Introduction
Osteosarcoma is a malignant neoplasm of the bone
that is prevalent in teenagers and young adults. Although it has
been reported that numerous factors are associated with the
increased risk of osteosarcoma including age, genetic inheritance,
chronic inflammation, viral infection and radiation exposure, the
cause of osteosarcoma remains undetermined (1). Most osteosarcoma patients are treated
with chemotherapy or radiation therapy. Yet, some patients remain
at high risk of relapse or metastasis, thus necessitating a
successful treatment strategy (2).
Since osteosarcoma has highly invasive and metastatic potential,
determining the factors promoting survival, migration and invasion
of osteosarcoma cells is imperative (3,4).
Elucidation of the molecular mechanisms involved in the promotion
of proliferation, migration and invasion in osteosarcoma may
further aid in the understanding of the pathogenesis of the
disease, but also may offer novel targets for effective
therapies.
Human glioma pathogenesis-related protein 1
(GLIPR1), also known as CRISP7, is a p53 target gene which
is downregulated in cancer and one of the reasons for its
deregulation is due to methylation of its promoter (5–8). Lack
of GLIPR1 was found to be associated with reduced tumor-free
survival in an animal model, and the orthotopic injection of
adenoviral vectors overexpressing GLIPR1 in a mouse model of
metastatic prostate cancer led to decreased microvessel density,
implying that GLIPR1 has anti-angiogenic ability and can increase
infiltration of cancer-associated macrophages and cytotoxic T cells
(9–13). These studies indicated that GLIPR1
is a tumor-suppressor. However, an understanding of the regulatory
mechanisms of GLIPR1 remain incomplete.
MicroRNAs (miRNAs/miRs) are regulatory, non-coding
RNAs ~18–25 nucleotides in length and are expressed at specific
stages of tissue development or cell differentiation. They have
large-scale effects on the expression of a variety of genes at the
post-transcriptional level (14–19).
The discovery of miRNAs has broadened our scope and understanding
of the mechanisms that can regulate gene expression (14–19).
miRNAs induce mRNA degradation or translational suppression through
base-pairing with its targeted mRNAs (14–19).
The aberrant expression of miRNAs has been linked to various human
types of cancer and has been proposed to have an oncogenic or a
tumor-suppressor role and they have been shown to play key roles in
cell survival, proliferation, apoptosis, migration, invasion,
angiogenesis and various other characteristic features which are
altered in cancers (20,21).
Recently, it has been reported that miR-16 is
downregulated in osteosarcoma cell lines and tissues (22). Overexpression of miR-16 was found to
suppress proliferation and tumor growth in an animal model of
osteosarcoma (22). Furthermore,
IGF1R, as a tumor-suppressive gene, is a direct target of miR-16
and its expression is inversely correlated with miR-16 levels in
osteosarcoma (22). Mechanistic
investigation further revealed that miR-16 overexpression inhibited
the Raf1/MEK1/2/ERK pathway. Yet, the identification of novel
target genes of miR-16 may help us to further understand its role
in osteosarcoma.
In the present study, we showed that GLIPR1 protein
was downregulated in osteosarcoma. Its overexpression inhibited
proliferation, migration and invasion and induced differentiation
of cancer-initiating cells (CICs) in osteosarcoma. Moreover, GLIPR1
overexpression upregulated miR-16 expression in osteosarcoma cells.
The upregulation suppressed the proliferation, migration and
invasion as well as induced differentiation of CICs in
osteosarcoma. Thus, we conclude that GLIPR1 inhibited the
proliferation, migration and invasion and induced the
differentiation of CICs by regulating miR-16 in osteosarcoma. The
present study provides direct evidence that GLIPR1 is a bona fide
tumor suppressor and identifies GLIPR1 and miR-16 as key components
for regulating the proliferation, migration, invasion and CICs in
osteosarcoma.
Materials and methods
Osteosarcoma tissues
Osteosarcoma and adjacent normal tissues were
obtained from the Department of Orthopedics, Linyi People's
Hospital Affiliated to Shandong University, Linyi, Shandong. All
tissues were histologically examined, and pathologists confirmed
the diagnosis. The Huabei Medical Ethics Committee of Linyi
People's Hospital approved the experiments undertaken. The use of
the human tissue samples followed internationally recognized
guidelines as well as local and national regulations. Informed
consent was obtained from each individual.
Cell culture
Osteosarcoma cell line MG63 was obtained from the
American Type Culture Collection (ATCC; Vanassas, MA, USA).
