Introduction
Lung cancer is one of the most commonly diagnosed
cancers and remains the leading cause of cancer-related death
worldwide (1,2). It has been traditionally subdivided
into two principal groups, namely, small cell lung cancer and
non-small cell lung cancer. The latter type is more common than the
former. Despite diverse treatment methods including surgery,
radiation therapy and chemotherapy, the overall 5-year survival
rate remains ~18.2% (3). The high
mortality rate of lung cancer is partly due to the lack of
effective prognostic biomarkers. Therefore, the identification of
novel prognostic factors as biomarkers that may be used in the
early detection of lung cancer is critical.
Long non-coding RNAs (lncRNAs) are mRNA-like
transcripts with more than 200 nucleotides that lack significant
protein-coding abilities (4,5).
Increasing evidence suggests that lncRNAs are a new class of
players involved in the development and progression of cancer
(6). More and more research
suggests that these transcripts are frequently aberrantly expressed
in cancers, and some have been implicated in the diagnosis and
prognostication (7) in
neuroblastoma (8), prostate
(9), breast (10–12),
ovarian (13,14), gastric (15) and colorectal cancer (16,17),
and multiple myeloma (18). Due to
the specific expression of lncRNAs in cancer, lncRNAs could become
biomarkers by which to diagnosis cancer or predict patient
survival. Thus, identification of various lncRNAs which are
specifically expressed in lung cancer may have predictive and
prognostic value for lung cancer patients.
Currently, massive lncRNA-specific probes are
presented on microarray platforms (Affymetrix U133 Plus 2.0); thus,
we are able to use previously published gene expression microarray
data from the Gene Expression Omnibus (GEO) database to identify
various prognostic signature lncRNAs. Furthermore, bioinformatic
analysis was used to identify the signaling pathways that involve
lncRNAs by Gene Set Enrichment Analysis (GSEA).
Materials and methods
Lung cancer datasets and patient
information
Lung cancer datasets were downloaded from the GEO
database. A total of 739 patients were utilized in the present
study after filtering out samples without clinical survival
information. It included 293 patients from GSE30219 (19), 226 patients from GSE31210 (20), 168 patients from GSE37745 (21), and 52 patients from GSE19188
(22). We selected these datasets
that included >50 patients with survival status information. We
followed the strategy of using the largest dataset (GSE30219) as
training set. Three independent datasets (GSE31210, GSE37745 and
GSE19188) were included in the present study as testing sets.
Microarray processing and lncRNA profile
mining
All the microarray raw data (CEl files) of four lung
cancer cohorts were processed using the robust multichip average
(RMA) algorithm for background adjustment (23). GATExplorer was used to process
microarrays on a local computer for gene expression of lncRNAs
(24). lncRNA mapper was obtained
from GATExplorer, which included the probes that do not map to any
coding region but that were mapped to a database for non-coding
RNAs of human and mouse (derived from RNAdb (25). The coding potential analysis of the
lncRNAs was carried out by CNCI to classify protein-coding or
non-coding transcripts (26). Each
lncRNA included at least a minimum of three probes mapping in the
corresponding lncRNA entity. We created a risk-score formula
according to the expression of these eight lncRNAs for survival
prediction. Patients having higher risk scores were expected to
have poorer survival outcomes.
Statistical analysis
The association between the lncRNA gene expression
and patient survival was assessed by univariable Cox proportional
hazards regression analysis along with a permutation test using
BRB-ArrayTools (Biometric Research Branch) package (27) in the training set. We identified
expression of several lncRNAs that were strongly correlated with
survival. Considering that a smaller number of genes in the model
would make the model more practical, we performed the random
survival forests variable hunting (RSFVH) algorithm (28). Using a smaller number of genes
selected fitted in a multivariable Cox regression model; we
constructed a formula to predict survival in the training set. Each
patient was assigned a risk score that is a linear combination of
the expression levels of the significant lncRNAs weighted by their
respective Cox regression coefficients (29). According to this risk score,
patients in the training set were divided into low-risk and
high-risk groups using the median risk score as the cut-off. The
Kaplan-Meier method was used to estimate survival time, and other
three independent testing groups were performed for validation.
Differences in survival times between the low-risk and high-risk
groups in each set were compared by the two-sided log-rank test,
respectively.
Bioinformatic analysis of lncRNA gene
function
GSEA was performed by the JAVA program (http://www.broadinstitute.org/gsea) using MSigDB
C2 CP: canonical pathway gene set collection (1,320 gene sets
available). Gene sets with a false discovery rate (FDR) value
<0.05 after performing 1,000 permutations were considered to be
significantly enriched (30).
Cytoscape (version 2.8.2) and the Enrichment Map software were used
to visualize the GSEA results (31). Encyclopedia of Genes and Genomes
(KEGG) enrichment analyses of the co-expressed protein-coding genes
with prognostic lncRNAs were performed to predict the biological
function of prognostic lncRNAs using the DAVID Bioinformatics Tool
(version 6.7) (32). Enrichment
analysis was carried out using the functional annotation clustering
options, and was limited to KEGG pathways in the 'Biological
Process' categories.
