |
| Positive regulation
of angiogenesis (n=2)a |
| IL1A | Interleukin 1, α | 11725198_at | 72.8526 |
| IL1B | Interleukin 1, β | 11719916_at | 54.0777 |
| Development
(n=6)a |
| CSF3 | Granulocyte
colony-stimulating factor 3 | 11729514_a_at | 45.4721 |
| TNFAIP2 | Tumor necrosis factor
α-induced protein 2 | 11717823_s_at | 23.9717 |
| VEGFA | Vascular endothelial
growth factor A | 11725372_x_at | 12.5152 |
| TYMP | Thymidine
phosphorylase | 11722449_x_at | 9.2008 |
| TNF | Tumor necrosis factor
superfamily, member 2 | 11721577_at | 3.8156 |
| VEGFC | vascular endothelial
growth factor C | 11720163_at | 2.4673 |
| Positive regulation
vascular endothelial growth factor production (n=2)a |
| IL1A | Interleukin 1, α | 11725198_at | 72.8526 |
| IL1B | Interleukin 1, β | 11719916_at | 54.0777 |
| Cell adhesion
(n=3)a |
| TNFAIP6 | Tumor necrosis
factor, α-induced protein 6 | 11743636_at | 186.9013 |
| LAMB3 | Laminin, β3 | 11719487_a_at | 126.4414 |
| CLDN1 | Claudin 1 | 11728234_a_at | 32.8574 |
| Signal transduction
(n=4)a |
| CCL20 | Chemokine(C-C
motif) ligand 20 | 11724828_at | 334.6801 |
| TNFAIP6 | Tumor necrosis
factor, α-induced protein 6 | 11743636_at | 186.9013 |
| CXCL5 | Chemokine (C-X-C
motif) ligand 5 | 11728716_x_at | 51.4412 |
| VEGFC | Vascular
endothelial growth factor C | 11720163_at | 2.4673 |
| Endothelial cell
differentiation (n=2)a |
| JAG1 | Jagged | 11720825_s_at | 4.3523 |
| S1PR1 | Sphingosine
1-phosphate receptor 1 | 11743816_s_at | 3.16 |
| Positive regulation
of cell proliferation (n=1)a |
| CXCL5 | Chemokine (C-X-C
motif) ligand 5 | 11728716_x_at | 51.4412 |
| Cell-cell signaling
(n=6)a |
| IL8 | Interleukin 8 | 11763226_x_at | 385.6941 |
| CCL20 | Chemokine (C-C
motif) ligand 20 | 11724828_at | 334.6801 |
| STC2 | Stanniocalcin
2 | 11721436_a_at | 63.1026 |
| CXCL6 | Chemokine (C-X-C
motif) ligand 6 | 11730801_at | 158.1146 |
| IL1B | Interleukin 1,
β | 11719916_at | 54.0777 |
| CXCL5 | Chemokine (C-X-C
motif) ligand 5 | 11728716_x_at | 51.4412 |
| Angiogenesis
(n=6)a |
| IL8 | Interleukin 8 | 11763226_x_at | 385.6941 |
| TNFAIP2 | Tumor necrosis
factor α-induced protein 2 | 11717823_s_at | 23.9717 |
| VEGFA | Vascular
endothelial growth factor A | 11725372_x_at | 12.5152 |
| TYMP | Thymidine
phosphorylase | 11722449_x_at | 9.2008 |
| S1PR1 | Sphingosine
1-phosphate receptor 1 | 11743816_s_at | 3.16 |
| VEGFC | Vascular
endothelial growth factor C | 11720163_at | 2.4673 |
| Cell
differentiation (n=5)a |
| TNFAIP2 | Tumor necrosis
factor α-induced protein 2 | 11717823_s_at | 23.9717 |
| TYMP | Thymidine
phosphorylase | 11722449_x_at | 9.2008 |
| VEGFC | Vascular
endothelial growth factor C | 11720163_at | 2.4673 |
| NOTCH2 | Notch | 11732189_at | 2.459 |
| MMP19 | Matrix
metalloproteinase-19 | 11719531_a_at | 2.3774 |
 | Table IIGenes differentially expressed in the
induced HUVECs vs. normal HUVECs based on the significantly changed
pathway. |
Table II
Genes differentially expressed in the
induced HUVECs vs. normal HUVECs based on the significantly changed
pathway.
