Introduction
Pancreatic cancer (PC) is the seventh leading cause
of cancer-related death among men and women worldwide (1). Despite improvements in surgical
techniques and adjuvant medical therapy, the prognosis of PC has
not been significantly improved in over four decades (2). It is one of the most devastating
malignant diseases with a median survival of 3–6 months and a
5-year survival rate of less than 5% (3,4). After
careful assessment, only 15% of patients are considered to be
candidates for surgical resection and undergo resection with
curative intent (5). In addition to
massive primary tumors, ~30% of patients die from locally
destructive PC, and the other 70% may have widespread metastatic
disease at the time of death (6).
Therefore, the discovery of new biomarkers and wider insights into
the mechanisms involved in pancreatic tumorigenesis and metastasis
are crucial.
Leukemia inhibitory factor receptor (LIFR) is an
integral component of the glycoprotein 130-LIFR complex and
participates in signal transduction through the interleukin-6
(IL-6) cytokine family, which includes IL-6, IL-11,
cardiotrophin-1, ciliary neurotrophic factor, oncostatin M and
cardiotrophin-like cytokine (7).
The biological roles of the IL-6 cytokine family are widely
different, ranging from glucose uptake, maintenance of stem cell
pluripotency, to modulation of cell proliferation. According to
Alisoltani et al and the Oncomine data-mining analysis,
downregulation of LIFR expression has been found in several types
of cancers, including breast, gastric, colorectal, liver and PC
(8). LIFR has been observed in
several human malignancies, including medulloblastoma,
nasopharyngeal carcinoma, lung, breast and liver cancer (9–13).
Chen et al identified LIFR as a metastasis suppressor which
exerted its function through the Hippo-YAP pathway (11). Luo et al demonstrated that
LIFR negatively regulated the metastasis of hepatocellular
carcinoma by regulating the phosphoinositide 3-kinase/AKT pathway
(14). However, the precise role of
LIFR in PC remains largely unexplored. The purpose of the present
study was to explore the possibility of LIFR as a potential
molecular marker and therapeutic target for PC.
Materials and methods
Patient samples, cell lines and tissue
microarray
From 2012 to 2014, 26 PC patients (15 males and 11
females), ranging from 34 to 72 years of age (mean age, 53.6
years), who underwent radical resections were recruited in the
current investigation with informed consent. Research consent was
approved by the Ethics Committee of Ruijin Hospital, Shanghai Jiao
Tong University School of Medicine. All patients were aware of the
potential risks and complications of the proposed treatment scheme.
The corresponding non-tumor tissues were collected at least 3 cm
away from the margin of the tumor. All the specimens including
tumor and paired non-tumor tissues were cut into small pieces and
placed in liquid nitrogen immediately. All of the samples were
submitted for routine pathologic evaluation and diagnostic
confirmation. The human PC cell lines Capan-1, CFPAC-1, SW-1990,
BxPC-3 and PANC-1 were purchased from the American Type Culture
Collection (ATCC; Manassas, VA, USA) and PATU-8988 was purchased
from Nanjing KeyGen Biotech Co., Ltd. (Nanjing, China). All tumor
cell lines were cultured in Dulbecco's modified Eagle's medium
(DMEM) or RPMI-1640 medium supplemented with 10% fetal bovine serum
(FBS) at 37°C in 5% CO2. Total RNA and genomic DNA were
isolated according to previously reported methods (15). Additionally, PC tissue array of 29
patients was purchased from Shanghai Outdo Biotech Co., Ltd
(Shanghai, China).
Vector construction and transfection
The full-length cDNA of LIFR was obtained using a
reverse transcription kit (Takara) and total RNA was extracted by
RT-PCR from human PC tissues using TRIzol reagent kit (Invitrogen,
Carlsbad, CA, USA). The primers for the coding sequence (CDS) of
double-strand DNA fragments of LIFR were: 5′-GGATCCAT
GATGGATATTTACGTATGT-3′ (forward) and 5′-ACGCGT
TTAATCGTTTGGTTTGTTCTG-3′ (reverse), and were subcloned into the
pWPI-GFP vector to generate pWPI-GFP/LIFR. Transfection of the
constructed plasmid and empty vector into PATU-8988 cells was
performed using Lipofectamine 2000 (Invitrogen) according to the
manufacturer's procedure. Cells that had been transfected with the
constructed plasmid were then selected by antibiotic resistance in
cell culture medium containing 1,500 µg/ml G418 to obtain
cell strains with stable expression of LIFR. After 6 weeks of
culture in the presence of G418, the remaining cells were isolated.
Positive stable clones with pWPI-GFP/LIFR were maintained with
pWPI-GFP empty vector transfection (PATU-8988/vector) as
control.
RNA interference
Based on human LIFR gene data, synthesized DNA
nucleotide fragments, encoding shRNA for knockdown of LIFR, were
inserted into pL/shRNA/F lentiviral vector to obtain
pL/shRNA/shR-LIFR. The constructions were further confirmed by DNA
sequencing. The packaging of the lentivirus and establishment of
stable knockdown cell clones were performed as mentioned above. The
sequences of these synthesized oligonucleotides for RNAi LIFR were
as follows:
5′-GATCCCCGCTGATTTCTCAACCTCTACATTCAAGAGATGTAGAGGTTGAGAAATCAGCTTTTTGGAA-3′
(forward) and
5′-AGCTTTTCCAAAAAGCTGATTTCTCAACCTCTACATCTCTTGAATGTAGAGGTTGAGAA
ATCAGCGGG-3′ (reverse). Then, the pL/shRNA/shR-LIFR lentiviral
vector was transfected into Capan-1 cells. Stable clones
(Capan-1/sh) were established by selection with 5 µg/ml
blasticidin. The irrelevant nucleotides in sh-NC did not target any
annotated human genes and served as a negative control.
Immunohistochemistry
Paraffin-embedded tissue samples from PC specimens
underwent a heat pre-treatment of 60°C for 1 h, then dewaxed in
xylene, rehydrated in a series of ethanol and treated with 0.01
mol/l citrate buffer (pH 6.0) for antigen retrieval. After
inhibition of endogenous peroxidase activity for 30 min with
methanol containing 0.3% H2O2, the sections
were stained with rabbit anti-LIFR antibody (C-19) (1:300; Santa
Cruz Biotechnology, Santa Cruz, CA, USA) or the anti-Ki67 antibody
(ab15580) (1:100; Abcam, Cambridge, MA, USA) at 4°C overnight. For
tissue arrays, after being dewaxed, hydrated and blocked of
non-specific binding sites, the microarray was incubated with
rabbit anti-LIFR antibody (C-19) (1:300; Santa Cruz Biotechnology)
at 4°C overnight. The following experimental procedure was
performed according to the manufacturer's instructions for the
LSAB+ kit (Dako, Carpinteria, CA, USA). Three pathologists who were
blinded to any patient data independently examined the cellular
location of LIFR and compared the staining between the tumor and
normal tissues. Immunohistochemistry staining score = positive cell
score + staining intensity score. The percentage of positive cells
was classified according to five grades (percentage scores):
<10% (grade 0), 10–25% (grade 1), >25–50% (grade 2),
>50–75% (grade 3) and >75% (grade 4). Immunohistochemical
staining intensity was graded as follows: 0 (no staining), 1
(bright yellow), 2 (orange) and 3 (brown). The total scores of ≤3,
>3–5, and ≥6 were defined as negative, weak and strong positive,
respectively.
