Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common cancer among males, and it is the ninth leading cause of
cancer-related deaths in females worldwide (1). HCC is the most common form of liver
cancer, accounting for between 85 and 90% of all primary liver
cancers (2). The leading risk
factor for developing HCC is cirrhosis and the rate is 80–90%
(3). It has been reported that
chronic hepatitis B virus (HBV) infection is the cause of the
majority of cirrhosis cases (4).
Despite the fact that patients can be successfully treated by
surgery, liver transplantation, chemotherapy and interventional
therapy (5,6), HCC is commonly diagnosed in the
advanced stage after related symptoms appear, and the 5-year
survival rate remains at 7% (7).
During the past 20 years, the mortality rate associated with HCC
has significantly increased and epidemiologic evidence indicates
that the burden on medical care costs may significantly increase
during the next decades (8). The
serum α-fetoprotein (AFP) level is used as a diagnostic marker for
HCC, yet AFP results may be negative in as many as 40% of cases
presenting with early stage HCC. Even in 15–30% of advanced
patients, AFP levels remain normal and imaging examination must be
utilized to discriminate between tumor and non-neoplastic lesions
(9). The lack of knowledge
regarding the mechanisms of the tumorigenesis of HCC results in
ineffective therapy and a high probability of relapse after
treatment (10). Therefore,
identification of reliable diagnostic markers for HCC is urgently
needed.
Long non-coding RNAs (lncRNAs), which were first
described by Brockdorff et al in 1992 (11), are molecules with a length longer
than 200 bp that cannot code protein products. Khachane and
Harrison (12) demonstrated that
the proportion of lncRNAs associated with cancer was 2-fold higher
than that of protein coding genes in the human genome. Increasing
evidence has pointed to a relationship between lncRNAs and cancers,
including metastasis, migration, apoptosis and clinical outcome
(13). For example, highly
upregulated in liver cancer (HULC) is a specific gene which is
markedly upregulated both in tissue and plasma in HCC (5). The H19 lncRNA was found to be
associated with tumorigenesis and invasion, partially via the
regulation of carcinogenic miRNA-675 which locates in its first
exon (14). lncRNA-ROR can
downregulate miR-145, causing cancer progression and drug
resistance (15). Urothelial
carcinoma-associated 1 (UCA1) can directly bind to miR-216b, and
the abnormal expression of UCA1 in HCC is correlated with
tumor-node-metastasis (TNM) stage, metastasis, survival and AFP
level (16). Unfortunately, the
functional role of lncRNAs in HCC remains largely unexplored.
lncRNA SPRY4 intronic transcript 1 (SPRY4-IT1) is a
703-bp molecule which maps to chromosome 5q31.3. This transcript
may mediate cell growth, proliferation, apoptosis and was found to
be upregulated in several different tumors, including glioma,
prostate, breast, non-small cell lung and esophageal squamous cell
cancer (17–21). Mazar et al (22) found that SPRY4-IT1 was upregulated
in melanoma cells, and demonstrated that RNAi-mediated knockdown of
SPRY4-IT1 may induce apoptosis via lipin 2-mediated alterations in
lipid metabolism leading to cellular lipotoxicity. Peng et
al (23) indicated that
upregulation of SPRY4-IT1 expression was significantly correlated
with tumor size, depth of invasion, distant metastasis and TNM
stage in gastric cancer, and knockdown of SPRY4-IT1 expression
suppressed cell migration, invasion, proliferation and colony
formation capabilities. Zhang et al (24) showed that overexpression of
SPRY4-IT1 was associated with the progression and development of
clear cell renal carcinoma. Much research concerning SPRY4-IT1 in
cancers has been carried out. However, the clinical and prognostic
significance of lncRNA SPRY4-IT1 expression in HCC has not been
reported.
The aim of the present study was to investigate the
gene expression of lncRNA SPRY4-IT1 in HCC patients and cell lines,
and then analyze the correlation between clinical characteristics
and SPRY4-IT1 levels. We also evaluated the diagnostic value of
SPRY4-IT1 in plasma. Moreover, we assessed whether SPRY4-IT1 could
serve as a new biomarker for HCC.
Materials and methods
Tissue and blood samples
We recruited 87 patients (81 men and 6 women, mean
age, 55±10) with HCC who underwent surgery without preoperative
chemotherapy or radiotherapy from 2011 to 2015 at Zhongnan Hospital
of Wuhan University, Wuhan, China. Tumor tissue specimens and
corresponding adjacent non-tumor tissues were collected and stored
at −8°C until use.
Whole blood samples of 145 patients were obtained
from Zhongnan Hospital of Wuhan University during 2015. The samples
were classified into three groups: pre-operation (48 men and 12
women; mean age, 57±12), 2 weeks after surgery (48 men and 12
women; mean age, 57±12), patients with hepatitis B and cirrhosis
(63 men and 22 women; mean age, 54±12), which were collected in
EDTA tubes and centrifuged at 2,000 × g for 5 min at 4°C to spin
down the blood cells. The supernatants were transferred to
microcentrifuge tubes and centrifuged at 12,000 × g for 5 min at
4°C. The plasma was then stored at −80°C until use. Patients who
underwent previous preoperative chemotherapy or radiotherapy were
excluded from the study. Then, from the Physical Examination
Center, we collected 63 controls (50 men and 13 women; mean age,
54±11), who were without hepatitis, hepatic diseases and abnormal
liver biochemical outcomes.
Ethical approval
Tissue and plasma specimens were collected after
obtaining informed consent of the patients in accordance with the
institutional ethical guidelines and approved by the Ethics
Committee of Zhongnan Hospital of Wuhan University (Wuhan, China)
for the use of these clinical materials.
Cell culture
The human HCC Hep-3B and HepG2 cell lines were
obtained from the China Center for Type Culture Collection (CCTCC;
Wuhan, China). HuH-7 and the normal human hepatocyte cell line L-02
were purchased from the Procell Inc. (Wuhan, China). MHCC97-L and
HCCLM-9 cells were maintained at our laboratory. MHCC97-L cells
were cultured in Dulbecco's modified Eagle's medium (DMEM), and the
other cell lines were cultured in RPMI-1640 medium (both from
Gibco, Grand Island, NY, USA). All cells were cultured at 37°C in
5% CO2 in culture media containing 10% fetal bovine
serum (FBS; Gibco).
RNA extraction
Total RNA was extracted from tissues and cells using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and we used total
RNA separate extraction kit (BioTeke, Beijing, China) for plasma
samples. RNA was reverse transcribed to cDNA using PrimeScript™ RT
reagent kit with gDNA Eraser (Takara, Japan). The conditions were
as follows: 42°C for 2 min, and then 37°C for 15 min, and 85°C for
5 sec.