| Gene ID | Gene
description | Probe set | Fold |
|---|
| Cytokine-cytokine
receptor interaction (n=11)a |
| IL8 | Interleukin 8 | 11763226_x_at | 385.6941 |
| CCL20 | Chemokine(C-C
motif) ligand 20 | 11724828_at | 334.6801 |
| CXCL3 | Chemokine (C-X-C
motif) ligand 3 | 11728476_a_at | 298.7616 |
| IL6 | Interleukin 6 | 11760425_a_at | 4.3931 |
| CSF2 | Colony
stimulatingfactor 2 | 11728876_at | 143.5679 |
| IL1A | Interleukin 1,
α | 11725198_at | 72.8526 |
| CXCL2 | Chemokine (C-X-C
motif) ligand 2 | 11744127_at | 70.206 |
| IL1B | interleukin 1,
β | 11719916_at | 54.0777 |
| CXCL5 | Chemokine (C-X-C
motif) ligand 5 | 11728716_x_at | 51.4412 |
| CSF3 | Colony stimulating
factor 3 | 11729514_a_at | 45.4721 |
| Toll-like receptor
signaling pathway (n=3)a |
| IL8 | Interleukin 8 | 11763226_x_at | 385.6941 |
| IL6 | Interleukin 6 | 11760425_a_at | 4.3931 |
| IL1B | Interleukin 1,
β | 11719916_at | 54.0777 |
| JAK-STAT signaling
pathway (n=3)a |
| IL6 | Interleukin 6 | 11760425_a_at | 4.3931 |
| CSF2 | Colony stimulating
factor 2 | 11728876_at | 143.5679 |
| CSF3 | Colony stimulating
factor 3 | 11729514_a_at | 45.4721 |
| VEGF signaling
pathway (n=3)a |
| PTGS2 |
Prostaglandin-endoperoxide synthase 2 | 11724038_a_at | 5.9638 |
| VEGFA | Vascular
endothelial growth factor A | 11725372_x_at | 12.5152 |
| PRKCA | Classical protein
kinase C | 11754557_s_at | 2.2595 |
| Wnt signaling
pathway (n=2)a |
| MYC | Myc proto-oncogene
protein | 11745021_a_at | 4.5211 |
| JUN | Transcription
factor AP-1 | 11718397_s_at | 3.6646 |
| TGF-β signaling
pathway (n=1)a |
| MYC | Myc proto-oncogene
protein | 11745021_a_at | 4.5211 |
| mTOR signaling
pathway (n=2)a |
| VEGFA | Vascular
endothelial growth factor A | 11725372_x_at | 12.5152 |
| VEGFC | Vascular
endothelial growth factor C | 11720163_at | 2.4673 |
| Cell adhesion
molecules (CAMs) (n=1)a |
| CLDN1 | Claudin 1 | 11728234_a_at | 32.8574 |