Western blotting
Tumor and cell samples were collected and lysed
using RIPA buffer (Solarbio, Beijing, China) in the presence of
protease inhibitor cocktail and protein concentration was measured
by BCA protein assay kit (both from Pierce, Rockford, IL, USA). An
equal amount of total cellular protein was electrophoresed by 10%
SDS-PAGE, then transferred to polyvinylidene difluoride (PVDF)
membranes. The membranes were blocked with 5% skim milk for 2 h,
and then incubated with primary antibodies overnight at 4°C.
Primary antibodies were as follows: LIFR (C-19) (1:1,000; Santa
Cruz Biotechnology), vimentin (D21H3) (1:1,000), N-cadherin (D4R1H)
(1:1,000), β-catenin (D10A8) (1:1,000), slug (C19G7) (1:1,000) (all
from Cell Signaling Technology, Danvers, MA, USA) and GAPDH
(Abcam). After the membranes were incubated with the secondary
antibody for 2 h at room temperature, the proteins were visualized
using an enhanced chemiluminescence detection system (Amersham
Biosciences, Piscataway, NJ, USA) according to the manufacturer's
protocol.
Colony formation assay
For the colony formation assay, 1,000 cells were
layered onto 6-well plates and cultured at 37°C for ~14 days in
order to let the colonies develop. After visible colonies of cells
were observed, the cells were washed twice with phosphate-buffered
saline (PBS), and fixed with 4% paraformaldehyde and stained with
crystal violet for 30 min. Colonies containing 50 cells or more
were counted.
Transwell migration and invasion
assays
For the cell migration assay, a total number of
1×105 cells were re-suspended in serum-free culture
solution and added to the Transwell upper chambers (8 µm;
24-well format; Corning, Lowell, MA, USA). To measure the invasion,
the upper chamber's membrane was pre-coated with Matrigel (BD
Biosciences, San Diego, CA, USA) according to the manufacturer's
protocols before cell solution was added to the upper chamber. The
lower chambers included 0.6 ml of medium containing 10% FBS as a
chemoattractant. After 24 h of culturing, the chambers were fixed
with 10% methanol and stained with 0.5% crystal violet solution for
15 min, and then washed by PBS. Cells in the lower chamber were
observed and counted in six random fields under an inverted
microscope.
Wound healing assay
Cells were layered onto 6-well plates and cultured
to confluency. The 200-µl pipette tips were used to scratch
three separate wounds on the monolayer of cells. Plates were washed
with fresh medium after the cells had been cultured for 0, 12 or 24
h, and then photographed. The distances between wound edges were
measured.
In vivo tumorigenesis and metastasis
assays by micro-PET/CT
PC xenografts were established in nude mice.
Four-week-old male BALB/c nude mice were purchased from the
Institute of Zoology, Chinese Academy of Sciences of Shanghai. The
animal research was approved by the Ethics Committee of Ruijin
Hospital, Shanghai Jiao Tong University School of Medicine. All
experiments were performed in accordance with the official
recommendations of the China Zoological Society and animals
received humane care according to the criteria outlined in the
‛Guide for the Care and Use of Laboratory Animals'. Briefly,
Capan-1/sh, Capan-1/nc, PATU-8988/vector and PATU-8988/LIFR cells
were re-suspended in PBS (pH 7.4). The suspension, containing
1×106 cells, was subcutaneously injected into the right
flank of nude mouse. The length (L) and width (W) of each tumor
were measured every 7 days with a digital caliper, and the tumor
volume was calculated using the formula: Tumor volume =
(Width2 × Length)/2. Mice were sacrificed by cervical
decapitation 5 weeks after injection. Tumors were weighed and fixed
for hematoxylin and eosin (H&E) and immunohistochemical
staining. The tumorigenic experiments in vivo were performed
with 5 mice in each group.
The tail vein injection assay was employed to
evaluate the role of LIFR in tumor metastases in vivo. The
stable clones of Capan-1/sh, Capan-1/nc, PATU-8988/vector and
PATU-8988/LIFR were injected into athymic nude mice via tail veins.
After 6 weeks, the lung metastasis lesions were detected by
micro-PET/CT. PET/CT imaging was performed by the Department of
Nuclear Medicine, Ruijin Hospital, Shanghai Jiao Tong University
School of Medicine according to Meng's method (16). Then, the mice were sacrificed and
all the suspicious lung metastasis sites were evaluated by
histological examination.
Statistical analyses
Statistical analyses were performed using SPSS 13.0
software (SPSS, Inc., Chicago, IL, USA). For comparison among
groups, an analysis of variance (ANOVA) and Student's t-test were
used. Differences between tumor volumes were assessed by
Mann-Whitney U test. The Chi-square test was used for the
comparison of categorical data. A P-value <0.05 was considered
to indicate a statistically significant result.
Results
LIFR expression is downregulated in PC
tissues and is associated with clinicopathological parameters
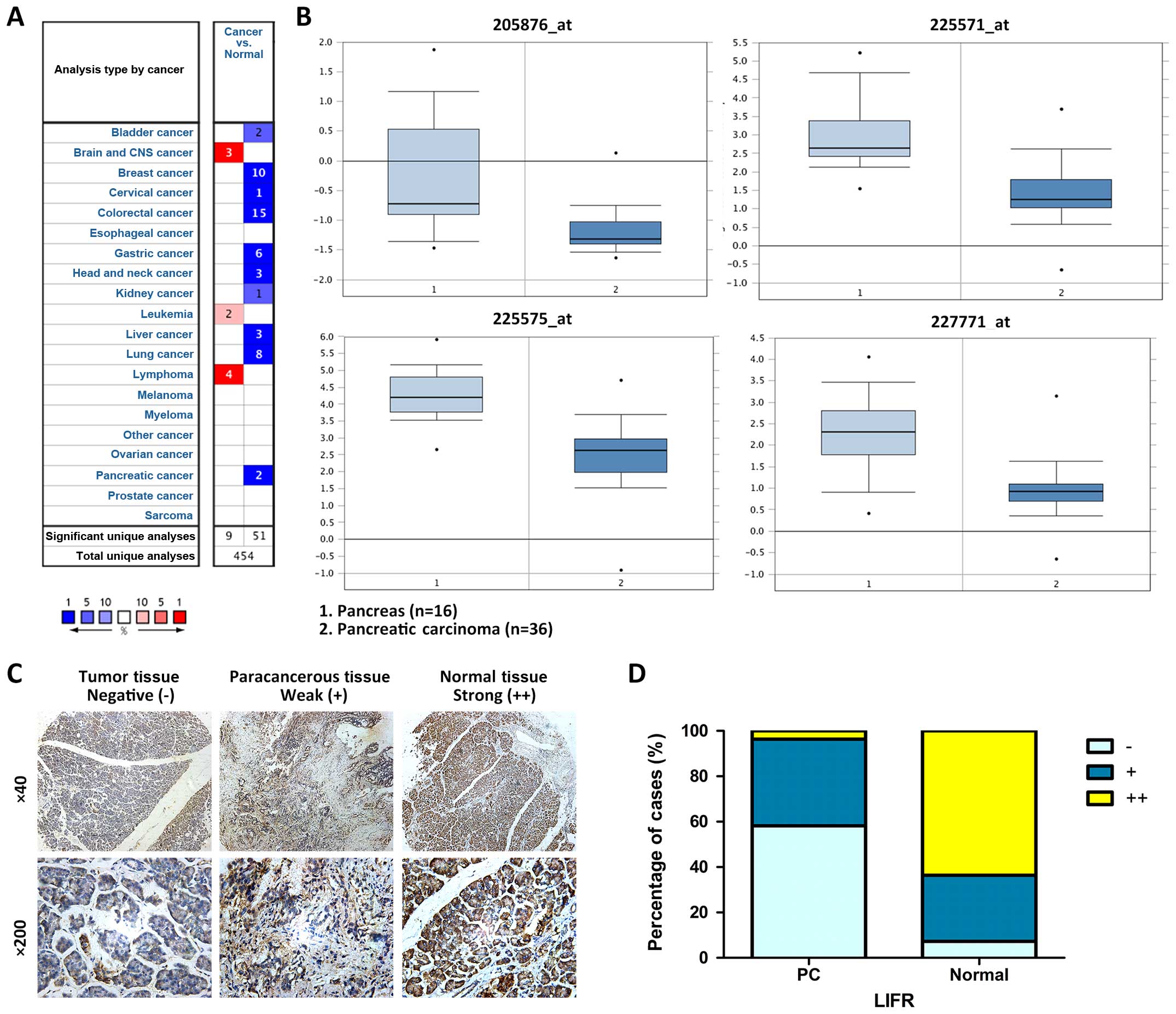
To illuminate the expression of LIFR in multiple
cancer tissues, Oncomine data-mining analysis analyses were first
performed. Pei Pancreas Statistics confirmed that with all four
probes testing it showed significantly decreased levels of LIFR
mRNA in PC compared with that in normal pancreas tissues (Fig. 1A and B, available at Oncomine
website).
