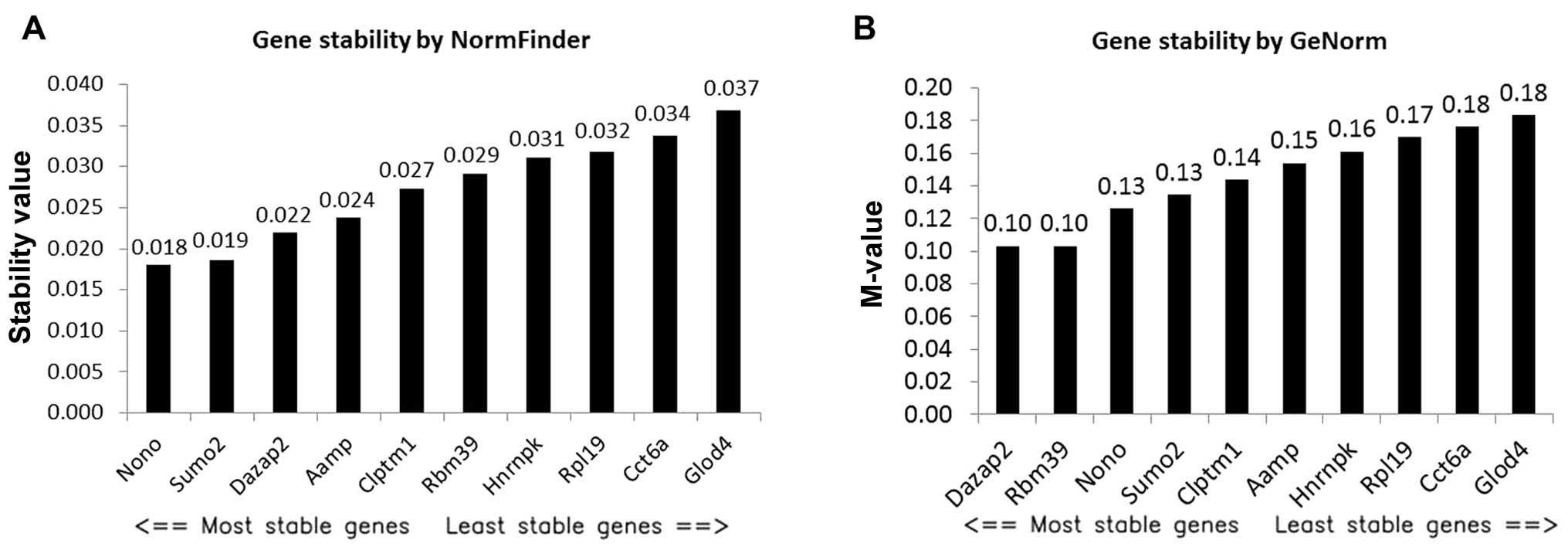
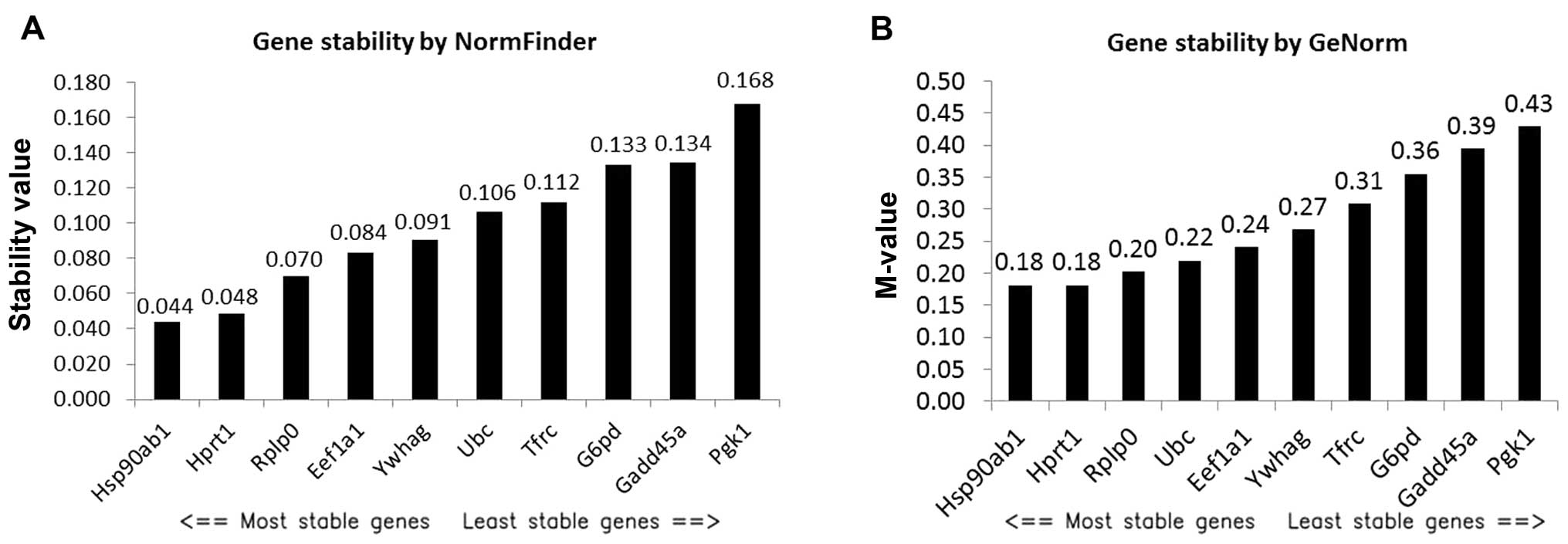
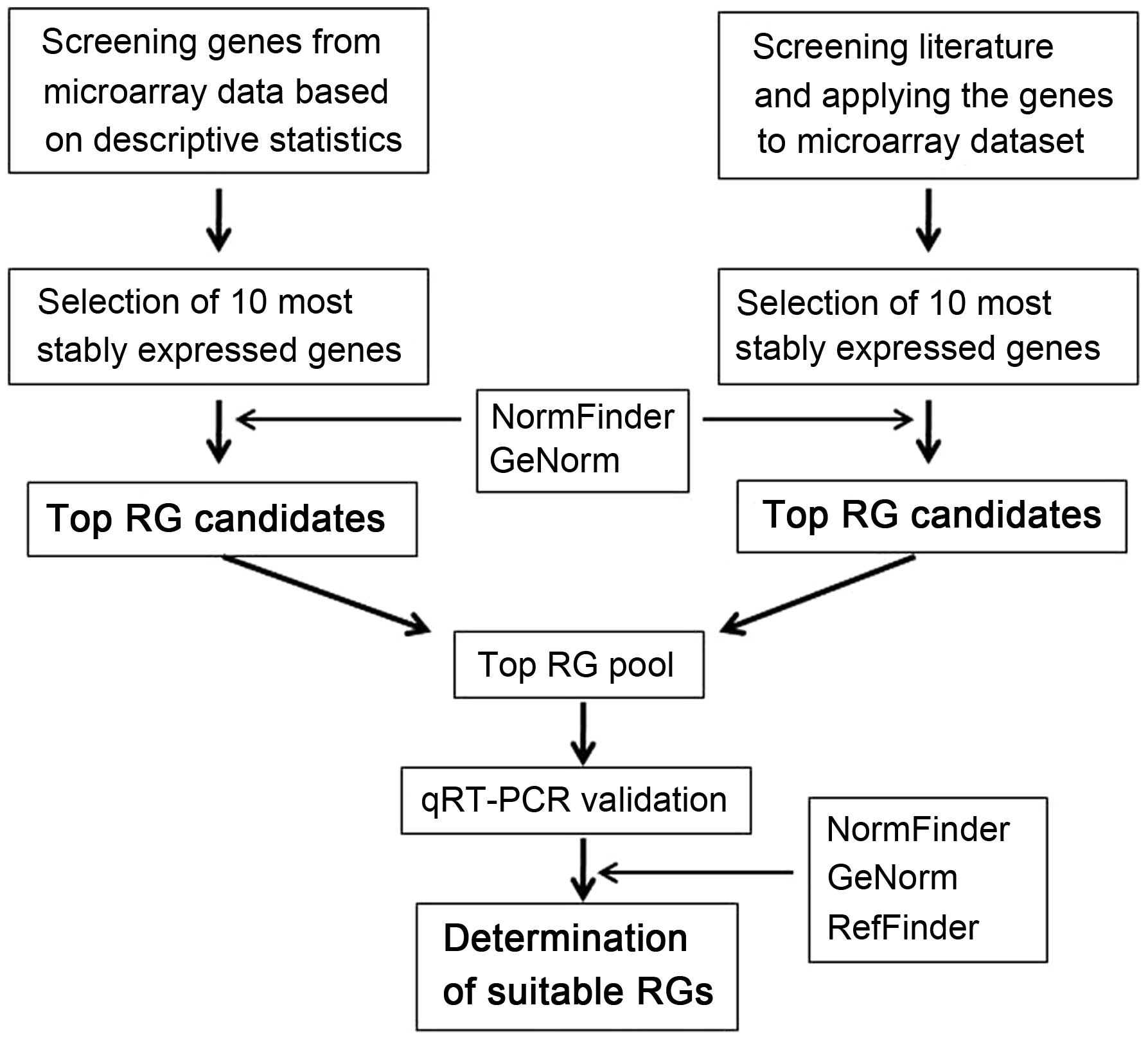
|
1
|
Kanojia D, Sawant SS, Borges AM, Ingle AD
and Vaidya MM: Alterations in keratins and associated proteins
during 4-nitro-quinoline-1-oxide induced rat oral carcinogenesis. J
Carcinog. 11:142012. View Article : Google Scholar
|
|
2
|
Yang Z, Guan B, Men T, Fujimoto J and Xu
X: Comparable molecular alterations in 4-nitroquinoline
1-oxide-induced oral and esophageal cancer in mice and in human
esophageal cancer, associated with poor prognosis of patients. In
Vivo. 27:473–484. 2013.PubMed/NCBI
|
|
3
|
Moon SM, Ahn MY, Kwon SM, Kim SA, Ahn SG
and Yoon JH: Homeobox C5 expression is associated with the
progression of 4-nitroquinoline 1-oxide-induced rat tongue
carcinogenesis. J Oral Pathol Med. 41:470–476. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Miranda SR, Noguti J, Carvalho JG, Oshima