| MAPK signaling
pathway (n=2)a |
| IL1A | Interleukin 1,
α | 11725198_at | 72.8526 |
| IL1B | Interleukin 1,
β | 11719916_at | 54.0777 |
| ECM-receptor
interaction(n=1)a |
| LAMB3 | Laminin, β3 | 11719487_a_at | 126.4414 |
Confirmation the changes in gene
expression by qRT-PCR
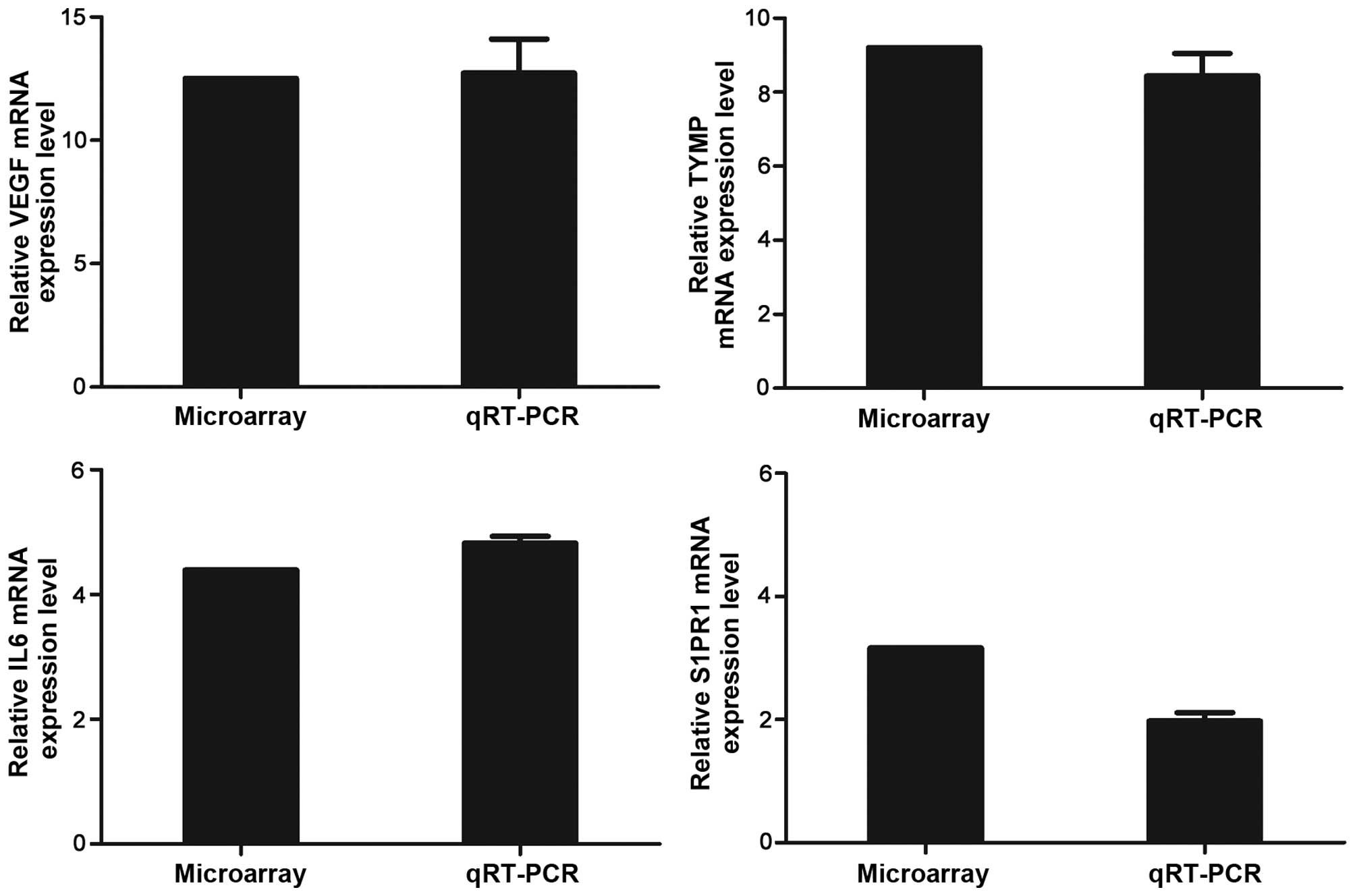
According to the relevant functional annotations and
their high fold-changed values, IL6 which is involved in the
JAK-STAT signaling pathway and Toll-like receptor signaling
pathway, VEGFA which is involved in VEGF signaling pathway and mTOR
signaling pathway, S1PR1 and TYMP which are involved in the
biological process of angiogenesis and cell differentiation were
chosen for qRT-PCR validation. The HUVECs used for qRT-PCR and
microarray analysis were induced by KYSE70 supernatant at the same
condition. For all the four genes, the qRT-PCR results were in
accordance with the microarray date (Fig. 4).
Differences in the mRNA levels of the
target genes analyzed in HUVECs induced by human esophageal
carcinoma tissue homogenates
KYSE70 human esophageal carcinoma conditioned medium
is different from the esophageal carcinoma tissue of patients.
qRT-PCR was further used in induced HUVECs by human esophageal
carcinoma tissue homogenates. The results of qRT-PCR showed that
IL6, VEGFA, S1PR1, TYMP mRNA increased in the induced HUVECs by
esophageal carcinoma tissue homogenates compared with induced
HUVECs by pericarcinoma tissue homogenates (p<0.001).
Discussion
Esophageal carcinoma is one of the most common
cancers in China. When the tumor grows beyond 1–2 mm in diameter,
angiogenesis is indispensable for supporting the expansive growth.
It has been proved that the tumor microenvironment governs
tumor-associated angiogenesis in multiple ways, and there are great
differences between tumor vascular endothelial cells and normal
vascular endothelial cells (6,16).