To confirm our above findings, we then examined the
expression level of LIFR in 26 PC clinical cases and 29 PC tissue
microarrays, including 53 pancreatic duct adenocarcinomas and 2
pancreatic adenosquamous carcinomas. Immunohistochemical staining
of 55 paired tissues showed that LIFR expression was significantly
lower in the tumor tissues than that in the non-tumor tissues
(negative, 32:4, positive, 21:16; strong positive, 2:35) (Table I) (Fig.
1C and D). Furthermore, LIFR was downregulated in 70.9% (39/55)
of the PC cases. These results were consistent with those from the
Oncomine analyses. PC tissue microarray was further employed to
examine the correlation between LIFR expression and
clinicopathological features. The results showed that downregulated
LIFR was associated with lymph node metastasis (P=0.014), local
invasion (P=0.047) and TNM stage (P=0.002), but not with other
clinicopathological factors including gender, age and tumor
location (Table II). Further
analysis showed that only TNM stage of the patients was correlated
to LIFR expression (P=0.002), not gender, age, tumor location or
tumor size (Table III). All these
findings suggested that downregulation of LIFR plays a critical
role in PC development and LIFR may be an independent prognostic
factor of PC.
 | Table IComparison of LIFR expression between
PC and normal pancreatic tissue (n=55). |
Table I
Comparison of LIFR expression between
PC and normal pancreatic tissue (n=55).
| LIFR
expression | Clinical cases
(n=26)
| Tissue microarrays
(n=29)
| All (n=55)
| P-value |
|---|
| PC | Normal | PC | Normal | PC | Normal |
|---|
| − | 16 | 1 | 16 | 3 | 32 | 4 | <0.01 |
| + | 9 | 7 | 12 | 9 | 21 | 16 | |
| ++ | 1 | 18 | 1 | 17 | 2 | 35 | |
 | Table IIAssociation between LIFR expression
and clinicospathological factors of the PC patients. |
Table II
Association between LIFR expression
and clinicospathological factors of the PC patients.
| Variables | No. of cases | LIFR immunostaining
| P-value |
|---|
Positive
(n=13) | Negative
(n=16) |
|---|
| Gender | | | | 0.730 |
| Male | 21 | 9 | 12 | |
| Female | 8 | 4 | 4 | |
| Age (years) | | | | 0.089 |
| >60 | 15 | 9 | 6 | |
| ≤60 | 14 | 4 | 10 | |
| Tumor location | | | | 0.244 |
| Head | 19 | 10 | 9 | |
| Tale | 10 | 3 | 7 | |
| Tumor size
(cm) | | | | 0.051 |
| >45 | 10 | 2 | 8 | |
| ≤45 | 19 | 11 | 8 | |
| T stage | | | | 0.033 |
| T1 | 6 | 5 | 1 | |
| T2+T3 | 23 | 8 | 15 | |
| Lymph node | | | | 0.014 |
| metastasis | | | | |
| Negative | 20 | 12 | 8 | |
| Positive | 9 | 1 | 8 | |
| Local invasion | | | | 0.047 |
| Negative | 12 | 8 | 4 | |
| Positive | 17 | 5 | 12 | |
| Distant
metastasis | | | | 0.119 |
| Negative | 23 | 12 | 11 | |
| Positive | 6 | 1 | 5 | |
| TNM stage | | | | 0.002 |
| IA-IB | 13 | 10 | 3 | |
| IIA-IV | 16 | 3 | 13 | |
 | Table IIIAssociation between TNM stage and
clinicopathological factors of the PC patients. |
Table III
Association between TNM stage and
clinicopathological factors of the PC patients.