CT and Ribeiro DA: Oxidative DNA damage is a preliminary step
during rat tongue carcinogenesis induced by 4-nitroquinoline
1-oxide. J Mol Histol. 42:181–186. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kitano M, Hatano H and Shisa H: Strain
difference of susceptibility to 4-nitroquinoline 1-oxide-induced
tongue carcinoma in rats. Jpn J Cancer Res. 83:843–850. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McCormick DL, Phillips JM, Horn TL,
Johnson WD, Steele VE and Lubet RA: Overexpression of
cyclooxygenase-2 in rat oral cancers and prevention of oral
carcinogenesis in rats by selective and nonselective COX
inhibitors. Cancer Prev Res (Phila). 3:73–81. 2010. View Article : Google Scholar
|
|
7
|
Nolan T, Hands RE and Bustin SA:
Quantification of mRNA using real-time RT-PCR. Nat Protoc.
1:1559–1582. 2006. View Article : Google Scholar
|
|
8
|
Taki FA, Abdel-Rahman AA and Zhang B: A
comprehensive approach to identify reliable reference gene
candidates to investigate the link between alcoholism and
endocrinology in Sprague-Dawley rats. PLoS One. 9:e943112014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gutierrez L, Mauriat M, Guénin S, Pelloux
J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F,
Bellini C, et al: The lack of a systematic validation of reference
genes: A serious pitfall undervalued in reverse
transcription-polymerase chain reaction (RT-PCR) analysis in
plants. Plant Biotechnol J. 6:609–618. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thellin O, Zorzi W, Lakaye B, De Borman B,
Coumans B, Hennen G, Grisar T, Igout A and Heinen E: Housekeeping
genes as internal standards: Use and limits. J Biotechnol.
75:291–295. 1999. View Article : Google Scholar
|
|
11
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:research0034.1. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper - Excel-based tool using pair-wise correlations.