Real-time PCR analysis
The expression of SPRY4-IT1 was determined on the
Bio-Rad CFX96 (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using
SYBR-Green I Premix Ex Taq according to the manufacturer's
instructions. The reactions started at 95°C for 5 min, followed by
45 cycles of 95°C for 30 sec, 61°C for 30 sec and 72°C for 30 sec.
In order to normalize the results for the qPCR, expression of 18s
was used. The synthesized primers were as follows: SPRY4-IT1 sense,
5′-GTTTTTGCTGAGCTGGTGGTT-3′ and antisense, 5′antisense,
5′-TAGTAGCGACGGGCGGTGTG-3′. The relative gene expression level was
calculated using the comparative Ct method formula
2−ΔCt. All experiments were carried out in duplicate and
each data point represents the mean results of the duplicate
experiments.
Statistical analysis
All statistical analyses were carried out using SPSS
version 17.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 5.0
(GraphPad Software, La Jolla, CA, USA). The data in the present
study were presented as mean ± standard deviation (SD) or median
(25 and 75 percentiles). p<0.05 was considered to indicate a
statistically significant result. The Shapiro-Wilk test was carried
out to check the normality of the distribution. The normally
distributed numeric variables were evaluated by Student's t-test,
while non-normally distributed variables were analyzed by
Kruskal-Wallis variance analysis. One-way ANOVA was used to
validate the different expression levels of SPRY4-IT1 among
subgroups. Chi-square test was used to analyze the categorical
variables. To estimate the diagnostic value of the biomarkers, area
under the corresponding ROC curve analysis was performed. Finally,
correlations were analyzed using Pearson correlation.
Results
SPRY4-IT1 expression is upregulated in
HCC tissues and cell lines
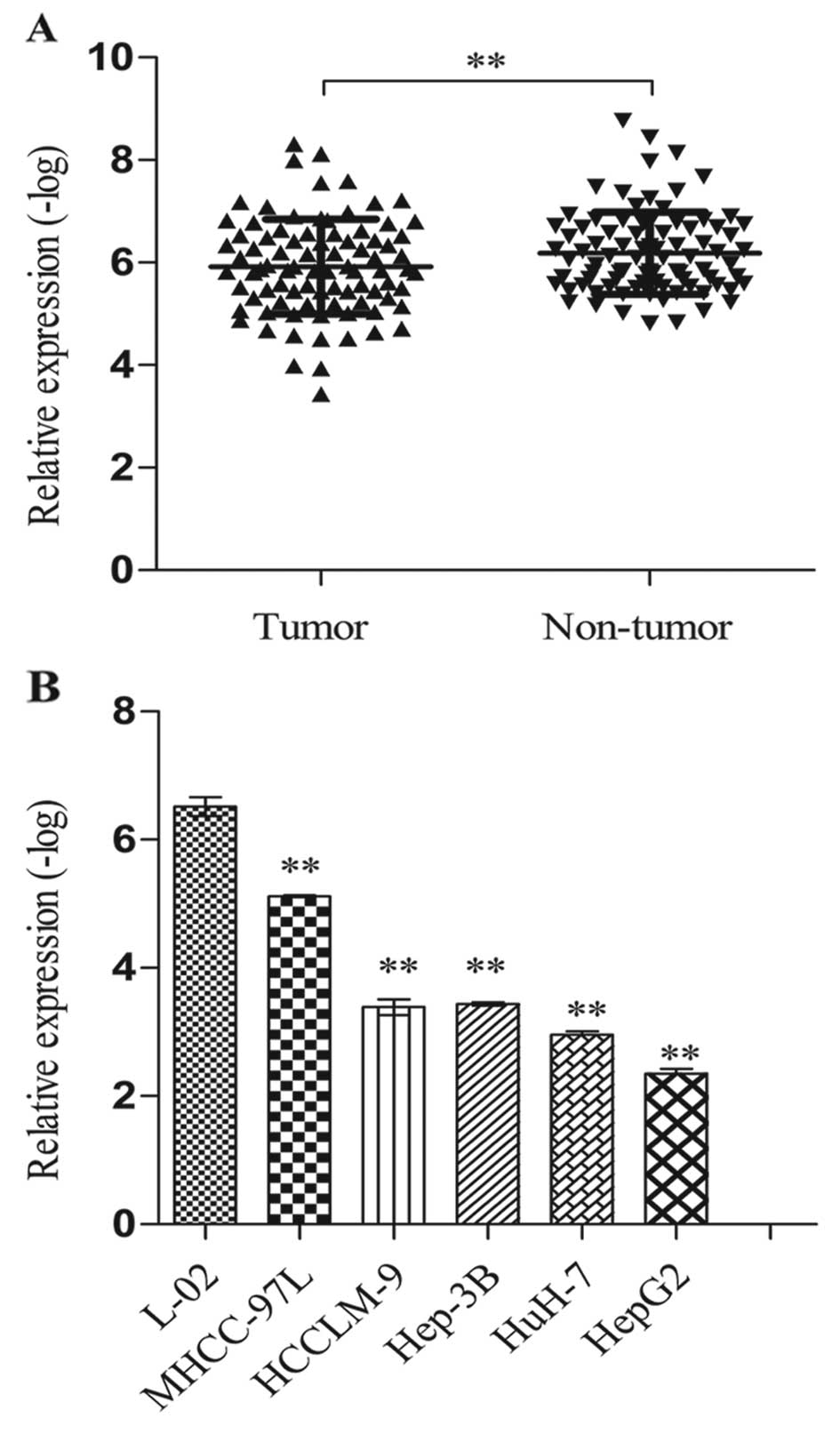
The relative expression levels of SPRY4-IT1 were
assessed by RT-qPCR. In 87 HCC and adjacent normal liver tissues,
the SPRY4-IT1 level in the tumor tissues was significantly
upregulated when compared to the level in the non-tumor tissues
(p<0.01; Fig. 1A). Expression of
SPRY4-IT1 relative to 18s in HCC cell lines was compared with a
normal human hepatocyte cell line. The results showed that the
expression of SPRY4-IT1 was much higher in the HCC cell lines than
that in the L-02 cells (MHCC97-L vs. L-02, p<0.01; HCCLM-9 vs.
L-02, p<0.01; Hep-3B vs. L-02, p<0.01; HuH-7 vs. L-02,
p<0.01; HepG2 vs. L-02, p<0.01; Fig. 1B).