| Variables | No. of cases | TNM stage
| P-value |
|---|
IA-IB
(n=13) | IIA-IV
(n=16) |
|---|
| Gender | | | | |
| Male | 21 | 10 | 11 | 0.624 |
| Female | 8 | 3 | 5 | |
| Age (years) | | | | |
| >60 | 15 | 8 | 7 | 0.340 |
| ≤60 | 14 | 5 | 9 | |
| Tumor location | | | | |
| Head | 19 | 11 | 8 | 0.051 |
| Tale | 10 | 2 | 8 | |
| Tumor size
(cm) | | | | |
| >45 | 10 | 2 | 8 | 0.051 |
| ≤45 | 19 | 11 | 8 | |
| LIFR
immunostaining | | | | |
| Positive | 13 | 10 | 3 | 0.002 |
| Negative | 16 | 3 | 13 | |
LIFR negatively regulates the
proliferation of PC cells
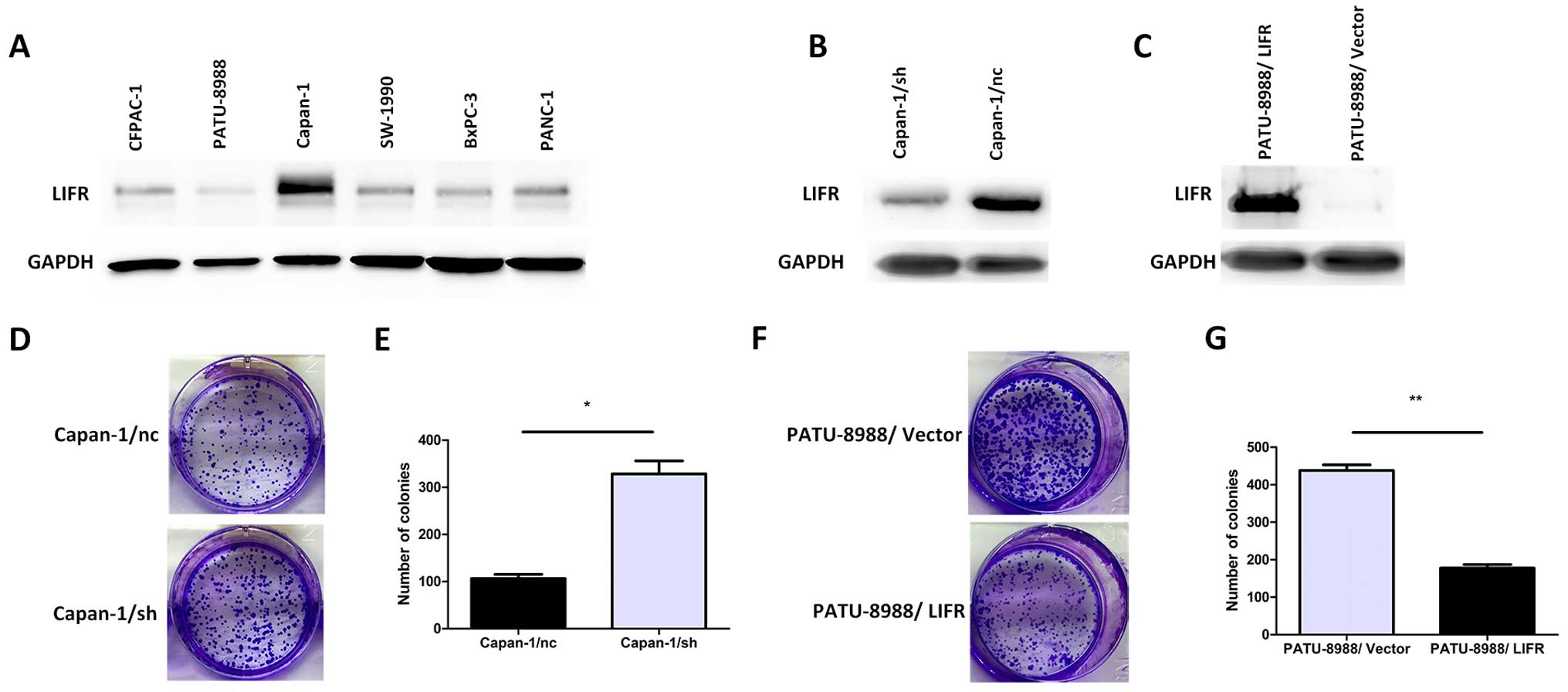
Based on the above results, to establish a paired
differential expression model for further study, we first
quantitatively analyzed the expression of LIFR in a series of PC
cell lines (CFPAC-1, PATU-8988, Capan-1, SW-1990, BxPC-3 and
PANC-1). Among these PC cells, the protein level of LIFR in Capan-1
cells was higher than that in the other PC cells and LIFR in highly
aggressive PATU-8988 cells was the lowest (Fig. 2A). Therefore, Capan-1 cells were
selected for the knockdown assay to reveal the role of LIFR in the
following study. The lentiviral-mediated shRNA was employed to
knockdown LIFR in the Capan-1 cells. Meanwhile, we used the
pWPI-GFP/LIFR vector to generate a PATU-8988/LIFR cell line
ectopically overexpressing LIFR. After the establishment of stable
clones of shRNA-mediated knockdown of LIFR in the Capan-1/sh cells
and effective overexpression of LIFR in the PATU-8988/LIFR cells,
western blotting was employed to confirm the expression level of
LIFR in the Capan-1/sh and PATU-8988/LIFR cells (Fig. 2B and C). Colony formation assays
indicated that silencing of endogenous LIFR in the Capan-1/sh cells
significantly increased the ability of colony formation compared
with the mock cells (328.5±27.5 vs. 106.5±8.5; P=0.016) (Fig. 2D and E). On the contrary,
overexpression of LIFR in the PATU-8988/LIFR cells markedly
impaired the ability of colony formation (177.5±9.5 vs. 438.0±15.0;
P=0.005) (Fig. 2F and G).
Collectively, these results indicated that LIFR may play a
functional role in pancreatic carcinogenesis.
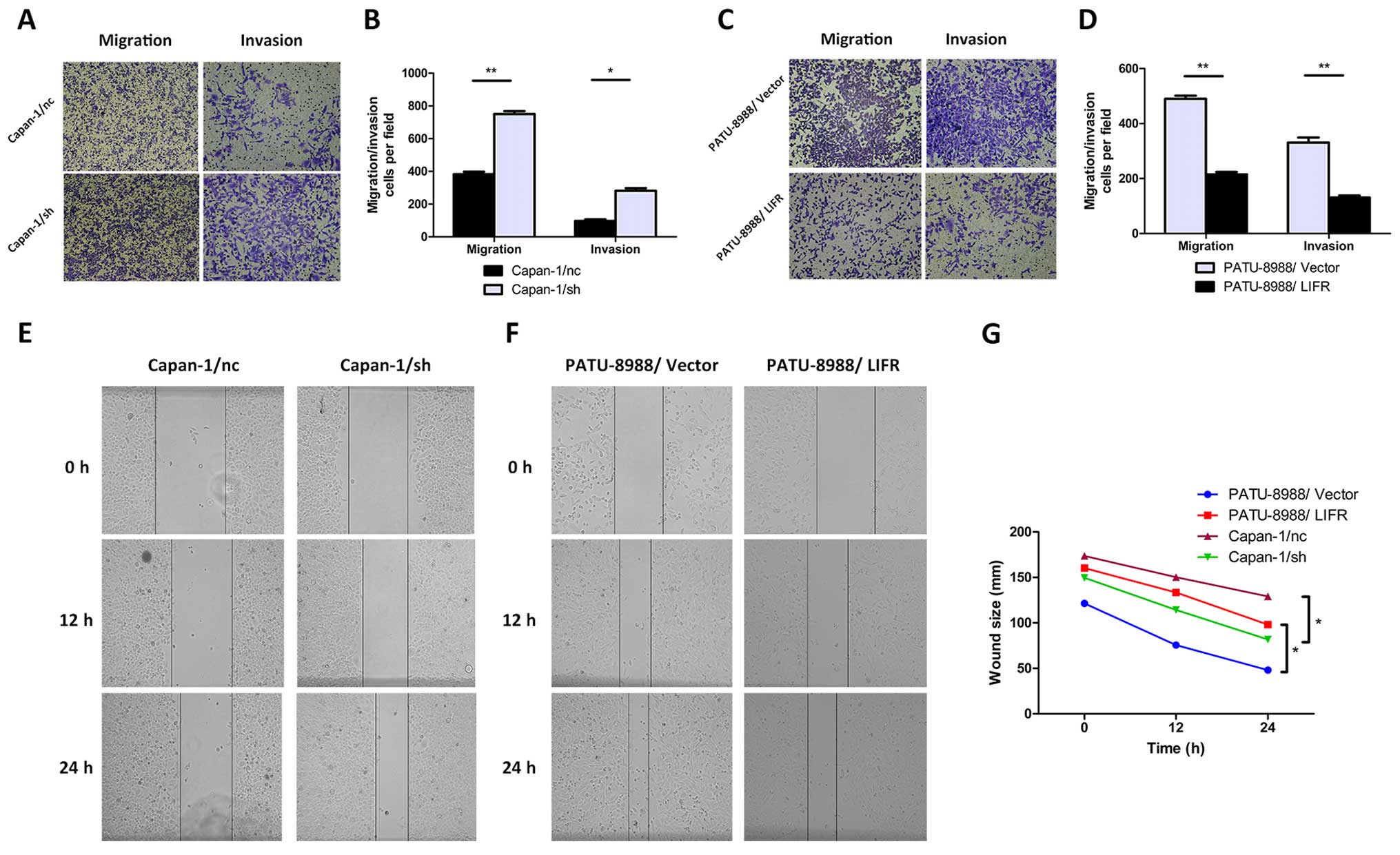
LIFR negatively regulates cell metastasis
and invasion of PC cells in vitro
A close correlation between clinical invasive
characteristics in PC patients and expression level of LIFR was
noted. This suggested that LIFR may play a negative regulatory role
in PC tumor metastasis. To further confirm the effect of LIFR on
the metastasis of PC cells, Transwell migration and invasion assays
were performed. As shown in Fig. 3A and
B, migration and invasion in the Capan-1/sh cells were
significantly enhanced after silencing of endogenous LIFR
(migration, 750.0±18.0 vs. 382.5±15.5, P=0.004; invasion,
281.0±16.0 vs. 97.0±10.0, P=0.010). In contrast, by overexpressing
LIFR in PC cells, the migratory and invasive abilities of the
PATU-8988/LIFR cells were apparently reduced (migration, 215.0±9.0
vs. 490.0±11.0, P=0.003; invasion, 130.5±7.5 vs. 330.5±18.5,
P=0.010) (Fig. 3C and D).