Results
Identification of prognostic lncRNA genes
from the training set
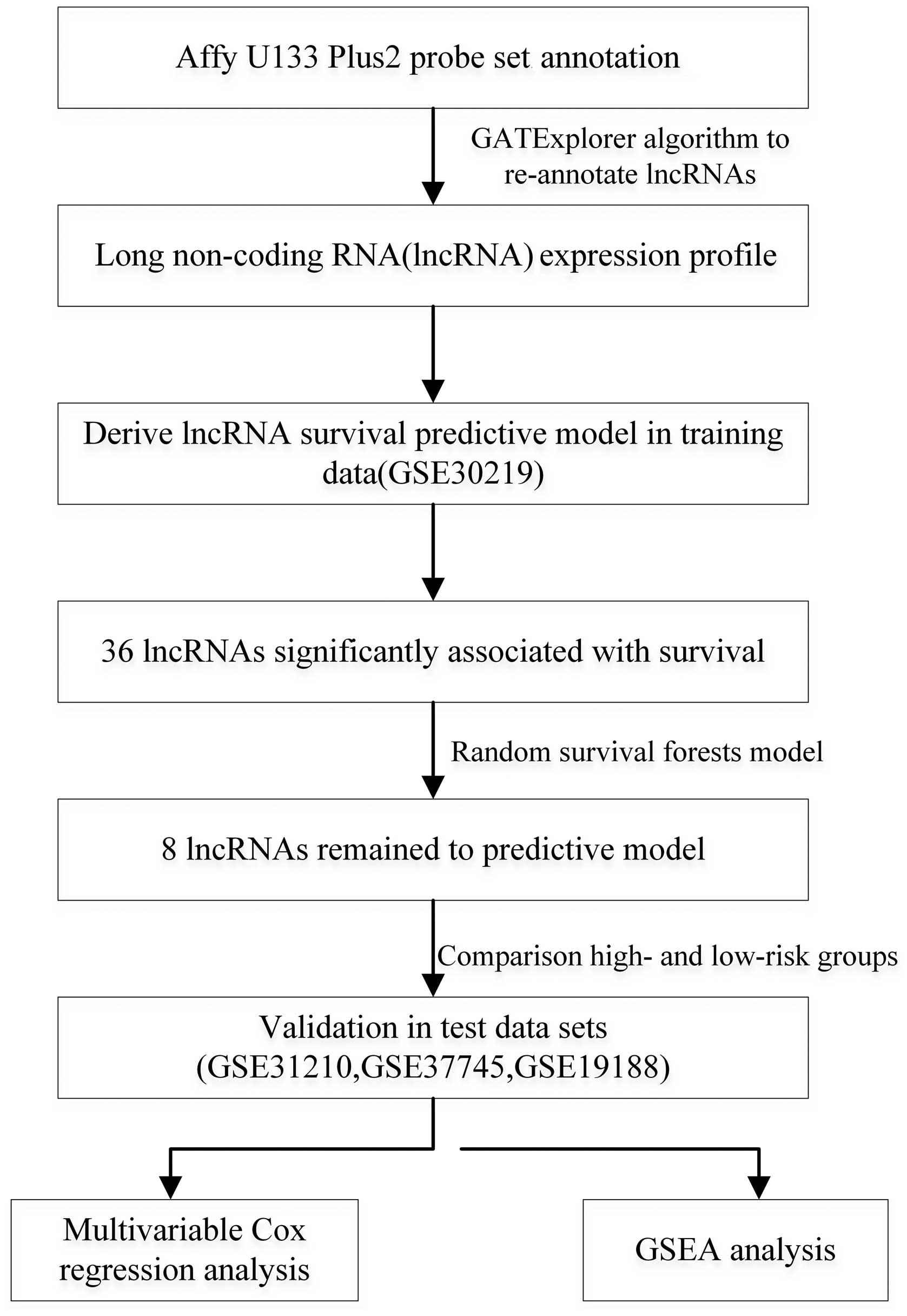
As summarized in the workflow (Fig. 1), all analyses were performed in the
training set (GSE30219) and validated in the testing set (GSE31210,
GSE37745 and GSE19188). The training set (n=293) was analyzed for
the detection of prognostic lncRNA genes. By subjecting the lncRNA
expression data derived from the training set to univariable Cox
proportional hazards regression analysis using the BBRB-ArrayTools,
we identified a set of 36 lncRNAs that were strongly correlated
with patient overall survival (p<0.001 and FDR <0.001) from a
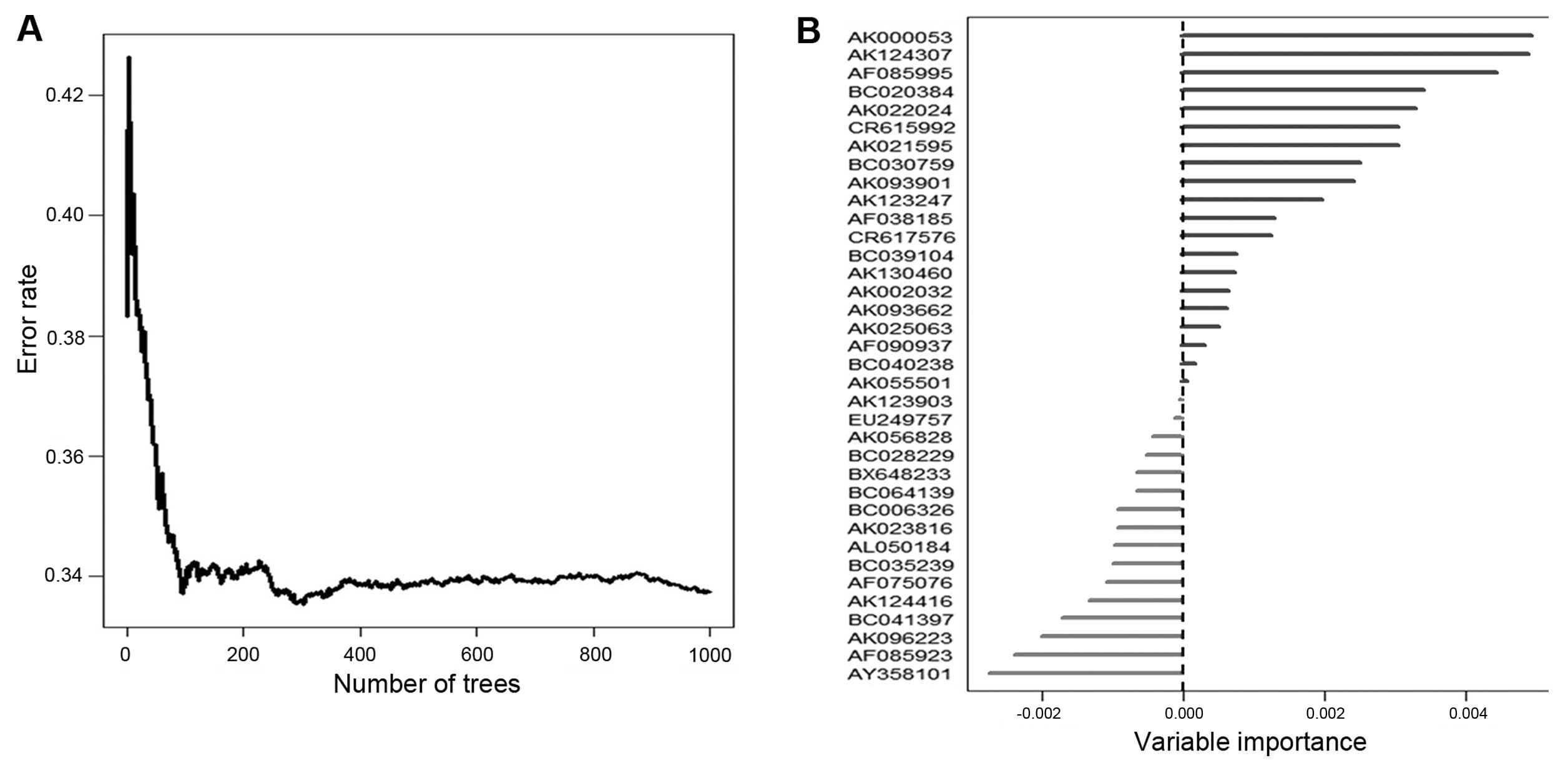
total of 5,635 lncRNAs. Based on the random survival forests model
(see Materials and methods), the eight lncRNAs (Table I) were selected as predictors
(Fig. 2). In Table I a list of these eight lncRNAs is
shown with their obtained coefficient and variable importance
values. Based on these results, BC030759 was the most relevant with
overall survival in the training set (HR=3.513); the positive
coefficients of the lncRNAs (AK021595, BC030759, AK000053, BC020384
and AK022024) indicated that their higher levels of expression were
associated with shorter survival, and the negative coefficients of
the other lncRNAs (AK124307, CR615992 and AF085995) indicated that
their higher levels of expression were associated with longer
survival. All of the eight lncRNAs have been verified in the ncRNA
Expression Database (www.nred.matticklab.com) and these eight transcripts
were classified as ncRNAs in this website (33). As coding potential analysis is
commonly used to classify whether a transcript is of coding
potential or not (34), we used
CNCI to test those eight transcripts (26). This tool also suggested that all the