Correlation between SPRY4-IT1 and
clinical variables
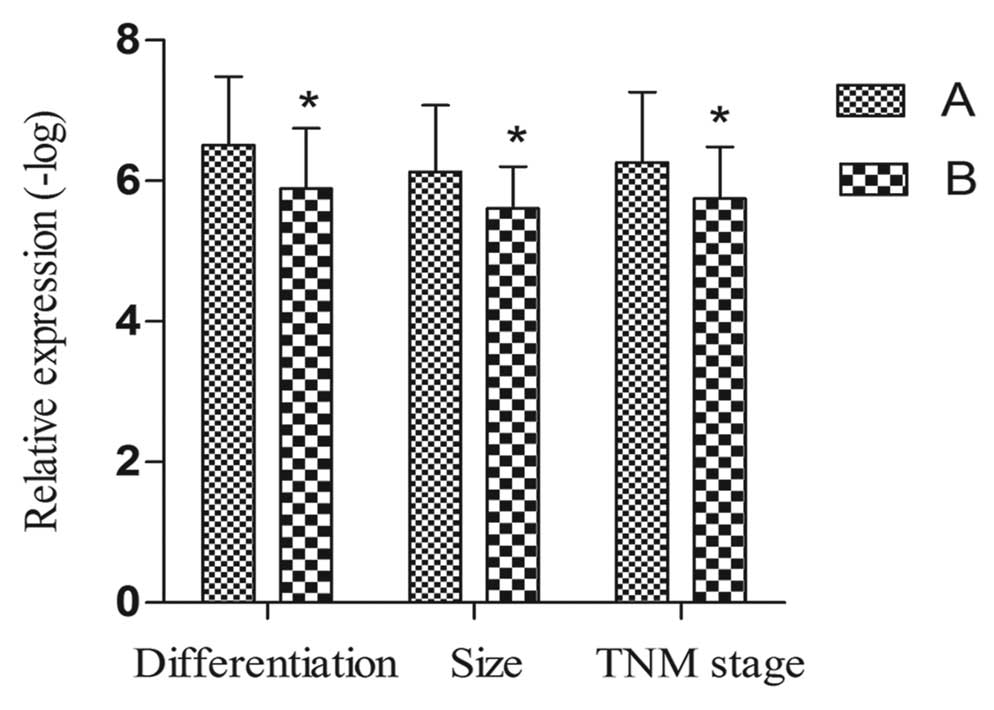
We detected the correlation between SPRY4-IT1 and
clinical parameters. As shown in Tables
I and II, the level of
SPRY4-IT1 expression was significantly correlated with
differentiation (r=0.249, p=0.039), tumor size (r=0.258, p=0.024)
and TNM stage (r=0.287, p=0.015) (Fig.
2). However, no correlations were found in regard to gender,
age, smoking, alcoholism, cirrhosis, AFP, HBV-DNA and other
biochemical indices.
 | Table IAssociation of SPRY4-IT1 expression
with clinical parameters in HCC. |
Table I
Association of SPRY4-IT1 expression
with clinical parameters in HCC.
|
Characteristics | n | SPRY4-IT1 relative
expression (−log)
|
|---|
| Mean ± SD | t | P-value |
|---|
| Tissue | | | −3.060 | 0.003b |
| HCC | 87 | 5.92±0.93 | | |
| Adjacent
non-cancerous liver | 87 | 6.19±0.80 | | |
| Gender | | | 0.394 | 0.694 |
| Male | 81 | 5.93±0.89 | | |
| Female | 6 | 5.78±1.39 | | |
| Age (years) | | | −0.509 | 0.612 |
| <55 | 40 | 5.87±0.96 | | |
| ≥55 | 47 | 5.97±0.90 | | |
| Smoking status | | | 0.488 | 0.627 |
| Negative | 52 | 5.96±0.89 | | |
| Positive | 35 | 5.86±0.99 | | |
| Alcoholism | | | 1.569 | 0.122 |
| Negative | 70 | 6.00±0.94 | | |
| Positive | 17 | 5.61±0.81 | | |
|
Differentiation | | | 2.106 | 0.039a |
| High | 10 | 6.51±0.97 | | |
| Moderate/low | 59 | 5.89±0.86 | | |
| Tumor size
(cm) | | | 2.298 | 0.024 |
| <10 | 55 | 6.13±0.95 | | |
| ≥10 | 21 | 5.61±0.59 | | |
| Tumor nodes | | | 0.678 | 0.500 |
| Single | 66 | 5.97±0.99 | | |
| Multi | 10 | 5.77±0.66 | | |
| TNM stage | | | 2.514 | 0.015a |
| I–II | 35 | 6.26±1.00 | | |
| III–IV | 41 | 5.75±0.73 | | |
| HBV DNA
(IU/ml) | | | 1.490 | 0.147 |
| <500 | 11 | 6.12±0.78 | | |
| ≥500 | 21 | 5.69±0.78 | | |
| Cirrhosis | | | −1.129 | 0.262 |
| Negative | 39 | 5.80±0.85 | | |
| Positive | 48 | 6.02±0.98 | | |
| AFP (ng/ml) | | | 0.085 | 0.932 |
| <200 | 38 | 5.98±0.88 | | |
| ≥200 | 31 | 5.96±0.95 | | |
| CEA
(µg/l) | | | 0.471 | 0.640 |
| <7.2 | 44 | 5.94±0.79 | | |
| ≥7.2 | 3 | 5.72±0.71 | | |
| ALT (U/l) | | | −0.385 | 0.701 |
| <46 | 49 | 5.89±0.85 | | |
| ≥46 | 38 | 5.97±1.00 | | |
| AST (U/l) | | | 0.676 | 0.501 |
| <46 | 45 | 5.99±0.87 | | |
| ≥46 | 42 | 5.85±1.00 | | |
| GGT (U/l) | | | 0.967 | 0.337 |
| <55 | 34 | 6.03±0.85 | | |
| ≥55 | 44 | 5.83±0.94 | | |
| 5′-NT (U/l) | | | 0.482 | 0.631 |
| <10 | 61 | 5.87±0.90 | | |
| ≥10 | 2 | 5.56±0.81 | | |
| GLU (mmol/l) | | | −0.054 | 0.957 |
| <6.2 | 64 | 5.84±0.85 | | |
| ≥6.2 | 23 | 5.86±0.68 | | |
 | Table IICorrelation analysis of SPRY4-IT1 in
relation to clinical parameters of the HCC cases. |
Table II
Correlation analysis of SPRY4-IT1 in
relation to clinical parameters of the HCC cases.
|
Characteristics | Correlation
coefficient |
|---|
| Differentiation
(high vs. moderate/low) | 0.249 |
| Size (<10 vs.