Furthermore, we quantitatively investigated the
effect of LIFR on migratory ability by wound healing assay. The
results in Fig. 3E and G showed
that silencing of LIFR significant shortened the distance between
the wound edge in the Capan-1/sh cells compared with the Capan-1/nc
cells (P=0.034). Consistently, we observed a longer distance in
wound healing after overexpressing LIFR in the PATU-8988/LIFR cells
(P=0.013) (Fig. 3F and G). Taken
together, these results indicated that the aggressive and highly
metastatic phenotype of PC cells could be regulated by LIFR in
vitro.
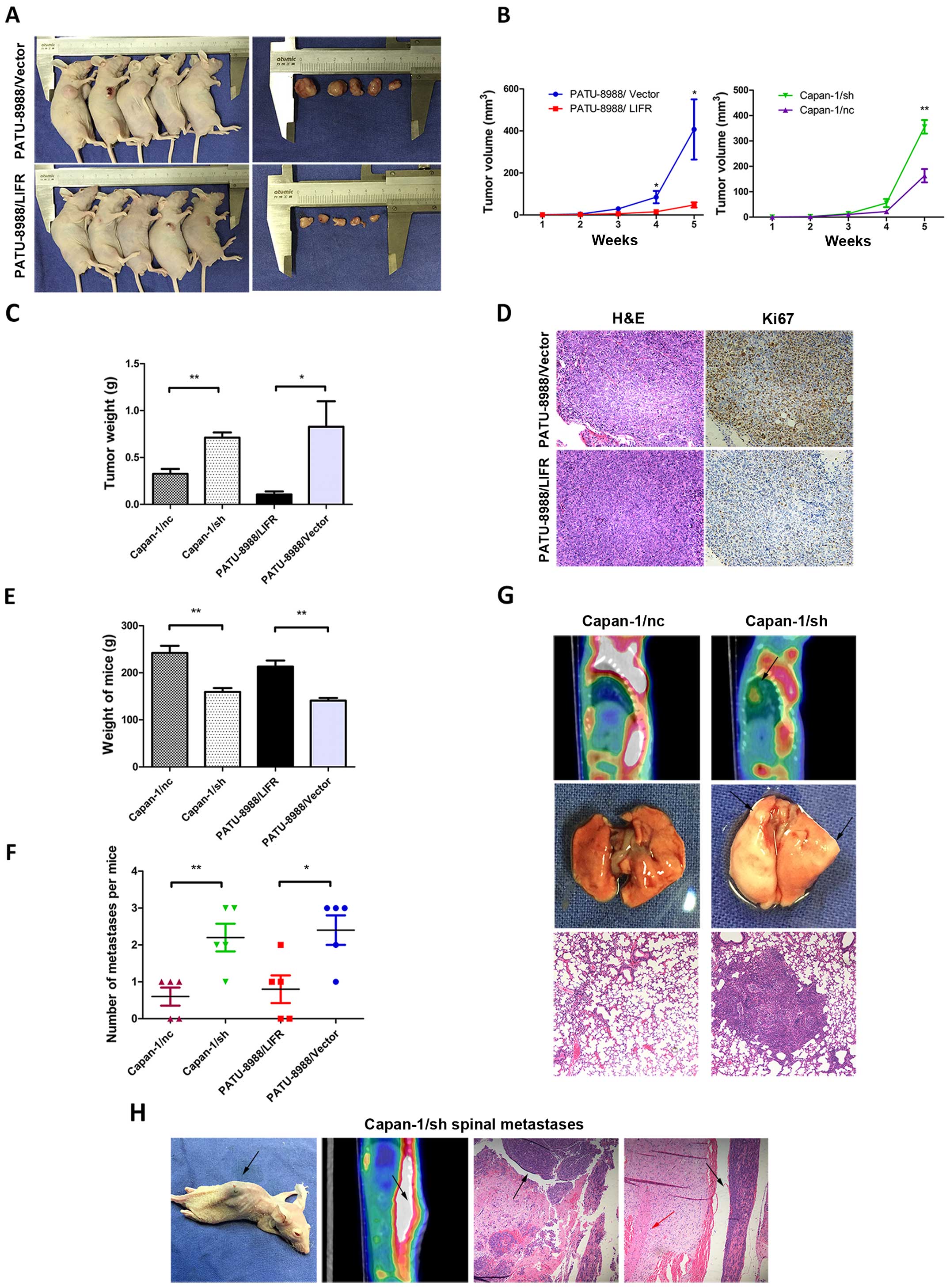
LIFR negatively regulates tumorigenesis
and metastasis of PC cells in vivo
For in vivo confirmation, a recombinant
lentivirus harboring LIFR was transfected into the PATU-8988 cells.
The stable clone expressing ectopic PATU-8988/LIFR was
subcutaneously injected into the flank of each athymic nude mouse,
and an equal volume of cells transfected with the empty vector was
injected into the opposite flank of the same mouse as the negative
control. As shown in Fig. 4A and B,
the PATU-8988/LIFR cells caused smaller tumor masses than the mock
vector control after 5 weeks of observation (PATU-8988/LIFR,
47.9±12.5 mm3; PATU-8988/vector, 407.2±143.2
mm3; P<0.05). Meanwhile, tumors derived from the
offspring subclones with LIFR-overexpressing cells were
significantly lighter than those in the control group
(PATU-8988/LIFR, 0.106±0.032 g; PATU-8988/vector, 0.828±0.273 g;
P<0.05) (Fig. 4C). Furthermore,
tumor sections from the nude mouse model were immunohistochemically
stained for Ki67. We observed that Ki67 expression was decreased in
the PATU-8988/LIFR group compared with the PATU-8988/vector group
(Fig. 4D). As expected, opposing
results were observed in the LIFR-silenced group compared with the
control group. Tumors derived from the offspring subclones with
inhibited LIFR were significantly larger and heavier than those in
the control after 5 weeks of observation (tumor volume, 355.5±26.6
vs. 163.1±25.3 mm3, P<0.01; tumor weight, 0.712±0.055
vs. 0.326±0.052 g, P<0.01) (Fig. 4B
and C). Thus, the data from the in vitro and in
vivo experiments revealed that LIFR negatively regulates the
tumorigenesis of PC cells.