eight transcripts were non-coding transcripts with no coding
potential.
 | Table IEight lncRNAs are significantly
associated with the overall survival in the training set
(n=293). |
Table I
Eight lncRNAs are significantly
associated with the overall survival in the training set
(n=293).
| lncRNA | Parametric
p-value | FDR | Hazard ratio | Coefficient | Variable
importance | Relative
importance |
|---|
| AK021595 | 3.00E-07 | 8.90E-05 | 1.621 | 0.212 | 0.003 | 0.6171 |
| BC030759 | 3.00E-07 | 8.90E-05 | 3.513 | 0.416 | 0.0025 | 0.5086 |
| AK000053 | 3.00E-07 | 8.90E-05 | 1.884 | 0.322 | 0.0049 | 1 |
| AK124307 | 6.00E-07 | 0.000154 | 0.696 | −0.165 | 0.0049 | 0.9943 |
| BC020384 | 1.40E-06 | 0.000303 | 1.716 | 0.301 | 0.0034 | 0.6914 |
| AK022024 | 1.60E-06 | 0.000334 | 2.127 | 0.423 | 0.0033 | 0.6686 |
| CR615992 | 2.60E-06 | 0.000488 | 0.527 | −0.084 | 0.003 | 0.6171 |
| AF085995 | 6.10E-06 | 0.000764 | 0.458 | −0.459 | 0.0044 | 0.9029 |
An eight-lncRNA signature predicts
survival of lung cancer patients in the training set
To investigate whether the eight-lncRNA signature
could provide an accurate prediction of survival in lung cancer
patients, we created a risk-score formula according to the
expression of these eight lncRNAs for survival prediction in the
training set GSE30219 (n=293), as follows: risk score,
0.212*AK021595+0.416*BC030759
+0.322*AK000053−0.165*AK124307+0.301*BC020384+0.42
3*AK022024−0.084*CR615992−0.459*AF085995. Then, we calculated the
eight-lncRNA signature risk score for each patient in the training
set. Patients were divided into a low-risk or high-risk group using
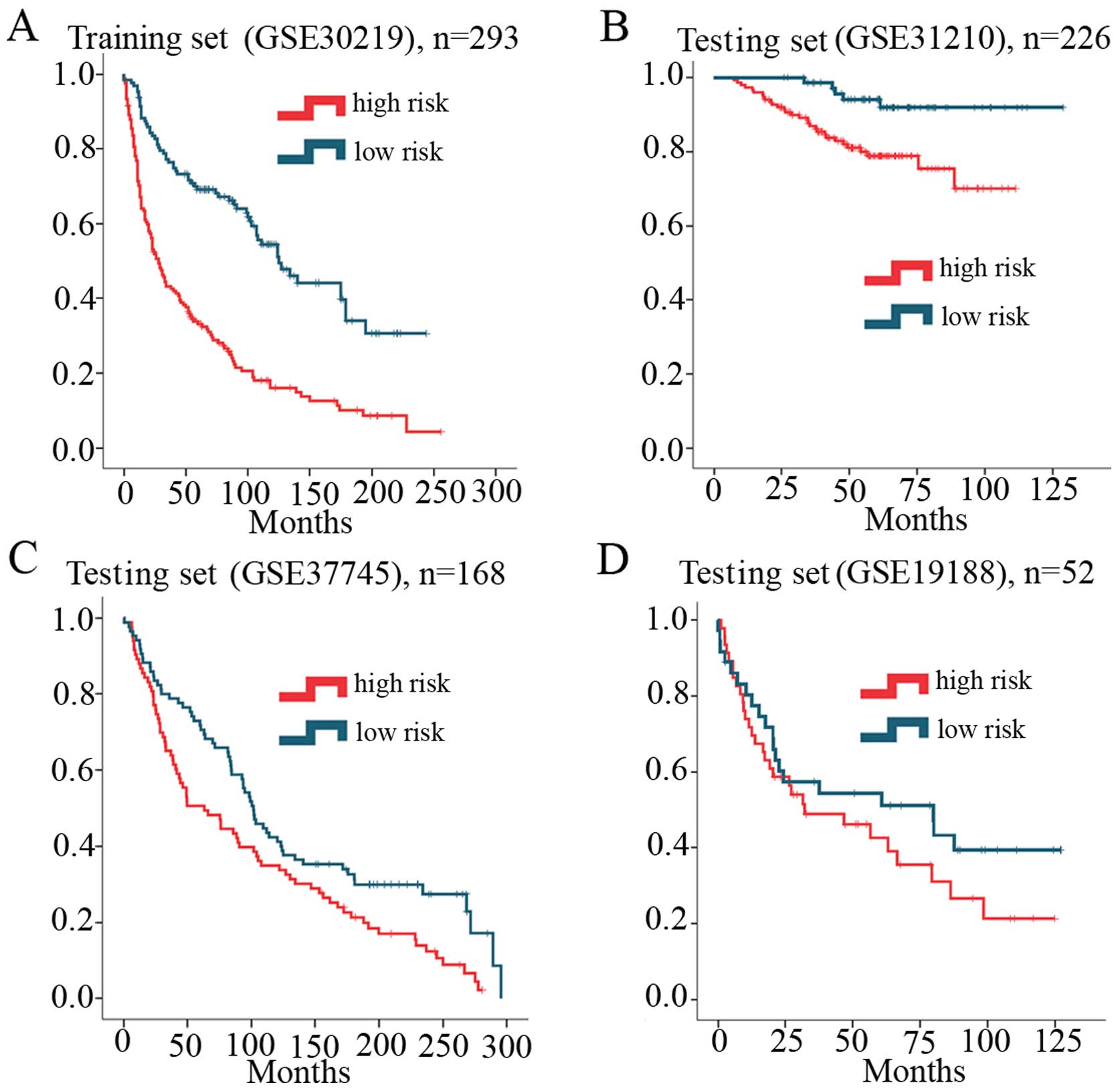
the median risk score as cut-off value. Patients in the high-risk
group had a shorter survival time than patients in the low-risk