≥10) | 0.258 |
| TNM stage (I–II vs.
III–IV) | 0.287 |
SPRY4-IT1 levels in plasma among
subgroups
The main demographic and clinical characteristics of
the studied subjects were illustrated in Table III. No difference was observed in
regard to important risk factors including gender, age, smoking,
alcoholism and glucose (GLU) in the three groups. There was a
significant difference in alanine aminotransferase (ALT), aspartate
aminotransferase (AST), total bilirubin (TBIL) and γ-glutamyl
transferase (GGT) among the groups.
 | Table IIICharacteristics of the studied
subjects. |
Table III
Characteristics of the studied
subjects.
|
Characteristics |
Pre-operation
n=60 | Hepatitis B and
cirrhosis
n=85 | Control
n=63 | P-value |
|---|
| Gender | | | | 0.639a |
| Male | 48 | 63 | 50 | |
| Female | 12 | 22 | 13 | |
| Age (years) | | | | 0.424a |
| <55 | 14 | 28 | 20 | |
| ≥55 | 46 | 57 | 43 | |
| Smoking | | | | 0.826a |
| Negative | 33 | 50 | 38 | |
| Positive | 27 | 35 | 25 | |
| Alcoholism | | | | 0.322a |
| Negative | 42 | 55 | 48 | |
| Positive | 18 | 30 | 15 | |
| ALT (U/l) | 44 (27, 88)c | 42 (24,
100)c | 20 (16, 27)c | <0.001b |
| AST (U/l) | 55 (30,
104)c | 46 (30,
102)c | 22 (20, 26)c | <0.001b |
| TBIL (µmol/l) | 23 (16, 39)c | 27 (16, 72)c | 19 (15, 21)c | <0.001b |
| GGT (U/l) | 78 (42,
204)c | 59 (29, 98)c | 17 (14, 24)c | <0.001b |
| GLU (mmol/l) | 4.9 (4.6,
5.8)c | 5.1 (4.4,
5.7)c | 4.9 (4.5,
5.3)c | 0.320b |
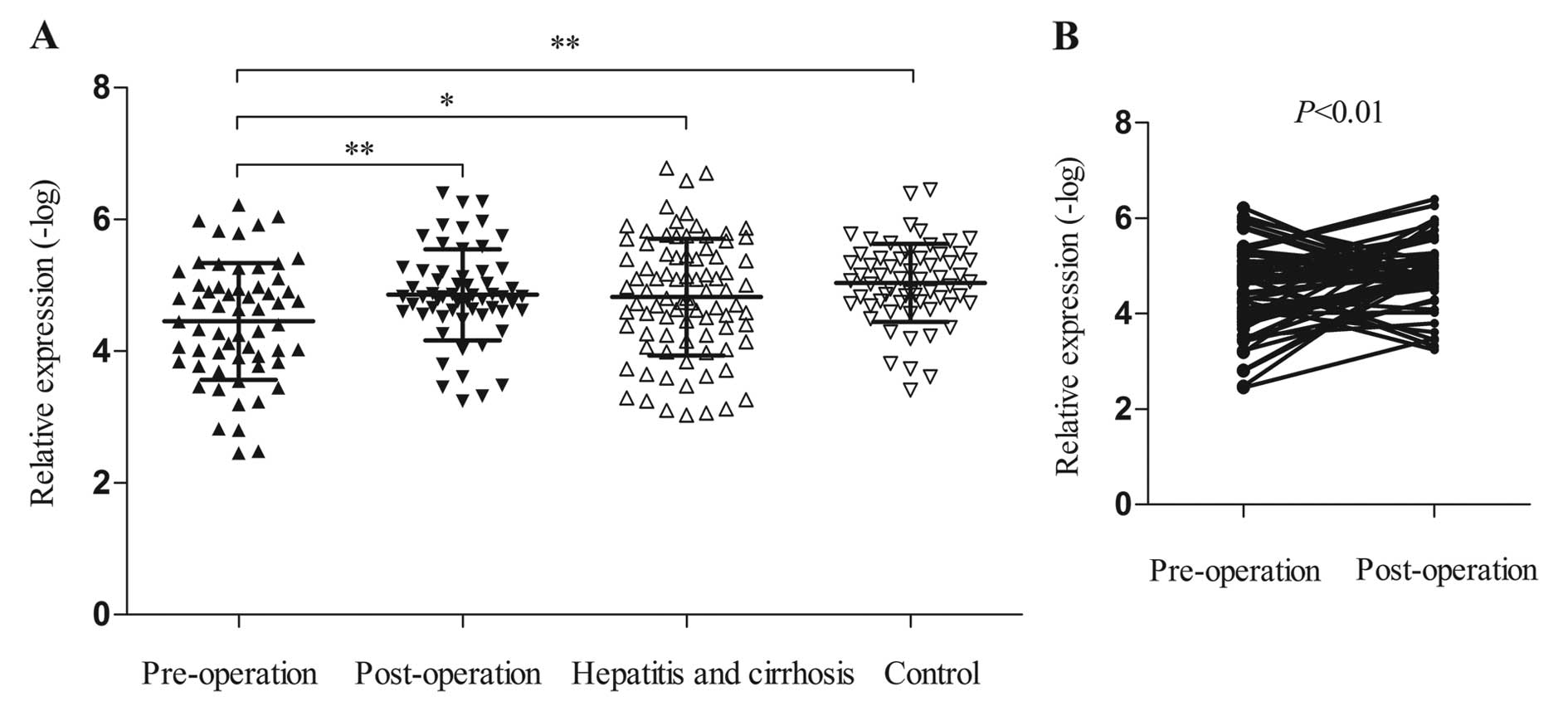
To observe the value of SPRY4-IT1 as a biomarker,
the levels of plasma target lncRNA in 60 HCC patients, 85 hepatitis
B and cirrhosis patients, and 63 control cases were measured by
RT-qPCR. The present study showed that the expression of SPRY4-IT1
at pre-operation was higher than that at post-operation, in
hepatitis B and cirrhosis and the control groups (pre-operation vs.
post-operation: p<0.01; pre-operation vs. hepatitis B and
cirrhosis: p<0.05; pre-operation vs. the controls: p<0.001)
(Fig. 3A). There were 60 paired
plasma samples in the present study, and the levels of SPRY4-IT1
were decreased in 40 of 60 HCC (66.7%) patients (Fig. 3B). However, upon comparison of the
levels in the other three subgroups (post-operation, hepatitis B
and cirrhosis, the controls), no marked differences were found.