Briefly, the cells were maintained in RPMI-1640 medium supplemented
with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA)
and penicillin/streptomycin at 37°C in a humidified atmosphere with
5% CO2.
Cell transfection
All expression plasmids were purchased from Tiangen
(Tianjin, China). miR-16/miR controls were purchased from Ambion,
Inc. (Ambion, Austin, TX, USA). Cells were seeded into 6-well
plates 24 h before transfection. When the cells reached 80%
confluency, the expression plasmids were transfected into the MG63
cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer's instructions.
Western blot analysis
Western blot analysis was performed as previously
described (22), mainly after
incubation with the primary antibodies anti-GLIPR1 (1:250);
anti-CD133 (1:250); anti-Nestin (1:250) and anti-β-actin (1:500)
(all from Abcam, Cambridge, MA, USA) overnight at 4°C. IRDye™
800-conjugated anti-rabbit secondary antibody (LI-COR, Biosciences,
Lincoln, NE, USA) was used for 30 min at room temperature. The
specific proteins were visualized by Odyssey™ Infrared Imaging
System (Gene Company, Lincoln, NE, USA).
Cell proliferation
The effect on the cell proliferation was assessed
using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium
bromide (MTT; Sigma, St. Louis, MO, USA) assay and it was performed
as previously described (23).
Absorbance was directly proportional to the number of surviving
cells.
Flow cytometric analysis
CD133 and Nestin expression analyses were performed
by flow cytometric analysis according to the instructions. Briefly,
the cells were dissociated into single-cell populations and labeled
with a phycoerythrin-conjugated CD133 or Nestin antibody (Abcam).
The expression level was calculated using the EPICS XL flow
cytometer with EXPO32 ADC software (Becton-Dickinson, San Jose, CA,
USA).
BrdU incorporation assay
The BrdU incorporation assay was performed as
previously described (22). Cells
grown on coverslips (Fisher, Pittsburgh, PA, USA) were incubated
with bromodeoxyuridine (BrdU) for 1 h and stained with the
anti-BrdU antibody (Upstate, Temecula, CA, USA) according to the
manufacturer's instructions. Images were captured under a laser
scanning microscope (Axioskop 2 plus; Carl Zeiss Co., Ltd., Jena,
Germany).
Colony formation assay
Colony formation assay was performed as previously
described (22). For the colony
formation assay, cells were transfected as indicated, and then
seeded into a 6-well plate. FBS was added (0.3 ml/well) on day 5.
After 9 days of incubation, the plates were washed with
phosphate-buffered saline (PBS) and stained with 0.1% crystal
violet. Colonies with over 50 cells were manually counted.
Real-time PCR for miRNAs
Total RNA from the cultured cells, with efficient
recovery of small RNAs, was isolated using the mirVana miRNA
isolation kit (cat no. AM2654; Ambion, Inc.). Detection of the
mature form of miRNAs was performed using the mirVana qRT-PCR miRNA
Detection kit (cat. no. AM7659; Ambion, Inc.), according to the
manufacturer's instructions. The U6 small nuclear RNA was used as
an internal control.
Sphere growth
Osteosarcoma cells (1×103/ml) in
serum-free RPMI-1640/1 mM Na pyruvate were seeded on 0.5% agar
precoated 6-well plates. After 7 days, one third of the medium was
exchanged every second day. Single spheres were chosen and
counted.
Invasion and wound healing assays, and
miRNA detection
Invasion and wound healing assays, and miRNA
detection were performed as previously described (24).
Statistical analysis
Data are presented as the mean ± SEM. Student's
t-test (two-tailed) was used to compare two groups (P<0.05 was
considered to indicate a statistically significant result).