group (Fig. 3A). The association of
the eight-lncRNA risk score and survival was also significant when
it was evaluated as a continuous variable in the univariable Cox
regression model (Table II).
 | Table IIUnivariable and multivariable Cox
regression analyses in the training and testing set. |
Table II
Univariable and multivariable Cox
regression analyses in the training and testing set.
| Variables | Univariable model
| Multivariable model
|
|---|
| HR | 95% CI of HR | P-value | HR | 95% CI of HR | P-value |
|---|
| Training set
(GSE30219) |
| Risk score | 2.718 | 2.171–3.404 | <0.0001 | 2.666 | 2.108–3.371 | <0.0001 |
| Age (years) | 1.038 | 1.024–1.052 | <0.0001 | 1.028 | 1.013–1.043 | <0.0001 |
| Gender | 0.589 | 0.375–0.926 | 0.022 | 0.786 | 0.499–1.238 | 0.299 |
| Testing set
(GSE31210) |
| Risk score | 2.637 | 1.306–5.326 | 0.007 | 2.569 | 1.223–5.399 | 0.013 |
| Age (years) | 1.025 | 0.977–1.075 | 0.306 | 1.031 | 0.983–1.082 | 0.211 |
| Gender | 0.658 | 0.338–1.281 | 0.219 | 0.774 | 0.386–1.553 | 0.471 |
| Testing set
(GSE37745) |
| Risk score | 1.667 | 1.138–2.443 | 0.009 | 1.571 | 1.056–2.338 | 0.025 |
| Age (years) | 1.011 | 0.994–1.03 | 0.210 | 1.007 | 0.988–1.025 | 0.485 |
| Gender | 0.807 | 0.576–1.132 | 0.214 | 0.888 | 0.628–1.257 | 0.504 |
| Testing set
(GSE19188) |
| Risk score | 1.665 | 0.94–2.948 | 0.081 | 1.42 | 0.782–2.579 | 0.250 |
| Gender | 0.471 | 0.235–0.943 | 0.034 | 0.53 | 0.258–1.09 | 0.084 |
Validation of the eight-lncRNA signature
for survival prediction in the testing sets
To confirm our findings, we calculated the risk
score in the testing sets including GSE31210 (n=226), GSE37745
(n=168) and GSE19188 (n=52). Similar to the training set findings,
patients in the high-risk group had a shorter survival time than
patients in the low-risk group (Fig.