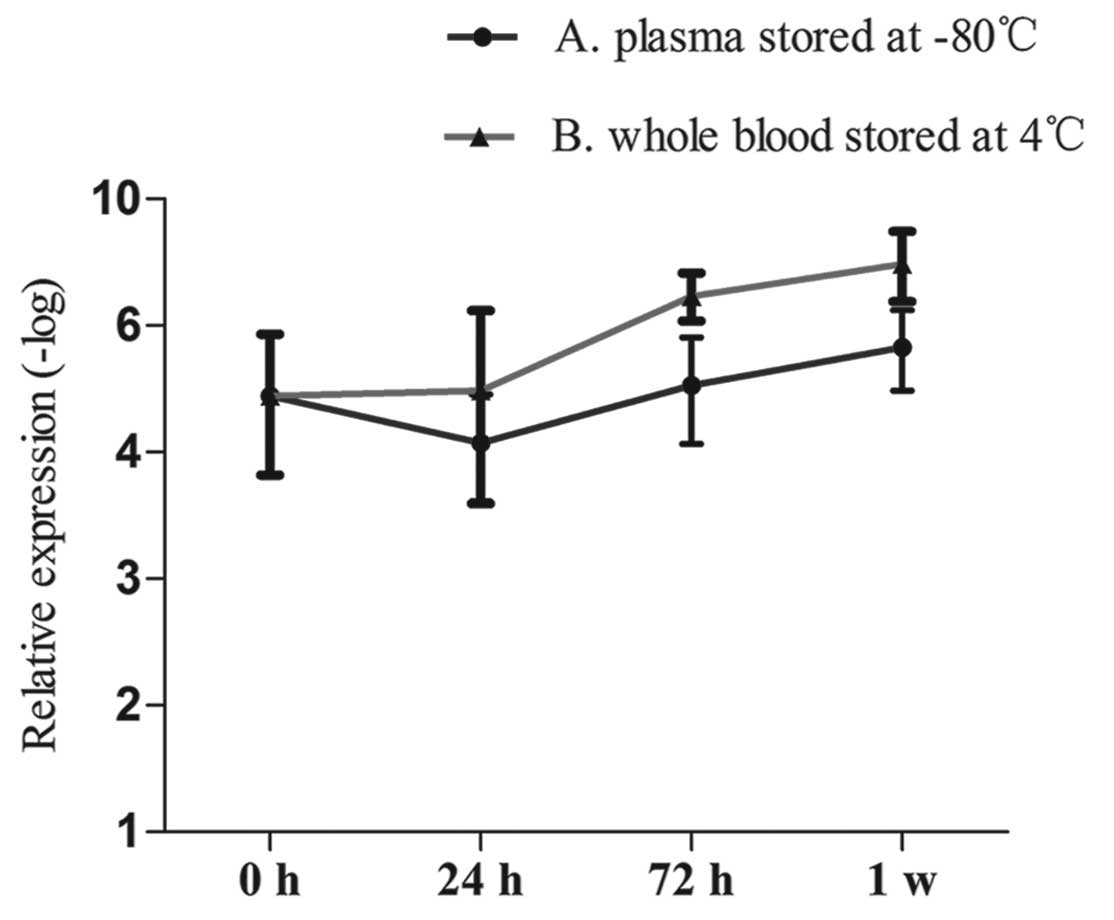
Stability detection of SPRY4-IT1 in human
plasma
The present study amplified SPRY4-IT1 in the plasma
of HCC patients for the first time. Then, we detected the stability
of this lncRNA. We collected plasma from 5 healthy individuals (4
men and 1 women) and stored the samples at −80°C for 0, 24 and 72 h
and 1 week. Meanwhile, we obtained whole blood from these
individuals, and stored the samples at 4°C. We then separated the
plasma at 0, 24 and 72 h and 1 week. The expression of SPRY4-IT1 in
the plasma was assessed by RT-qPCR. The results showed that the
entire process had minimal effects on the levels of SPRY4-IT1
(Fig. 4).
Diagnostic value analysis
To estimate the diagnostic value of SPRY4-IT1 in
plasma, ROC was constructed using 3 models: pre-operation vs. the
controls, and pre-operation vs. post-operation, and pre-operation
vs. hepatitis B and cirrhosis. The area under the ROC (AUCROC)
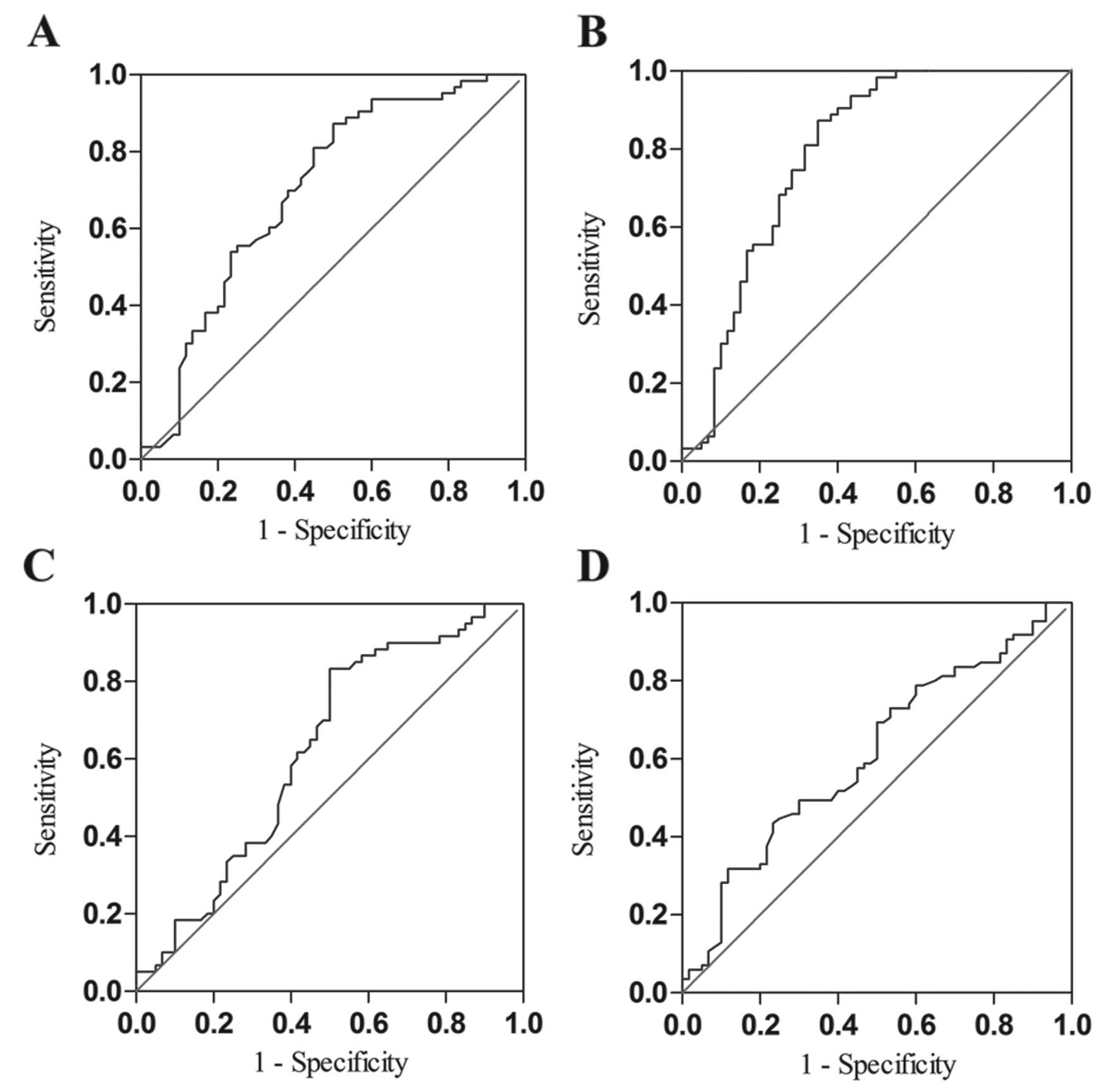
indicated that SPRY4-IT1 had adequate diagnostic value for
differentiating HCC patients from the controls (Fig. 5A). Combination of SPRY4-IT1 and AFP
(the cut-off value of AFP was at 200 ng/ml) possessed a moderate
ability for discrimination between HCC patients and controls; the
area was equal to 0.80 (Fig. 5B).
However, compared with the group of pre-operation vs. the controls,
the diagnostic value of SPRY4-IT1 in the other two groups was not
obvious (Fig. 5C and D and Table IV).
 | Table IVComparisons of the AUC of the
expression of SPRY4-IT1 in the subgroups. |
Table IV
Comparisons of the AUC of the
expression of SPRY4-IT1 in the subgroups.
| Group | AUC | 95% CI | P-value | Se (%) | Sp (%) |
|---|
| Pre vs. C | 0.702 | 0.609–0.796 | <0.001 | 87.3 | 50.0 |
| Pre vs. Ca | 0.800 | 0.706–0.874 | <0.001 | 87.3 | 65.0 |
| Pre vs. Post | 0.624 | 0.523–0.726 | 0.019 | 83.3 | 49.0 |
| Pre vs. HC | 0.611 | 0.518–0.703 | 0.024 | 43.5 | 86.7 |
Discussion
For decades, much research has investigated
potential biomarkers for hepatocellular carcinoma (HCC) (25). The prognosis of HCC remains quite
poor, since most patients are diagnosed at an advanced stage, when
treatments are less effective (26). Clinically, the most frequently used
factor for the diagnosis of HCC is AFP, but the sensitivity is low
(27). Using CT and MRI to diagnose
the early stage of HCC, the sensitivity (55–91%) and specificity
(77–96%) are higher (28). However,
due to the different ability among doctors to identify the lesion,
the accuracy is different. Recently, lncRNAs have been found to
play an important role in the occurrence, invasion and metastasis
of cancer (29). Only a few diverse