Results
GLIPR1 protein is downregulated in
osteosarcoma
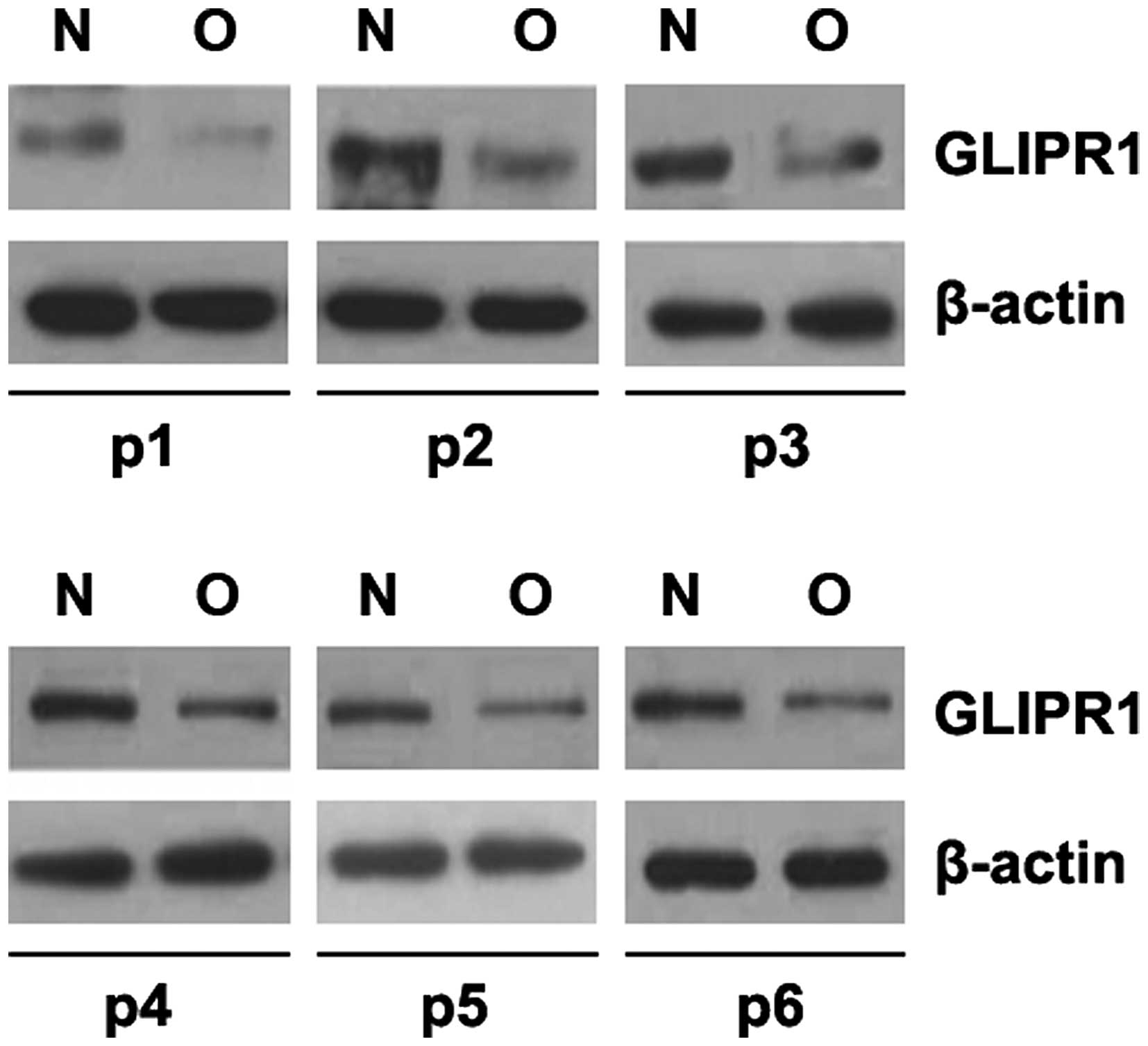
In an attempt to identify GLIPR1 expression between
osteosarcoma and adjacent normal tissues, we performed western
blotting in osteosarcoma tissues vs. normal tissues. Protein was
isolated from 6 pairs of osteosarcoma and normal tissues (patients
no. 1–6). We found that GLIPR1 protein was significantly decreased
in the sarcoma tissues, compared with that noted in the adjacent
normal tissues (Fig. 1). This
implies that GLIPR1 could be a tumor-suppressor gene in
osteosarcoma.