3B–D). Meanwhile, patient survival throughout the follow-up in
the low-risk group was better when compared to survival in the
high-risk group. In the univariable Cox regression model, the risk
score was similar with the high-risk group which had a shorter
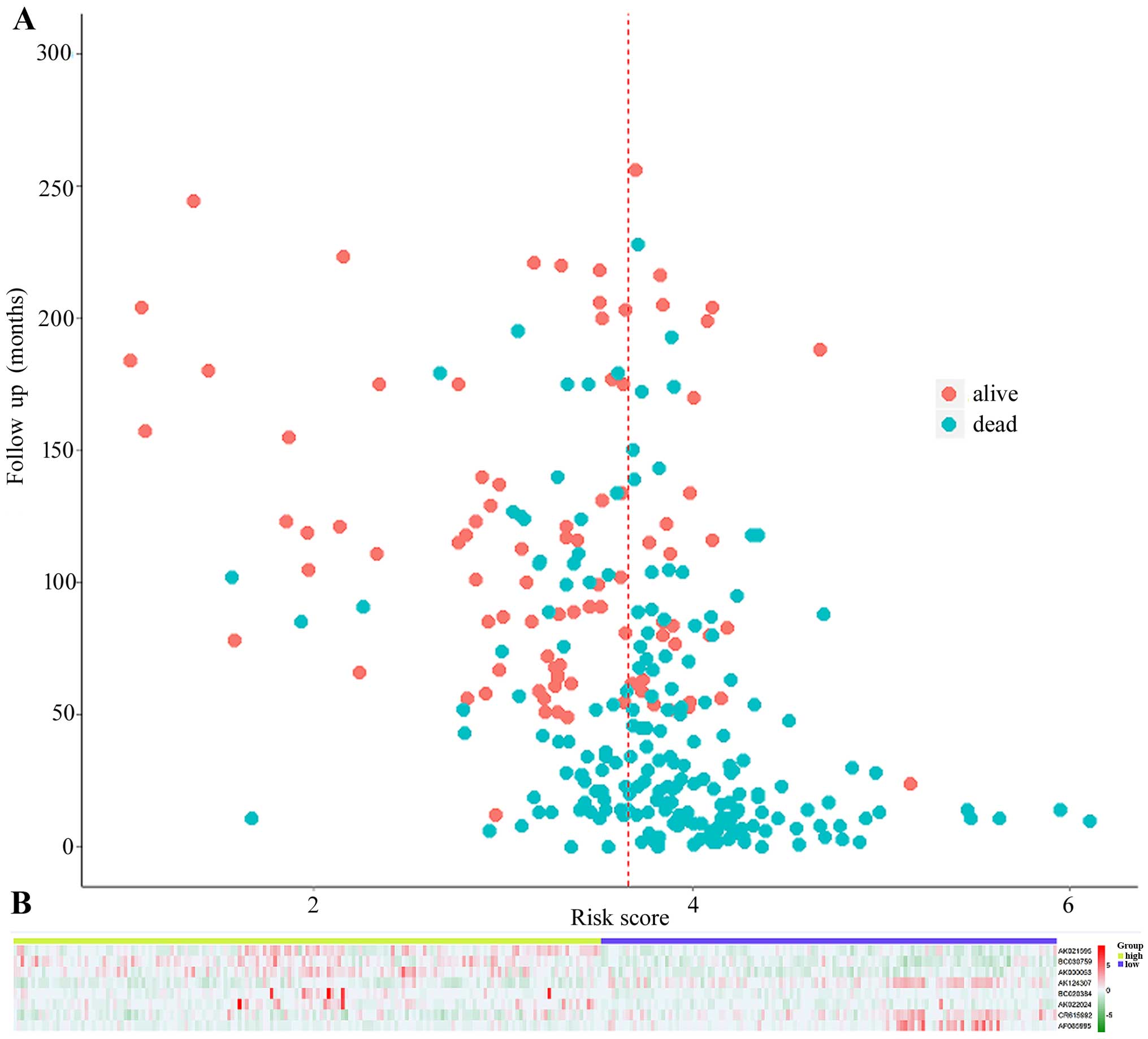
overall survival. The patient survival status (Fig. 4A) and lncRNA values (Fig. 4B) were analyzed independently in the
training set. Some of the clinical information (stage and subtype)
was not available for a substantial proportion of cases, thus we
performed multivariate Cox regression analysis concerning age and
gender. The result showed that the eight-lncRNA expression
signature was independent of age and gender. Eight-lncRNA risk
score, age (available in GSE31210 and GSE37745) and gender
(available in GSE31210, GSE37745 and GSE19188) were defined as
covariates. These results showed that risk score was an independent
predictor of lung cancer patient survival (Table II).