Herein, for the first time, we found a significant alteration of
gene expression in induced HUVECs by tumor microenvironment which
was simulated by esophageal cancer cell supernatant (KYSE70). This
study introduces a new prospect that contributes to understanding
the influence of tumor microenvironment on angiogenesis, and has an
important clinical significance in inhibiting the developing of
neovascularization, which would be a promising therapeutic strategy
for cancer treatment.
In the present study, we used gene chip and
bioinformatics technology to study the gene expression profile of
induced HUVECs by KYSE70 supernatant. Further statistical analysis
of differentially expressed genes revealed 3769 differentially
expressed genes in induced HUVECs, including 1609 up regulated
genes and 2160 downregulated genes (Fig. 1A). These genes were classified
according to their cellular component, molecular function,
biological process and pathway. The results indicate that there are
different molecular mechanisms governing the process of
angiogenesis affected by tumor microenvironment.
The analysis of gene annotations in this study
implied that the differently expressed genes mainly play important
roles in cell differentiation and angiogenesis, and are involved in
the MAPK signaling pathway, JAK-STAT signaling pathway, Toll-like
receptor signaling pathway, Hematopoietic cell lineage,
Cytokine-cytokine receptor interaction, VEGF signaling pathway,
mTOR signaling pathway, and Wnt signaling pathway. These pathways
or related genes have been previously confirmed to have great
relation with angiogenesis and cell differentiation (6,17,18).
VEGF signaling pathway is involved in proliferation,
survival, migration and integrity of endothelial cells, which play
a key role in cancer-induced angiogenic processes (6). Moreover, mTOR signaling pathway has
equal importance in tumorigenesis, development and the
differentiation of tumor stem cells (19). In this research, VEGFA which was
associated with these pathways was confirmed highly expressed in
induced HUVECs by qRT-PCR and the fold-change was consistent with
the microarray data. Further study indicated that induced HUVECs by
esophageal carcinoma tissue homogenate also expressed VEGFA at a
high level. These results may illustrate that tumor
microenvironment has influence on tumor angiogenesis by the way of
normal endothelial cell differentiation toward tumor endothelial
cells, and VEGFA may play an important role in this process.
Although many kinds of agents based on VEGF or its receptor have
been created, there is a need for further in-depth study (17).
Another upregulated gene, IL6, which is an
angiogenic member of the CXC chemokine family was also identified
by qRT-PCR. It has been proved that IL6 produced by endothelial
cells with the tumor contributes to tumor development through
neovascularization (20). IL6 was
also involved in the JAK-STAT signaling pathway and Toll-like
receptor signaling pathway which were significantly changed and
play an important role in angiogenesis (21). As the expression level of IL6 is
higher in induced HUVECs, we predict that there may be some
pro-angiogenic constituents in the tumor micro environment
promoting normal endothelial cells differentiation toward tumor
endothelial cells.
Another pathway that was significantly changed was
Wnt signaling pathway. It has been report that the Wnt signaling
promotes neovascularization of the retina in patients with diabetic
retinopathy, and has an important role in angiogenic activity of
endothelial cells (18). The Wnt
signaling pathway controls the proliferation, migration and
differentiation of vascular cells as well as the expression of
angiogenic factors, such as VEGF and IL8 (18,22).
The Wnt signaling pathway may promote neovascularization by
promoting normal endothelial cell differentiation into tumor
vascular endothelial cells.
Another gene involved in angiogenesis and
endothelial cell differentiation is S1PR1 (sphingosine 1-phosphate
receptor 1) which is one of the five G protein-coupled receptors
for S1P1 (23). Previous studies
have suggested that S1PR1 is required for stabilization of nascent
blood vessels during embryonic development (24). Further study using the Levis lung
carcinoma model of tumor growth showed that micro vessels within
the tumor expressed S1PR1. Immunofluorescence analysis with the
S1PR1 antibody also confirmed that S1PR1 is induced in endothelial
cells during tumor angiogenesis (25). In this study, the high S1PR1
expression level in induced HUVECs illustrated that the HUVECs
already have the characteristic of tumor vessel endothelial cells.
S1PR1 may have the potential to be a new biomarker in antitumor
angiogenesis.
TYMP is an enzyme involved in pyrimidine nucleoside
metabolism. It can catalyze the reversible phosphorolysis of
thymidine, deoxyuridine and their analogs to their bases and
2-deoxyribose-1-phosphate (26). It
has been reported that the TYMP expression is significantly higher
in the patient with endometriosis, which indicates it may play a
key role in angiogenesis (27).