Based on the results above, an in vivo model
was employed to further confirm the effect of LIFR on tumor
metastasis. Stably transfected Capan-1/sh cells (1×106
cells) were injected into the nude mouse via tail vein to observe
the distant metastasis of PC cells in vivo. After 6 weeks,
we observed that the weight of mice in the Capan-1/sh group was
significantly lower than that in the Capan-1/nc group (Fig. 4E; P<0.01). Lung metastasis was
then examined by micro/PET-CT and confirmed by pathological study.
Intriguingly, 5/5 mice injected with Capan-1/sh cells developed
larger and greater numbers of nodules of metastatic lung tumors
whereas 3 metastatic tumors out of 5 were observed in the
Capan-1/nc mice (Fig. 4F;
P<0.01). The presence of lung metastases in these mice was also
confirmed by histological analysis (Fig. 4G). Opposing results were observed in
the LIFR-overexpressing groups compared with the control groups
(Fig. 4E and F). In addition, we
identified spinal metastases in one mouse from the Capan-1/sh
group, and none in the Capan-1/nc group (Fig. 4H). Based on the in vitro and
in vivo data, the results supported that LIFR negatively
regulated the metastasis of PC cells both in vitro and in
vivo.
LIFR negatively regulates cell metastasis
and invasion of PC via epithelial to mesenchymal transition
Research has demonstrated that
epithelial-mesenchymal transition (EMT) is associated with cancer
metastasis. Based on the results above, we further studied the
possible molecular mechanisms by which LIFR contributes to PC cell
proliferation and migration. As downstream regulation pathways of
EMT, the activation of vimentin, β-catenin, slug and E-cadherin was
detected to explore the underlying regulatory network. As shown in
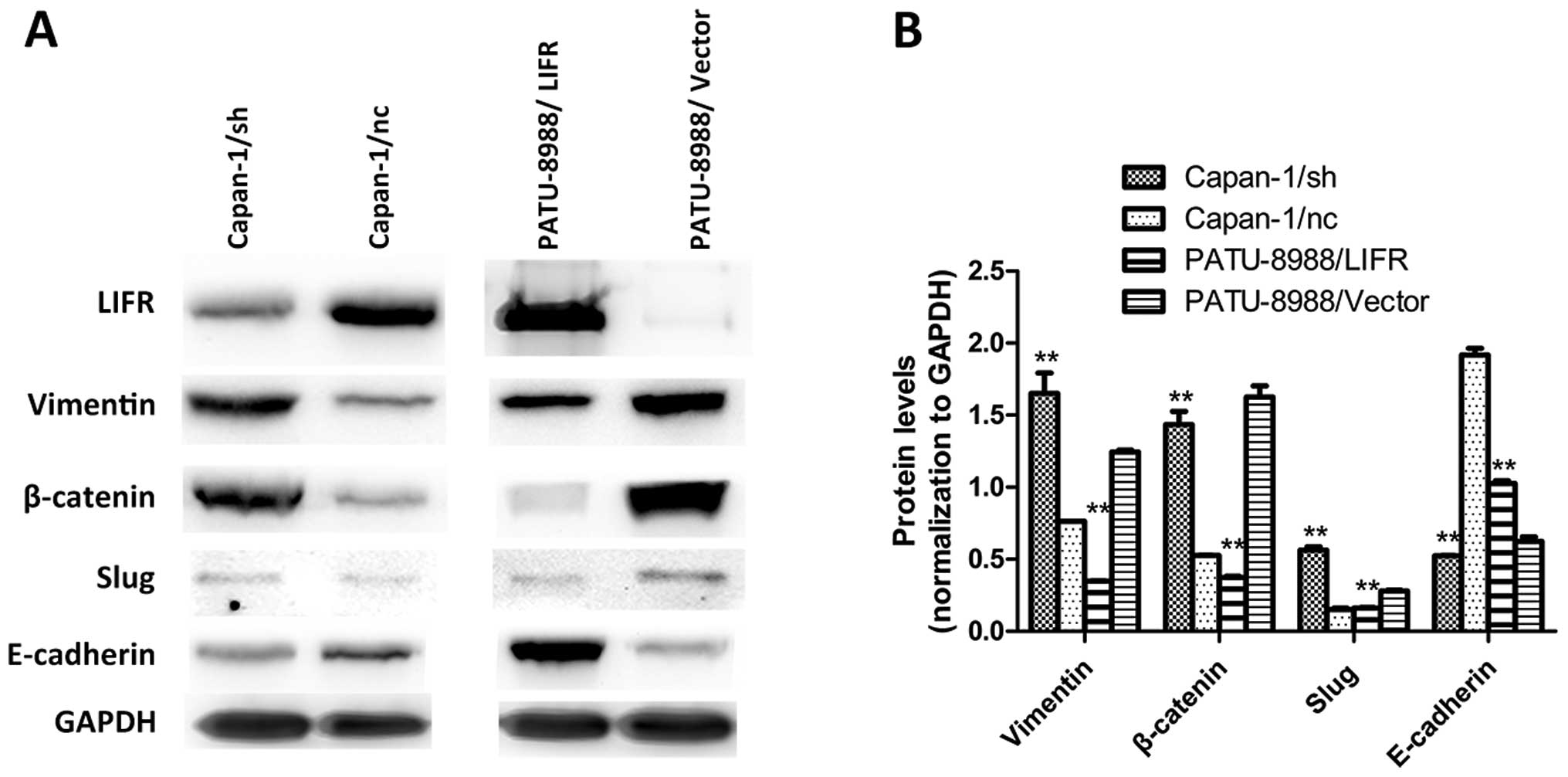
Fig. 5A and B, overexpression of
LIFR in the PATU-8988/LIFR cells significantly inhibited the
expression of vimentin, β-catenin and slug and induced the
expression of E-cadherin. Opposing results were observed in the
LIFR-silenced groups compared with the control groups (Fig. 5A and B). Collectively, these
findings suggest that LIFR negatively regulates the metastasis of
PC cells via EMT.
Discussion
Gearing et al originally isolated cDNA clones
encoding leukemia inhibitory factor receptor (LIFR) by expression
screening of a cDNA library using radioiodinated LIF as a probe
(17). LIFR was structurally
related to the IL-6 signal transducer and was found to belong to
the gp130 receptor family (18).