Identification of eight-lncRNA
signature-associated biological pathways and processes
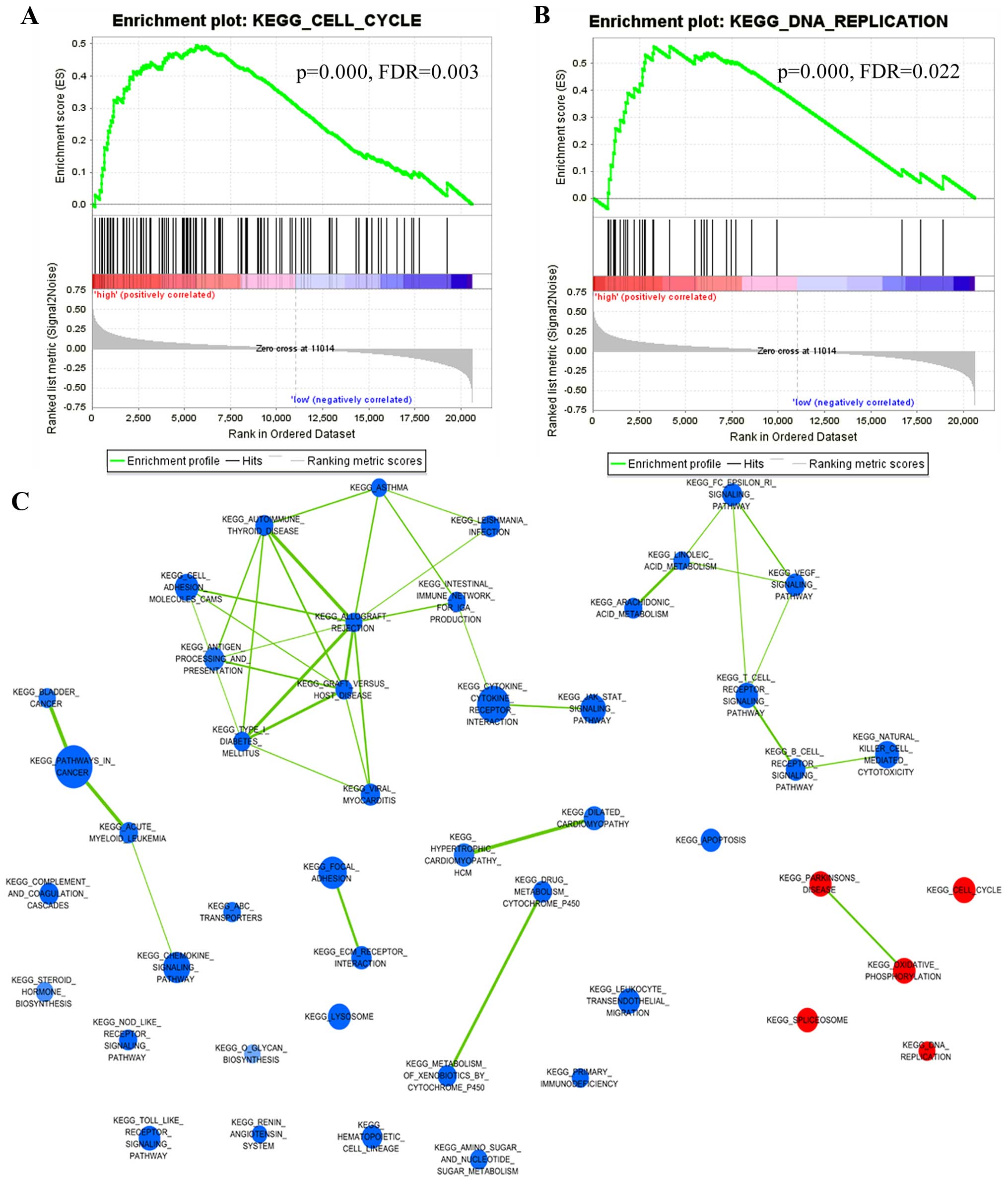
GSEA was carried out to identify the associated
biological processes and signaling pathways (30). We compared the gene expression
profile of lung cancer patients in the low-risk and high-risk
groups classified by the eight-lncRNA gene signature in the
training set (GSE 30219). The gene sets with significantly
different expression (FDR <0.01; p<0.005) were picked up,
which implied that the signature may be involved in the cell cycle
and DNA replication-related pathways (Fig. 5A and B), and it was visualized as an
interaction network with Cytoscape (Fig. 5C). These related pathways were
reported to affect cancer cell proliferation (35–37).
Discussion
As a new class of ncRNAs, lncRNAs were demonstrated
to be dysregulated in a variety of diseases, particularly in
cancers (38). Numerous studies of
abnormal lncRNA expression in various types of cancer suggest that
they play an important role in tumorigenesis, and lncRNAs may serve
as independent biomarkers for diagnosis and prognosis (39,40).
In lung cancer, numerous studies have investigated lncRNAs to
predict lung cancer patient survival (41–44).
Nevertheless, a single factor to predict the prognosis of tumors is
not accurate, since high specificity and sensitivity are lacking
for most lncRNAs. Currently, research has found that lncRNA
expression profiles can be obtained from publicly available,
custom-designed DNA microarrays by re-annotating the array probes
(12,17,34,45,46).
In the present study, in order to construct a risk
score model, we downloaded four datasets (GSE30219, GSE31210,
GSE37745 and GSE19188) from GEO databases, and obtained the lncRNA
profiling of lung cancer patients. We identified a prognostic,
eight-lncRNA signature from the training set. Furthermore,
examination of associated molecular pathways revealed that the
eight-lncRNA signature was more likely to involve the cell cycle
and DNA replication signaling pathways. Cell cycle disorder and DNA
replication induce cell proliferation and affect genome
instability, further increasing the possibility of canceration of
unstable cells, which participates in tumor occurrence and
development (35,47–49).
Thus, our findings suggest that lncRNA signatures may provide an
efficient classification tool for the clinical prognosis of lung
cancer.