Another study demonstrated that TYMP and VEGF expression were
correlated to patient with colorectal cancer, and TYMP plays an
important role in angiogenesis, ECM remodeling, and in the
prognosis of patients with colorectal cancer (28). Further studies are needed to define
its role, but it is clear that TYMP has a potential role as a new
biomarker in anti-angiogenesis.
In conclusion, the comparison of the gene expression
profiles of induced HUVECs by KYSE70 supernatant and normal HUVECs
suggested that the gene expression of endothelial cells were
altered according to their microenvironment. We believe that the
results pave the way to more functional studies that are needed to
elucidate their possible role in tumor angiogenesis. Specifically,
major signaling pathways particularly the VEGF signaling pathway,
mTOR signaling pathway, cytokine-cytokine receptor interaction and
Wnt signaling pathway play key roles in angiogenesis induced by
tumor microenvironment. Moreover, several genes such as VEGFA, IL6,
TYMP, S1PR1, especially TYMP and S1PR1 are suggested to serve as
new biomarkers of angiogenesis, and can be used as targets to
against esophageal carcinoma.
Acknowledgments
This work was supported by National Natural Science
Foundation of China (no. 81572972), Science and Technology
Innovation Talents Support Plan of University in Henan Province
(no. 15HASTIT038), and Science Foundation of Zhengzhou University
for the Excellent young teacher (no. 1421328057).
References
|
1
|
Miura S, Mitsui K, Heishi T, Shukunami C,
Sekiguchi K, Kondo J, Sato Y and Hiraki Y: Impairment of
VEGF-A-stimulated lamellipodial extensions and motility of vascular
endothelial cells by chondromodulin-I, a cartilage-derived
angiogenesis inhibitor. Exp Cell Res. 316:775–788. 2010. View Article : Google Scholar
|
|
2
|
Park HJ, Kim MN, Kim JG, Bae YH, Bae MK,
Wee HJ, Kim TW, Kim BS, Kim JB, Bae SK, et al: Up-regulation of
VEGF expression by NGF that enhances reparative angiogenesis during
thymic regeneration in adult rat. Biochim Biophys Acta.
1773:1462–1472. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Folkman J: Anti-angiogenesis: New concept
for therapy of solid tumors. Ann Surg. 175:409–416. 1972.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Abajo A, Bitarte N, Zarate R, Boni V,
Lopez I, Gonzalez-Huarriz M, Rodriguez J, Bandres E and
Garcia-Foncillas J: Identification of colorectal cancer metastasis
markers by an angiogenesis-related cytokine-antibody array. World J
Gastroenterol. 18:637–645. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu YR, Guan YY, Luan X, Lu Q, Wang C, Liu
HJ, Gao YG, Yang SC, Dong X, Chen HZ, et al: Delta-like ligand
4-targeted nanomedicine for antiangiogenic cancer therapy.
Biomaterials. 42:161–171. 2015. View Article : Google Scholar
|
|
6
|
Weis SM and Cheresh DA: Tumor
angiogenesis: Molecular pathways and therapeutic targets. Nat Med.
17:1359–1370. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Weis SM and Cheresh DA: Pathophysiological
consequences of VEGF-induced vascular permeability. Nature.
437:497–504. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu J, Zhao J, Liu K, Zhao J, Yang H, Huang
Y, Qin Z, Bai R, Li P, Ma J, et al: MAPK/ERK1/2 signaling mediates
endothelial-like differentiation of immature DCs in the
microenvironment of esophageal squamous cell carcinoma. Cell Mol
Life Sci. 67:2091–2106. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu J, Bai R, Qin Z, Zhang Y, Zhang X,
Jiang Y, Yang H, Huang Y, Li G, Zhao M, et al: Differentiation of
immature DCs into endothelial-like cells in human esophageal
carcinoma tissue homogenates. Oncol Rep. 30:739–744.
2013.PubMed/NCBI
|
|
10
|
Hillen F and Griffioen AW: Tumour
vascularization: Sprouting angiogenesis and beyond. Cancer
Metastasis Rev. 26:489–502. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang XX, Wang K, Li XZ, Zhai LQ, Qu CX,
Zhao Y, Liu ZR, Wang HZ, An QJ, Jing LW, et al: Targeted knockdown
of IQGAP1 inhibits the progression of esophageal squamous cell
carcinoma in vitro and in vivo. PLoS One. 9:e965012014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bisacchi D, Benelli R, Vanzetto C, Ferrari
N, Tosetti F and Albini A: Anti-angiogenesis and angioprevention:
Mechanisms, problems and perspectives. Cancer Detect Prev.