hypothetical mechanisms have been presented to explain how lncRNAs
exert their effects. These include interfering in the expression of
the adjacent encoding protein gene (30); participating in transcription,
chromatin-modifying and DNA methyltransferases to specific genomic
(31); binding with functional
protein (32); as precursors of
miRNAs and affecting target genes of miRNAs (33,34);
regulating signaling pathway via combining with chromosome
(35,36). Bussemakers et al (37) demonstrated that lncRNA PCA3 is a
prostate cancer-specific gene. In addition, HULC has been detected
in the plasma of HCC patients with high expression (5). Thus, lncRNAs may be considered as
novel diagnostic targets for HCC.
In the present study, for the first time, we
investigated the clinical value of SPRY4-IT1 in HCC patients. We
found that SPRY4-IT1 was upregulated in HCC tissues and cell lines
to a greater extent than levels in corresponding non-cancerous
tissues and normal human hepatocyte cell line L-02. Meanwhile, our
data showed that SPRY4-IT1 expression was related to
differentiation, tumor size and TNM stage. Kumarswamy et al
(38) elaborated that circulating
lncRNAs were from the tissues. Therefore, we also detected the
expression of SPRY4-IT1 in plasma. The results showed that the
levels of SPRY4-IT1 in HCC were much higher than levels in the
controls, post-operation and hepatitis B and cirrhosis tissues.
According to the present study, the expression of SPRY4-IT1 in
plasma could be used to evaluate the effect after surgery in HCC.
Due to a sharp decline in post-operative patients after 2 weeks, we
supposed that the removal of tumor tissues did effect the contents
of circulating lncRNAs. In 2014, Liu et al (39) found that plasma lncRNA FER1L4 levels
underwent a sharp decline in GC patients 2 weeks after surgery
(p=0.028). Therefore, we hypothesized that SPRY4-IT1 played a role
in the prognostic evaluation after surgery in the plasma of HCC
patients. The occurrence of HCC involves a multi-factor, multi-step
and complex process. This concept was validated by the different
expression levels of SPRY4-IT1 in the pre-operation group vs. the
hepatitis B and cirrhosis group. However, the exact explanation
needs further research.
Due to the existence of RNases, circulating lncRNAs
are thought to be unstable (40).
In the present study, we detected the expression of SPRY4-IT1 in
plasma stored at 4 and −80°C to assess the stability of SPRY4-IT1
in human plasma. No significant difference was observed in the
process. Hu et al (41) also
demonstrated the same result using another method. We confirmed
that circulating lncRNAs were markedly stable.