Zhou et al (46) also identified an eight-lncRNA
signature which may be an effective independent prognostic
molecular biomarker in the prediction of non-small lung cancer
patient survival, and our findings support the characteristics of
the eight-lncRNA. The overexpression of lncRNAs (AK021595,
BC030759, AK000053, BC020384 and AK022024) were found to be
correlated with shorter survival while other lncRNAs (AK124307,
CR615992 and AF085995) were downregulated in the high-risk group
compared to the low-risk group. Most importantly, the functional
study in cancer of these eight lncRNAs has not been reported to
date.
The limitations should be acknowledged for the
present study. First, in the present study, only 5,635 (out of
15,000+) human lncRNAs were included. The prognostic lncRNAs
identified here may not represent all the lncRNA candidates that
were potentially correlated with lung cancer overall survival.
Secondly, the longest survival time in the model was 250 months,
Thus, the patients in GSE37745 whose survival time was >300
months were removed. Thirdly, stage was not included in the present
study, since this information was not available for a substantial
proportion of cases. Meanwhile, the functions of these eight
lncRNAs were inferred by bioinformatics analysis, and these
biological roles in tumorigenesis were not clear and should be
investigated in experimental studies.
In summary, we identified a signature of a set of
eight lncRNAs, which predicted the overall survival in three
independent testing sets. Further bioinformatic analysis revealed
that the prognostic value was independent of age and gender.
Moreover, these lncRNAs are involved in cell cycle and DNA
replication signaling pathways. These lncRNAs may have clinical
implications as diagnostic markers. However, the biological roles
of these eight lncRNAs in tumorigenesis require further study.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (nos. 81400121, 81270607,
81541027 and 81501352).
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
GEO
|
Gene Expression Omnibus
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ling H, Vincent K, Pichler M, Fodde R,
Berindan-Neagoe I, Slack FJ and Calin GA: Junk DNA and the long
non-coding RNA twist in cancer genetics. Oncogene. 34:5003–5011.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lipovich L, Johnson R and Lin CY: MacroRNA
underdogs in a microRNA world: Evolutionary, regulatory, and
biomedical significance of mammalian long non-protein-coding RNA.
Biochim Biophys Acta. 1799:597–615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar
|
|
7
|
Qi P and Du X: The long non-coding RNAs, a
new cancer diagnostic and therapeutic gold mine. Mod Pathol.
26:155–165. 2013. View Article : Google Scholar
|
|
8
|
Yarmishyn AA, Batagov AO, Tan JZ, Sundaram
GM, Sampath P, Kuznetsov VA and Kurochkin IV: HOXD-AS1 is a novel
lncRNA encoded in HOXD cluster and a marker of neuroblas-toma
progression revealed via integrative analysis of noncoding
transcriptome. BMC Genomics. 15(Suppl 9): S72014. View Article : Google Scholar
|
|
9
|
Crea F, Watahiki A, Quagliata L, Xue H,
Pikor L, Parolia A, Wang Y, Lin D, Lam WL, Farrar WL, et al:
Identification of a long non-coding RNA as a novel biomarker and
potential therapeutic target for metastatic prostate cancer.
Oncotarget. 5:764–774. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shen X, Xie B, Ma Z, Yu W, Wang W, Xu D,
Yan X, Chen B, Yu L, Li J, et al: Identification of novel long
non-coding RNAs in triple-negative breast cancer. Oncotarget.
6:21730–21739. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun J, Chen X, Wang Z, Guo M, Shi H, Wang
X, Cheng L and Zhou M: A potential prognostic long non-coding RNA
signature to predict metastasis-free survival of breast cancer
patients. Sci Rep. 5:165532015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Meng J, Li P, Zhang Q, Yang Z and Fu S: A
four-long non-coding RNA signature in predicting breast cancer
survival. J Exp Clin Cancer Res. 33:842014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou M, Wang X, Shi H, Cheng L, Wang Z,
Zhao H, Yang L and Sun J: Characterization of long non-coding
RNA-associated ceRNA network to reveal potential prognostic lncRNA
biomarkers in human ovarian cancer. Oncotarget. Feb 3–2016.Epub
ahead of print. View Article : Google Scholar
|
|
14
|
Malek E, Jagannathan S and Driscoll JJ:
Correlation of long non-coding RNA expression with metastasis, drug
resistance and clinical outcome in cancer. Oncotarget. 5:8027–8038.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen F, Tian Y, Pang EJ, Wang Y and Li L:
MAlAT2-activated long noncoding RNA indicates a biomarker of poor
prognosis in gastric cancer. Cancer Gene Ther. Feb 27–2015.Epub
ahead of print. View Article : Google Scholar
|
|
16
|
Xue Y, Ma G, Gu D, Zhu L, Hua Q, Du M, Chu
H, Tong N, Chen J, Zhang Z, et al: Genome-wide analysis of long
noncoding RNA signature in human colorectal cancer. Gene.
556:227–234. 2015. View Article : Google Scholar
|
|
17
|
Hu Y, Chen HY, Yu CY, Xu J, Wang JL, Qian
J, Zhang X and Fang JY: A long non-coding RNA signature to improve
prognosis prediction of colorectal cancer. Oncotarget. 5:2230–2242.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou M, Zhao H, Wang Z, et al:
Identification and validation of potential prognostic lncRNA
biomarkers for predicting survival in patients with multiple
myeloma. J Exp Clin Cancer Res. 34:1022015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte Al, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:186ra662013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamauchi M, Yamaguchi R, Nakata A, Kohno
T, Nagasaki M, Shimamura T, Imoto S, Saito A, Ueno K, Hatanaka Y,
et al: Epidermal growth factor receptor tyrosine kinase defines
critical prognostic genes of stage I lung adenocarcinoma. PloS One.