Therefore, LIFR plays broad roles in cell proliferation, cell
differentiation and maintenance of stem cell pluripotency. LIFR was
identified in several human malignancies and was found to be a
metastasis suppressor in hepatocellular carcinoma and breast cancer
(11,14). Nandy et al reported that
inhibition of LIFR induced by miR-125a activated the JAK2-STAT3
pathway stimulating a pro-carcinogenic molecular event in
non-malignant breast epithelial stem cells along with inactivation
of Hippo-TAZ signaling (19). In
the present study, we firstly observed that the expression level of
LIFR was significantly lower in the pancreatic cancer (PC) tumor
tissues than that in the non-tumor tissues in 55 pairs of PC
tissues. The results showed that LIFR expression was significantly
lower in tumor tissues than that in non-tumor tissues. Furthermore,
the downregulation of LIFR was associated with local invasion,
lymph node metastasis and TNM stage, but not with the other
clinicopathological factors. Although the primary results above
indicated that LIFR may function in regulating the metastasis of
PC, the detailed role and related mechanism require further
elucidation.
Compared to other cancers, the metastases of PC,
such as perineural invasion, bone metastasis, are more common and
can be observed in up to 80% of cases (20–23).
As a result, regional invasion and distant metastasis are the most
common reasons causing the poor prognosis of PC (24,25).
As a possible metastasis suppressor in several cancers, LIFR plays
crucial roles in cell proliferation and differentiation. To
pinpoint the role of LIFR in PC, we initially investigated the
influence of LIFR overexpression on colony formation ability in PC
cells of both in vitro and in vivo. The results
showed that overexpression of LIFR significantly inhibited the
growth of PATU-8988/LIFR cells. In contrast, silencing of LIFR
promoted propagation ability in Capan-1/sh PC cells. Our results
were consistent with those found in hepatocellular carcinoma
(13). Considering the importance
of colony formation in tumorigenesis, our results suggested that
LIFR may play an important role in the malignant transformation of
PC. In contrast, wound healing was decreased significantly in the
PATU-8988/LIFR cells and accelerated in the Capan-1/sh cells.
Meanwhile, more Capan-1/sh cells infiltrated the Transwell gel and
membrane to the lower chamber than the control groups.
Consistently, only a small portion of PATU-8988/LIFR cells
penetrated the membrane, compared with the control PATU-8988/vector
cells. Based on the results in vitro, the nude mouse model
was employed to confirm these results. The xenograft and lung
metastasis models were established by subcutaneous and tail vein
injection. Evidenced by digital caliper and micro-PET/CT, the tumor
volume, weight and the lung metastatic nodules were increased by
LIFR silencing and inhibited by overexpression. Thus, the results
in vivo and in vitro clearly supported the notion
that LIFR negatively regulates the tumorigenesis and metastasis of
PC cells. Notably, in the present study, one out of five mice in
the LIFR silenced group showed spinal metastases after eight-week
injection via the tail vein, emphasizing the role of LIFR as a
metastasis suppressor. Altogether, our findings revealed that LIFR
could regulate the metastasis of PC cells and LIFR may serve as a
potential marker for aggressive phenotype and as a therapeutic
target.
Numerous studies have linked both migration and
invasion to epithelial-mesenchymal (EMT)-like transition (26–28).
Early steps of metastasis, particularly the early steps of
hematogenous metastasis and lymphatic metastasis, have been linked
to EMT (29). EMT markers are found
at the invading front of several cancers, including colorectal,
gastric, mammary and endometrial cancers (30–32).
In the present study, we found that silencing of LIFR significantly
increased the expression of vimentin, β-catenin and slug.
Meanwhile, the expression of E-cadherin was significantly
decreased. Opposing results were observed in the
LIFR-overexpressing groups compared with the control groups. Slug
triggers the steps of partial separation at cell-cell borders, cell
spreading and desmosomal disruption, which are initial and
essential parts of the EMT process (33), while β-catenin is a key component in
the E-cadherin cell adhesion complex and the microtubule network
(34). The morphological transition
of cancer cells from an epithelial to a fibroblastic appearance is
accompanied by scattering and directional migration toward serum
factors, a loss of epithelial markers, such as E-cadherin, a gain
of mesenchymal cell markers, such as vimentin, which essentially
lead to metastasis. Together the above findings suggest that LIFR
negatively regulates the metastasis of PC via EMT-like
transition.
Furthermore, Shah et al (35) and Wang et al (36) found that the Notch signaling pathway
was linked with the EMT phenotype of gemcitabine-resistant PC
cells. This suggests that LIFR plays an important role in the
chemoresistance of PC and could be a potential therapeutic approach
for the treatment of metastatic chemoresistant PC. Our ongoing
study will investigate the exact mechanism of LIFR as a tumor
suppressor.
In conclusion, LIFR functions importantly in the
tumorigenesis and metastasis of PC and the EMT regulation pathway
may underlie the mechanism. Our research thereby provides new
insight into PC metastasis and the function of LIFR.
Acknowledgments
The present study was supported by the Natural
Science Foundation of China (81172326), and the Shanghai Charity
Foundation for Cancer Research.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ryan DP, Hong TS and Bardeesy N:
Pancreatic adenocarcinoma. N Engl J Med. 371:1039–1049. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Quaresma M, Coleman MP and Rachet B:
40-year trends in an index of survival for all cancers combined and
survival adjusted for age and sex for each cancer in England and
Wales, 1971–2011: A population-based study. Lancet. 385:1206–1218.
2015. View Article : Google Scholar
|
|
5
|
Konstantinidis IT, Warshaw AL, Allen JN,
Blaszkowsky LS, Castillo CF, Deshpande V, Hong TS, Kwak EL, Lauwers
GY, Ryan DP, et al: Pancreatic ductal adenocarcinoma: Is there a
survival difference for R1 resections versus locally advanced
unresectable tumors? What is a 'true' R0 resection? Ann Surg.
257:731–736. 2013. View Article : Google Scholar
|
|
6
|
Iacobuzio-Donahue CA, Fu B, Yachida S, Luo
M, Abe H, Henderson CM, Vilardell F, Wang Z, Keller JW, Banerjee P,
et al: DPC4 gene status of the primary carcinoma correlates with
patterns of failure in patients with pancreatic cancer. J Clin
Oncol. 27:1806–1813. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kishimoto T, Akira S, Narazaki M and Taga
T: Interleukin-6 family of cytokines and gp130. Blood.
86:1243–1254. 1995.PubMed/NCBI
|
|
8
|
Alisoltani A, Fallahi H, Ebrahimi M,
Ebrahimi M and Ebrahimie E: Prediction of potential cancer-risk
regions based on transcriptome data: Towards a comprehensive view.
PLoS One. 9:e963202014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Salm F, Dimitrova V, von Bueren AO, Ćwiek
P, Rehrauer H, Djonov V, Anderle P and Arcaro A: The
phosphoinositide 3-kinase p110α isoform regulates leukemia
inhibitory factor receptor expression via c-Myc and miR-125b to
promote cell proliferation in medulloblastoma. PLoS One.
10:e01239582015. View Article : Google Scholar
|
|
10
|
Liu SC, Tsang NM, Chiang WC, Chang KP,
Hsueh C, Liang Y, Juang JL, Chow KP and Chang YS: Leukemia
inhibitory factor promotes nasopharyngeal carcinoma progression and
radioresistance. J Clin Invest. 123:5269–5283. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen D, Sun Y, Wei Y, Zhang P, Rezaeian
AH, Teruya-Feldstein J, Gupta S, Liang H, Lin HK, Hung MC, et al:
LIFR is a breast cancer metastasis suppressor upstream of the
Hippo-YAP pathway and a prognostic marker. Nat Med. 18:1511–1517.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tan X and Chen M: MYLK and MYL9 expression
in non-small cell lung cancer identified by bioinformatics analysis
of public expression data. Tumour Biol. 12:12189–12200. 2014.