For the first time, we detected the expression of
SPRY4-IT1 in plasma to analysis the diagnostic value. The area
under the AUC ROC showed that SPRY4-IT1 had a better diagnostic
value to differentiate HCC patients from the control, but the
diagnostic value of SPRY4-IT1 in the other two groups was not
obvious. Further analysis showed that the combination of SPRY4-IT1
and AFP achieved a better diagnostic accuracy. Thus, SPRY4-IT1 may
represent a promising target for HCC diagnosis. However, one
limitation of the study was that the sample size was small, thus
the present findings should be validated in trials with more
cases.
Xie et al (42) indicated that SPRY4-IT1 led to
gastric cancer cell metastasis partly via regulating
epithelial-mesenchymal transition (EMT) and DNA methylation may be
a key factor in controlling SPRY4-IT1 expression. Khaitan et
al (43) found that SPRY4-IT1
could connect with multiple molecules in the mitogen-activated
protein kinase (MAPK) signaling pathway promoting reduced
apoptosis, induced proliferation and enhanced metastasis. In HCC, a
number of studies reveal that EMT and the MAPK signaling pathway
play important roles, thus SPRY4-IT1 may have an effect on the
development of HCC (44,45). Lee et al (46) demonstrated that in prostate cancer,
siRNA knockdown of SPRY4-IT1 in PC3 cells inhibited cell
proliferation and invasion and increased cell apoptosis. These
studies indicated that SPRY4-IT1 played an important role in
tumorigenesis. Unfortunately, the exact function of SPRY4-IT1 in
HCC is still unknown.
In conclusion, the present study provides insights
into the expression levels of SPRY4-IT1 in HCC patients for the
first time. We demonstrated that the expression of SPRY4-IT1 was
significantly higher in HCC than that in corresponding
non-cancerous tissues and was correlated to differentiation, tumor
size and TNM stage. In vitro, our results also showed that
SPRY4-IT1 was upregulated in HCC cell lines to a greater extent
than that in a normal human hepatocyte cell line. We then detected
the levels in HCC plasma, and found that SPRY4-IT1 expression was
upregulated in the HCC group, and SPRY4-IT1 exhibited good
diagnostic value. These findings suggest for the first time that
the expression of SPRY4-IT1 could be used as a novel diagnostic
marker for HCC.
Acknowledgments
The present study was supported by the National
Basic Research Program of China (973 Program) (2012CB720605).
References
|
1
|
Au JS and Frenette CT: Management of
hepatocellular carcinoma: Current status and future directions. Gut
Liver. 9:437–448. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong
M, Dang Y, Feng Z and Chen G: Clinical implication of long
non-coding RNA NEAT1 expression in hepatocellular carcinoma
patients. Int J Clin Exp Pathol. 8:5395–5402. 2015.PubMed/NCBI
|
|
3
|
Waghray A, Murali AR and Menon KN:
Hepatocellular carcinoma: From diagnosis to treatment. World J
Hepatol. 7:1020–1029. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen KW, Ou TM, Hsu CW, Horng CT, Lee CC,
Tsai YY, Tsai CC, Liou YS, Yang CC, Hsueh CW, et al: Current
systemic treatment of hepatocellular carcinoma: A review of the
literature. World J Hepatol. 7:1412–1420. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xie H, Ma H and Zhou D: Plasma HULC as a
promising novel biomarker for the detection of hepatocellular
carcinoma. Biomed Res Int. 2013:1361062013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu WH, Zhang JB, Dang Z, Li X, Zhou T, Liu
J, Wang DS, Song WJ and Dou KF: Long non-coding RNA URHC regulates
cell proliferation and apoptosis via ZAK through the ERK/MAPK
signaling pathway in hepatocellular carcinoma. Int J Biol Sci.
10:664–676. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ilikhan SU, Bilici M, Sahin H, Akca AS,
Can M, Oz II, Guven B, Buyukuysal MC and Ustundag Y: Assessment of
the correlation between serum prolidase and alpha-fetoprotein
levels in patients with hepatocellular carcinoma. World J
Gastroenterol. 21:6999–7007. 2015.PubMed/NCBI
|
|
8
|
Quagliata L, Matter MS, Piscuoglio S,
Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z,
Boldanova T, et al: Long noncoding RNA HOTTIP/HOXA13 expression is
associated with disease progression and predicts outcome in
hepatocellular carcinoma patients. Hepatology. 59:911–923. 2014.
View Article : Google Scholar :
|
|
9
|
Zhang G, Ha SA, Kim HK, Yoo J, Kim S, Lee
YS, Hur SY, Kim YW, Kim TE, Park YG, et al: Combined analysis of
AFP and HCCR-1 as an useful serological marker for small
hepatocellular carcinoma: A prospective cohort study. Dis Markers.
32:265–271. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fang TT, Sun XJ, Chen J, Zhao Y, Sun RX,
Ren N and Liu BB: Long non-coding RNAs are differentially expressed
in hepatocellular carcinoma cell lines with differing metastatic
potential. Asian Pac J Cancer Prev. 15:10513–10524. 2014.
View Article : Google Scholar
|
|
11
|
Brockdorff N, Ashworth A, Kay GF, McCabe
VM, Norris DP, Cooper PJ, Swift S and Rastan S: The product of the
mouse Xist gene is a 15 kb inactive X-specific transcript
containing no conserved ORF and located in the nucleus. Cell.
71:515–526. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Khachane AN and Harrison PM: Mining
mammalian transcript data for functional long non-coding RNAs. PLoS
One. 5:e103162010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang TH, Lin YS, Chen Y, Yeh CT, Huang YL,
Hsieh TH, Shieh TM, Hsueh C and Chen TC: Long non-coding RNA AOC4P
suppresses hepatocellular carcinoma metastasis by enhancing
vimentin degradation and inhibiting epithelial-mesenchymal
transition. Oncotarget. 6:23342–23357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang JL, Zheng L, Hu YW and Wang Q:
Characteristics of long non-coding RNA and its relation to
hepatocellular carcinoma. Carcinogenesis. 35:507–514. 2014.
View Article : Google Scholar
|
|
15
|
Takahashi K, Yan IK, Kogure T, Haga H and
Patel T: Extracellular vesicle-mediated transfer of long non-coding
RNA ROR modulates chemosensitivity in human hepatocellular cancer.