Biotechnol Lett. 26:509–515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Czechowski T, Stitt M, Altmann T, Udvardi
MK and Scheible WR: Genome-wide identification and testing of
superior reference genes for transcript normalization in
Arabidopsis. Plant Physiol. 139:5–17. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peng X, Li W, Johnson WD, Torres KE and
McCormick DL: Overexpression of lipocalins and pro-inflammatory
chemokines and altered methylation of PTGS2 and APC2 in oral
squamous cell carcinomas induced in rats by
4-nitroquinoline-1-oxide. PLoS One. 10:e01162852015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peng X, Li W, Yuan L, Mehta RG, Kopelovich
L and McCormick DL: Inhibition of proliferation and induction of
autophagy by atorvastatin in PC3 prostate cancer cells correlate
with downregulation of Bcl2 and upregulation of miR-182 and p21.
PLoS One. 8:e704422013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Saviozzi S, Cordero F, Lo Iacono M,
Novello S, Scagliotti GV and Calogero RA: Selection of suitable
reference genes for accurate normalization of gene expression
profile studies in non-small cell lung cancer. BMC Cancer.
6:2002006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie F, Xiao P, Chen D, Xu L and Zhang B:
miRDeepFinder: A miRNA analysis tool for deep sequencing of plant
small RNAs. Plant Mol Biol. 80:75–84. 2012. View Article : Google Scholar
|
|
19
|
Sun JH, Nan LH, Gao CR and Wang YY:
Validation of reference genes for estimating wound age in contused
rat skeletal muscle by quantitative real-time PCR. Int J Legal Med.
126:113–120. 2012. View Article : Google Scholar
|
|
20
|
Li B, Matter EK, Hoppert HT, Grayson BE,
Seeley RJ and Sandoval DA: Identification of optimal reference
genes for RT-qPCR in the rat hypothalamus and intestine for the
study of obesity. Int J Obes. 38:192–197. 2014. View Article : Google Scholar
|
|
21
|
Sappayatosok K, Maneerat Y, Swasdison S,
Viriyavejakul P, Dhanuthai K, Zwang J and Chaisri U: Expression of
pro-inflammatory protein, iNOS, VEGF and COX-2 in oral squamous
cell carcinoma (OSCC), relationship with angiogenesis and their
clinico-pathological correlation. Med Oral Patol Oral Cir Bucal.
14:E319–E324. 2009.PubMed/NCBI
|
|
22
|
Morelatto R, Itoiz ME, Guiñazú N, Piccini
D, Gea S and López-de Blanc S: Nitric oxide synthase 2 (NOS2)
expression in histologically normal margins of oral squamous cell
carcinoma. Med Oral Patol Oral Cir Bucal. 19:e242–e247. 2014.
View Article : Google Scholar :
|
|
23
|
Brennan PA, Palacios-Callender M, Zaki GA,
Spedding AV and Langdon JD: Does type II nitric oxide synthase
expression correlate with cellular proliferation in oral squamous
cell carcinoma and dysplasia? Head Neck. 23:217–222. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dupasquier S, Delmarcelle AS, Marbaix E,
Cosyns JP, Courtoy PJ and Pierreux CE: Validation of housekeeping
gene and impact on normalized gene expression in clear cell renal
cell carcinoma: Critical reassessment of YBX3/ZONAB/CSDA
expression. BMC Mol Biol. 15:92014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo Y, Chen JX, Yang S, Fu XP, Zhang Z,
Chen KH, Huang Y, Li Y, Xie Y and Mao YM: Selection of reliable
reference genes for gene expression study in nasopharyngeal
carcinoma. Acta Pharmacol Sin. 31:1487–1494. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meyer FR, Grausgruber H, Binter C, Mair
GE, Guelly C, Vogl C and Steinborn R: Cross-platform microarray
meta-analysis for the mouse jejunum selects novel reference genes
with highly uniform levels of expression. PLoS One. 8:e631252013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fu J, Bian L, Zhao L, Dong Z, Gao X, Luan
H, Sun Y and Song H: Identification of genes for normalization of
quantitative real-time PCR data in ovarian tissues. Acta Biochim
Biophys Sin (Shanghai). 42:568–574. 2010. View Article : Google Scholar
|
|
28
|
Schroder AL, Pelch KE and Nagel SC:
Estrogen modulates expression of putative housekeeping genes in the
mouse uterus. Endocrine. 35:211–219. 2009. View Article : Google Scholar : PubMed/NCBI
|