7:e439232012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Botling J, Edlund K, Lohr M, Hellwig B,
Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Pontén F, et
al: Biomarker discovery in non-small cell lung cancer: Integrating
gene expression profiling, meta-analysis, and tissue microarray
validation. Clin Cancer Res. 19:194–204. 2013. View Article : Google Scholar
|
|
22
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of Affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Risueño A, Fontanillo C, Dinger ME and De
las Rivas J: GATExplorer: Genomic and transcriptomic explorer;
mapping expression probes to gene loci, transcripts, exons and
ncRNAs. BMC Bioinformatics. 11:2212010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pang KC, Stephen S, Engström PG,
Tajul-Arifin K, Chen W, Wahlestedt C, Lenhard B, Hayashizaki Y and
Mattick JS: RNAdb - a comprehensive mammalian noncoding RNA
database. Nucleic Acids Res. 33:D125–D130. 2005. View Article : Google Scholar
|
|
26
|
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C,
Liu Y, Chen R and Zhao Y: Utilizing sequence intrinsic composition
to classify protein-coding and long non-coding transcripts. Nucleic
Acids Res. 41:e1662013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Simon R, Lam A, Li MC, Ngan M, Menenzes S
and Zhao Y: Analysis of gene expression data using BRB-ArrayTools.
Cancer Inform. 3:11–17. 2007.PubMed/NCBI
|
|
28
|
Ishwaran H and Kogalur UB: Consistency of
random survival forests. Stat Probab lett. 80:1056–1064. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bralten LB and French PJ: Genetic
alterations in glioma. Cancers. 3:1129–1140. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Merico D, Isserlin R, Stueker O, Emili A
and Bader GD: Enrichment map: A network-based method for gene-set
enrichment visualization and interpretation. PloS One.
5:e139842010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar :
|
|
33
|
Dinger ME, Pang KC, Mercer TR, Crowe ML,
Grimmond SM and Mattick JS: NRED: A database of long noncoding RNA
expression. Nucleic Acids Res. 37:D122–D126. 2009. View Article : Google Scholar :
|
|
34
|
Du Z, Fei T, Verhaak RG, Su Z, Zhang Y,
Brown M, Chen Y and Liu XS: Integrative genomic analyses reveal
clinically relevant long noncoding RNAs in human cancer. Nat Struct
Mol Biol. 20:908–913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Williams GH and Stoeber K: The cell cycle
and cancer. J Pathol. 226:352–364. 2012. View Article : Google Scholar
|
|
36
|
Kim T, Jeon YJ, Cui R, Lee JH, Peng Y, Kim
SH, Tili E, Alder H and Croce CM: Role of MYC-regulated long
noncoding RNAs in cell cycle regulation and tumorigenesis. J Natl
Cancer Inst. 107:pii: dju505. 2015. View Article : Google Scholar
|
|
37
|
Macheret M and Halazonetis TD: DNA
replication stress as a hallmark of cancer. Annu Rev Pathol.
10:425–448. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang G, Lu X and Yuan L: lncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li J, Chen Z, Tian L, Zhou C, He MY, Gao
Y, Wang S, Zhou F, Shi S, Feng X, et al: LncRNA profile study
reveals a three-lncRNA signature associated with the survival of
patients with oesophageal squamous cell carcinoma. Gut.
63:1700–1710. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fernando TR, Rodriguez-Malave NI, Waters
EV, Yan W, Casero D, Basso G, Pigazzi M and Rao DS: lncRNA
expression discriminates karyotype and predicts survival in
B-lymphoblastic leukemia. Mol Cancer Res. 13:839–851. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS
and Feng XJ: Increased expression of the lncRNA PVT1 promotes
tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol.
7:6929–6935. 2014.PubMed/NCBI
|
|
42
|
Xie X, Liu HT, Mei J, Ding FB, Xiao HB, Hu
FQ, Hu R and Wang MS: LncRNA HMlincRNA717 is downregulated in
non-small cell lung cancer and associated with poor prognosis. Int
J Clin Exp Pathol. 7:8881–8886. 2014.
|
|
43
|
Han L, Zhang EB, Yin DD, Kong R, Xu TP,
Chen WM, Xia R, Shu YQ and De W: Low expression of long noncoding
RNA PANDAR predicts a poor prognosis of non-small cell lung cancer
and affects cell apoptosis by regulating Bcl-2. Cell Death Dis.
6:e16652015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lin L, Gu ZT, Chen WH and Cao KJ:
Increased expression of the long non-coding RNA ANRIL promotes lung
cancer cell metastasis and correlates with poor prognosis. Diagn
Pathol. 10:142015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou M, Guo M, He D, Wang X, Cui Y, Yang
H, Hao D and Sun J: A potential signature of eight long non-coding
RNAs predicts survival in patients with non-small cell lung cancer.
J Transl Med. 13:2312015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Desjobert C, El Maï M, Gérard-Hirne T,
Guianvarc'h D, Carrier A, Pottier C, Arimondo PB and Riond J:
Combined analysis of DNA methylation and cell cycle in cancer
cells. Epigenetics. 10:82–91. 2015. View Article : Google Scholar :
|
|
48
|
Tachibana KE, Gonzalez MA and Coleman N:
Cell-cycle-dependent regulation of DNA replication and its
relevance to cancer pathology. J Pathol. 205:123–129. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Falaschi A, Abdurashidova G and Biamonti
G: DNA replication, development and cancer: A homeotic connection?
Crit Rev Biochem Mol Biol. 45:14–22. 2010. View Article : Google Scholar
|