FEBS Open Bio. 4:458–467. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang F, Ying HQ, He BS, Pan YQ, Deng QW,
Sun HL, Chen J, Liu X and Wang SK: Upregulated lncRNA-UCA1
contributes to progression of hepatocellular carcinoma through
inhibition of miR-216b and activation of FGFR1/ERK signaling
pathway. Oncotarget. 6:7899–7917. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu H, Lv Z and Guo E: Knockdown of long
noncoding RNA SPRY4-IT1 suppresses glioma cell proliferation,
metastasis and epithelial-mesenchymal transition. Int J Clin Exp
Pathol. 8:9140–9146. 2015.PubMed/NCBI
|
|
18
|
Mouraviev V, Lee B, Patel V, Albala D,
Johansen TE, Partin A, Ross A and Perera RJ: Clinical prospects of
long noncoding RNAs as novel biomarkers and therapeutic targets in
prostate cancer. Prostate Cancer Prostatic Dis. 19:14–20. 2016.
View Article : Google Scholar
|
|
19
|
Shi Y, Li J, Liu Y, Ding J, Fan Y, Tian Y,
Wang L, Lian Y, Wang K and Shu Y: The long noncoding RNA SPRY4-IT1
increases the proliferation of human breast cancer cells by
upregulating ZNF703 expression. Mol Cancer. 14:512015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong
R, Yang JS, Xu TP, Liu YW, Zou YF, et al: EZH2-mediated epigenetic
suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell
proliferation and metastasis by affecting the
epithelial-mesenchymal transition. Cell Death Dis. 5:e12982014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xie HW, Wu QQ, Zhu B, Chen FJ, Ji L, Li
SQ, Wang CM, Tong YS, Tuo L, Wu M, et al: Long noncoding RNA
SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and
associated with poor prognosis. Tumour Biol. 35:7743–7754. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mazar J, Zhao W, Khalil AM, Lee B, Shelley
J, Govindarajan SS, Yamamoto F, Ratnam M, Aftab MN, Collins S, et
al: The functional characterization of long noncoding RNA SPRY4-IT1
in human melanoma cells. Oncotarget. 5:8959–8969. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Peng W, Wu G, Fan H, Wu J and Feng J: Long
noncoding RNA SPRY4-IT1 predicts poor patient prognosis and
promotes tumorigenesis in gastric cancer. Tumour Biol.
36:6751–6758. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang HM, Yang FQ, Yan Y, Che JP and Zheng
JH: High expression of long non-coding RNA SPRY4-IT1 predicts poor
prognosis of clear cell renal cell carcinoma. Int J Clin Exp
Pathol. 7:5801–5809. 2014.PubMed/NCBI
|
|
25
|
Tang J, Jiang R, Deng L, Zhang X, Wang K
and Sun B: Circulation long non-coding RNAs act as biomarkers for
predicting tumorigenesis and metastasis in hepatocellular
carcinoma. Oncotarget. 6:4505–4515. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tu ZQ, Li RJ, Mei JZ and Li XH:
Down-regulation of long non-coding RNA GAS5 is associated with the
prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol.
7:4303–4309. 2014.PubMed/NCBI
|
|
27
|
Yamashita T, Kitao A, Matsui O, Hayashi T,
Nio K, Kondo M, Ohno N, Miyati T, Okada H, Yamashita T, et al:
Gd-EOB-DTPA-enhanced magnetic resonance imaging and
alpha-fetoprotein predict prognosis of early-stage hepatocellular
carcinoma. Hepatology. 60:1674–1685. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jain D: Tissue diagnosis of hepatocellular
carcinoma. J Clin Exp Hepatol. 4(Suppl 3): S67–S73. 2014.
View Article : Google Scholar
|
|
29
|
Huang M, Chen WM, Qi FZ, Xia R, Sun M, Xu
TP, Yin L, Zhang EB, De W and Shu YQ: Long non-coding RNA ANRIL is
up-regulated in hepatocellular carcinoma and promotes cell
apoptosis by epigenetically silencing of KLF2. J Hematol Oncol.
8:572015. View Article : Google Scholar
|
|
30
|
Khalil AM and Rinn JL: RNA-protein
interactions in human health and disease. Semin Cell Dev Biol.
22:359–365. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao Q, Li T, Qi J, Liu J and Qin C: The
miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in
HBV-related hepatocellular carcinoma and promotes tumorigenesis and
tumor progression. PLoS One. 9:e1097822014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen LL and Carmichael GG: Decoding the
function of nuclear long non-coding RNAs. Curr Opin Cell Biol.
22:357–364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bussemakers MJ, van Bokhoven A, Verhaegh
GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N and Isaacs
WB: DD3: A new prostate-specific gene, highly overexpressed in
prostate cancer. Cancer Res. 59:5975–5979. 1999.PubMed/NCBI
|
|
38
|
Kumarswamy R, Bauters C, Volkmann I, Maury
F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F and Thum
T: Circulating long noncoding RNA, LIPCAR, predicts survival in
patients with heart failure. Circ Res. 114:1569–1575. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu Z, Shao Y, Tan L, Shi H, Chen S and
Guo J: Clinical significance of the low expression of FER1L4 in
gastric cancer patients. Tumour Biol. 35:9613–9617. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tsui NB, Ng EK and Lo YM: Stability of
endogenous and added RNA in blood specimens, serum, and plasma.
Clin Chem. 48:1647–1653. 2002.PubMed/NCBI
|
|
41
|
Hu X, Bao J, Wang Z, Zhang Z, Gu P, Tao F,
Cui D and Jiang W: The plasma lncRNA acting as fingerprint in
non-small-cell lung cancer. Tumour Biol. Oct 9–2015.Epub ahead of
print.
|
|
42
|
Xie M, Nie FQ, Sun M, Xia R, Liu YW, Zhou
P, De W and Liu XH: Decreased long noncoding RNA SPRY4-IT1
contributing to gastric cancer cell metastasis partly via affecting
epithelial-mesenchymal transition. J Transl Med. 13:2502015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang L, Yang F, Yuan JH, Yuan SX, Zhou
WP, Huo XS, Xu D, Bi HS, Wang F and Sun SH: Epigenetic activation
of the MiR-200 family contributes to H19-mediated metastasis
suppression in hepatocellular carcinoma. Carcinogenesis.
34:577–586. 2013. View Article : Google Scholar
|
|
45
|
Ma L, Ji L, Yu Y and Wang J: Novel
molecular targets for diagnosis and treatment of hepatocellular
carcinoma. Discov Med. 19:7–14. 2015.PubMed/NCBI
|
|
46
|
Lee B, Mazar J, Aftab MN, Qi F, Shelley J,
Li JL, Govindarajan S, Valerio F, Rivera I, Thurn T, et al: Long
noncoding RNAs as putative biomarkers for prostate cancer
detection. J Mol Diagn. 16:615–626. 2014. View Article : Google Scholar : PubMed/NCBI
|