View Article : Google Scholar
|
|
13
|
Luo Q, Zhang Y, Wang N, Jin G, Jin H, Gu
D, Tao X, Huo X, Ge T, Cong W, et al: Leukemia inhibitory factor
receptor is a novel immunomarker in distinction of
well-differentiated HCC from dysplastic nodules. Oncotarget.
6:6989–6999. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo Q, Wang C, Jin G, Gu D, Wang N, Song
J, Jin H, Hu F, Zhang Y, Ge T, et al: LIFR functions as a
metastasis suppressor in hepatocellular carcinoma by negatively
regulating phosphoinositide 3-kinase/AKT pathway. Carcinogenesis.
36:1201–1212. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Seewoo V, Yang W, Du H, Wang J, Lin A,
Shen B, Peng C, Li H and Qiu W: The different induction mechanisms
of growth arrest DNA damage inducible gene 45 β in human hepatoma
cell lines. Chemotherapy. 58:165–174. 2012. View Article : Google Scholar
|
|
16
|
Hong X, Bu L, Wang Y, Xu J, Wu J, Huang Y,
Liu J, Suo H, Yang L, Shi Y, et al: Increases in the risk of
cognitive impairment and alterations of cerebral β-amyloid
metabolism in mouse model of heart failure. PLoS One. 8:e638292013.
View Article : Google Scholar
|
|
17
|
Gearing DP, Thut CJ, VandeBos T, Gimpel
SD, Delaney PB, King J, Price V, Cosman D and Beckmann MP: Leukemia
inhibitory factor receptor is structurally related to the IL-6
signal transducer, gp130. EMBO J. 10:2839–2848. 1991.PubMed/NCBI
|
|
18
|
Taga T, Narazaki M, Yasukawa K, Saito T,
Miki D, Hamaguchi M, Davis S, Shoyab M, Yancopoulos GD and
Kishimoto T: Functional inhibition of hematopoietic and
neurotrophic cytokines by blocking the interleukin 6 signal
transducer gp130. Proc Natl Acad Sci USA. 89:10998–11001. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nandy SB, Arumugam A, Subramani R, Pedroza
D, Hernandez K, Saltzstein E and Lakshmanaswamy R: MicroRNA-125a
influences breast cancer stem cells by targeting leukemia
inhibitory factor receptor which regulates the Hippo signaling
pathway. Oncotarget. 6:17366–17378. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li J, Ma Q, Liu H, Guo K, Li F, Li W, Han
L, Wang F and Wu E: Relationship between neural alteration and
perineural invasion in pancreatic cancer patients with
hyperglycemia. PLoS One. 6:e173852011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakao A, Harada A, Nonami T, Kaneko T and
Takagi H: Clinical significance of carcinoma invasion of the
extrapancreatic nerve plexus in pancreatic cancer. Pancreas.
12:357–361. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dai H, Li R, Wheeler T, Ozen M, Ittmann M,
Anderson M, Wang Y, Rowley D, Younes M and Ayala GE: Enhanced
survival in perineural invasion of pancreatic cancer: An in vitro
approach. Hum Pathol. 38:299–307. 2007. View Article : Google Scholar
|
|
23
|
Keleg S, Büchler P, Ludwig R, Büchler MW
and Friess H: Invasion and metastasis in pancreatic cancer. Mol
Cancer. 2:142003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shimada K, Nara S, Esaki M, Sakamoto Y,
Kosuge T and Hiraoka N: Intrapancreatic nerve invasion as a
predictor for recurrence after pancreaticoduodenectomy in patients
with invasive ductal carcinoma of the pancreas. Pancreas.
40:464–468. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Takahashi H, Ohigashi H, Ishikawa O, Gotoh
K, Yamada T, Nagata S, Tomita Y, Eguchi H, Doki Y and Yano M:
Perineural invasion and lymph node involvement as indicators of
surgical outcome and pattern of recurrence in the setting of
preoperative gemcitabine-based chemoradiation therapy for
resectable pancreatic cancer. Ann Surg. 255:95–102. 2012.
View Article : Google Scholar
|
|
26
|
Pasquier J, Abu-Kaoud N, Al Thani H and
Rafii A: Epithelial to mesenchymal transition in a clinical
perspective. J Oncol. 2015:7921822015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu X, Yun F, Shi L, Li ZH, Luo NR and Jia
YF: Roles of signaling pathways in the epithelial-mesenchymal
transition in cancer. Asian Pac J Cancer Prev. 16:6201–6206. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cohen EN, Gao H, Anfossi S, Mego M, Reddy
NG, Debeb B, Giordano A, Tin S, Wu Q, Garza RJ, et al: Inflammation
mediated metastasis: Immune induced epithelial-to-mesenchymal
transition in inflammatory breast cancer cells. PLoS One.
10:e01327102015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee JM, Dedhar S, Kalluri R and Thompson
EW: The epithelial-mesenchymal transition: New insights in
signaling, development, and disease. J Cell Biol. 172:973–981.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Domínguez D, Montserrat-Sentís B,
Virgós-Soler A, Guaita S, Grueso J, Porta M, Puig I, Baulida J,
Francí C and García de Herreros A: Phosphorylation regulates the
subcellular location and activity of the snail transcriptional
repressor. Mol Cell Biol. 23:5078–5089. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rosivatz E, Becker KF, Kremmer E, Schott
C, Blechschmidt K, Höfler H and Sarbia M: Expression and nuclear
localization of Snail, an E-cadherin repressor, in adenocarcinomas
of the upper gastrointestinal tract. Virchows Arch. 448:277–287.
2006. View Article : Google Scholar
|
|
32
|
Brabletz T, Jung A, Reu S, Porzner M,
Hlubek F, Kunz- Schughart LA, Knuechel R and Kirchner T: Variable
beta-catenin expression in colorectal cancers indicates tumor
progression driven by the tumor environment. Proc Natl Acad Sci
USA. 98:10356–10361. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Savagner P, Yamada KM and Thiery JP: The
zinc-finger protein slug causes desmosome dissociation, an initial
and necessary step for growth factor-induced epithelial-mesenchymal
transition. J Cell Biol. 137:1403–1419. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
MacDonald BT, Tamai K and He X:
Wnt/beta-catenin signaling: Components, mechanisms, and diseases.
Dev Cell. 17:9–26. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shah AN, Summy JM, Zhang J, Park SI,
Parikh NU and Gallick GE: Development and characterization of
gemcitabine-resistant pancreatic tumor cells. Ann Surg Oncol.
14:3629–3637. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Z, Li Y, Kong D, Banerjee S, Ahmad A,
Azmi AS, Ali S, Abbruzzese JL, Gallick GE and Sarkar FH:
Acquisition of epithelial-mesenchymal transition phenotype of
gemcitabine-resistant pancreatic cancer cells is linked with
activation of the notch signaling pathway. Cancer Res.
69:2400–2407. 2009. View Article : Google Scholar : PubMed/NCBI
|