27:229–238. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carmeliet P and Jain RK: Molecular
mechanisms and clinical applications of angiogenesis. Nature.
473:298–307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim BH, Lee Y, Yoo H, Cui M, Lee S, Kim
SY, Cho JU, Lee H, Yang BS, Kwon YG, et al: Anti-angiogenic
activity of thienopyridine derivative LCB03-0110 by targeting
VEGFR-2 and JAK/STAT3 signalling. Exp Dermatol. 24:503–509. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu J, Bai R, Qin Z, Zhang Y, Zhang X,
Jiang Y, Yang H, Huang Y, Li G, Zhao M, et al: Differentiation of
immature DCs into endothelial-like cells in human esophageal
carcinoma tissue homogenates. Oncol Rep. 30:739–744.
2013.PubMed/NCBI
|
|
16
|
St Croix B, Rago C, Velculescu V, Traverso
G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C,
Vogelstein B, et al: Genes expressed in human tumor endothelium.
Science. 289:1197–1202. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Finley SD and Popel AS: Effect of tumor
microenvironment on tumor VEGF during anti-VEGF treatment: Systems
biology predictions. J Natl Cancer Inst. 105:802–811. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang L, Yin M, Wei X, Liu J, Wang X, Niu
C, Kang X, Xu J, Zhou Z, Sun S, et al: Bach1 represses
Wnt/β-catenin signaling and angiogenesis. Circ Res. 117:364–375.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Severi T, van Malenstein H, Verslype C and
van Pelt JF: Tumor initiation and progression in hepatocellular
carcinoma: Risk factors, classification, and therapeutic targets.
Acta Pharmacol Sin. 31:1409–1420. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gijsbers K, Gouwy M, Struyf S, Wuyts A,
Proost P, Opdenakker G, Penninckx F, Ectors N, Geboes K and Van
Damme J: GCP-2/CXCL6 synergizes with other endothelial cell-derived
chemokines in neutrophil mobilization and is associated with
angiogenesis in gastrointestinal tumors. Exp Cell Res. 303:331–342.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin ZY, Chuang WL and Chuang YH:
Amphotericin B up-regulates angiogenic genes in hepatocellular
carcinoma cell lines. Eur J Clin Invest. 39:239–245. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou Y and Nathans J: Gpr124 controls CNS
angiogenesis and blood-brain barrier integrity by promoting
ligand-specific canonical wnt signaling. Dev Cell. 31:248–256.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arnon TI, Xu Y, Lo C, Pham T, An J,
Coughlin S, Dorn GW and Cyster JG: GRK2-dependent S1PR1
desensitization is required for lymphocytes to overcome their
attraction to blood. Science. 333:1898–1903. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jain RK: Molecular regulation of vessel
maturation. Nat Med. 9:685–693. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chae SS, Paik JH, Furneaux H and Hla T:
Requirement for sphingosine 1-phosphate receptor-1 in tumor
angiogenesis demonstrated by in vivo RNA interference. J Clin
Invest. 114:1082–1089. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ciaparrone M, Quirino M, Schinzari G,
Zannoni G, Corsi DC, Vecchio FM, Cassano A, La Torre G and Barone
C: Predictive role of thymidylate synthase, dihydropyrimidine
dehydrogenase and thymidine phosphorylase expression in colorectal
cancer patients receiving adjuvant 5-fluorouracil. Oncology.
70:366–377. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Laudanski P, Charkiewicz R, Kuzmicki M,
Szamatowicz J, Świątecka J, Mroczko B and Niklinski J: Profiling of
selected angiogenesis-related genes in proliferative eutopic
endometrium of women with endometriosis. Eur J Obstet Gynecol
Reprod Biol. 172:85–92. 2014. View Article : Google Scholar
|
|
28
|
Mitselou A, Ioachim E, Skoufi U, Tsironis
C, Tsimogiannis KE, Skoufi C, Vougiouklakis T and Briasoulis E:
Predictive role of thymidine phosphorylase expression in patients
with colorectal cancer and its association with
angiogenesis-related proteins and extracellular matrix components.
In Vivo. 26:1057–1067. 2012.PubMed/NCBI
|