GLIPR1 inhibits proliferation, migration
and invasion in osteosarcoma
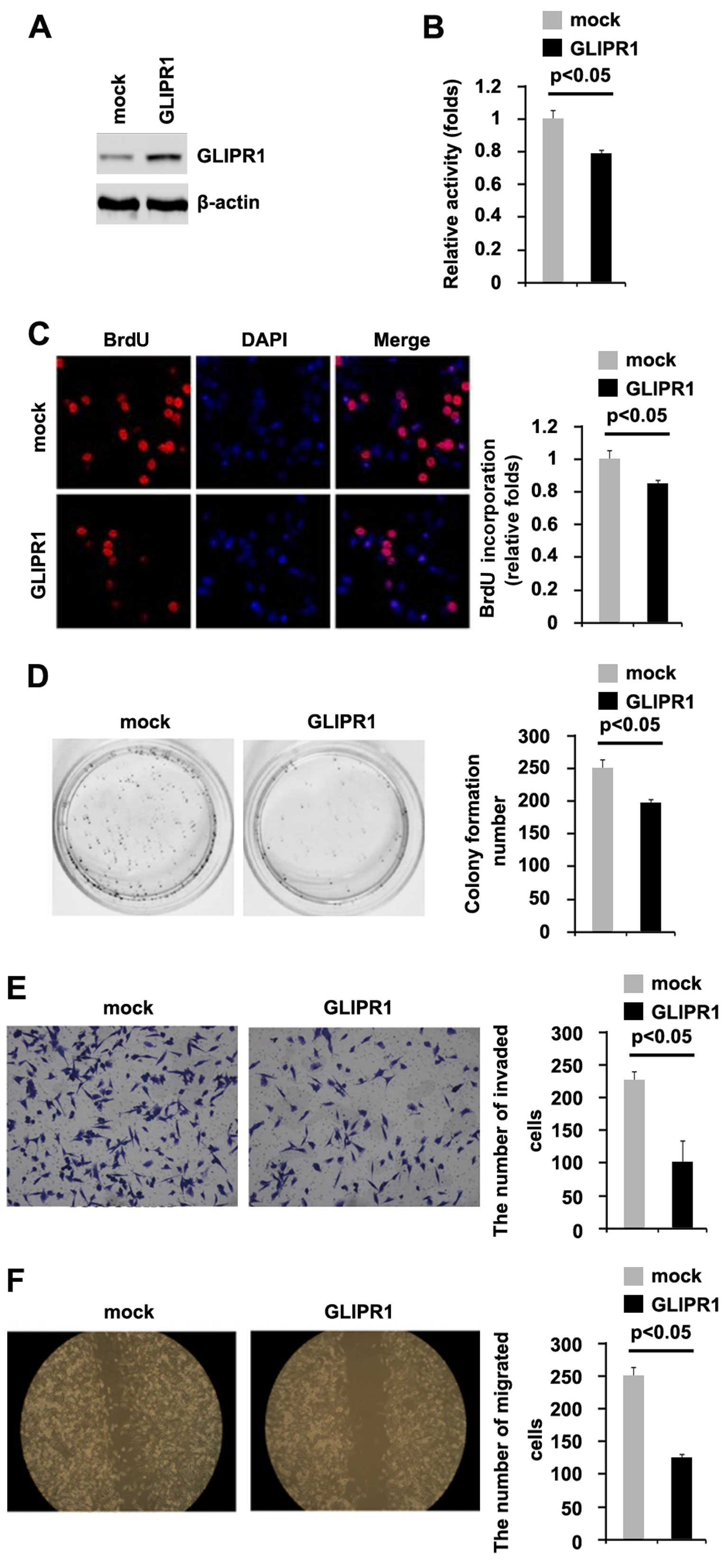
To investigate whether GLIPR1 can affect the
proliferation of osteosarcoma cells, firstly using western
blotting, we tested whether GLIPR1-expressing plasmids could stably
express GLIPR1 protein in MG63 cells. The results showed that
GLIPR1 protein was significantly increased by the GLIPR1-expressing
plasmids in the cells (Fig. 2A). In
addition, we performed MTT assay to detect proliferation of the
MG63 cells following transfection with the GLIPR1-expressing
plasmids. The results showed that GLIPR1 inhibited the
proliferation of the MG63 cells after 48 h of transfection
(Fig. 2B). To further study the
effects of GLIPR1 on proliferation, we performed BrdU incorporation
assay to detect DNA synthesis in the cells. The results confirmed
that GLIPR1 significantly inhibited DNA synthesis in the cells
(Fig. 2C). In order to identify the
effect of GLIPR1 on colony formation, we performed a colony
formation assay. The results showed that overexpression of GLIPR1
significantly suppressed the colony formation rate of the MG63
cells following transfection (Fig.
2D).
In an attempt to identify the role of GLIPR1 in
regulating invasion and migration of MG63 cells, we performed
invasion and would healing assays to detect the invasion and
migration of the MG63 cells following transfection with the
GLIPR1-expressing plasmids and empty vectors. Ectopic GLIPR1 did
inhibit the invasion and motility by ~2-fold in the cells (Fig. 2E and F).
GLIPR1 induces differentiation of CICs in
osteosarcoma
Malignant tumors tend to relapse after surgical
resection, and this behavior is believed to be largely attributable
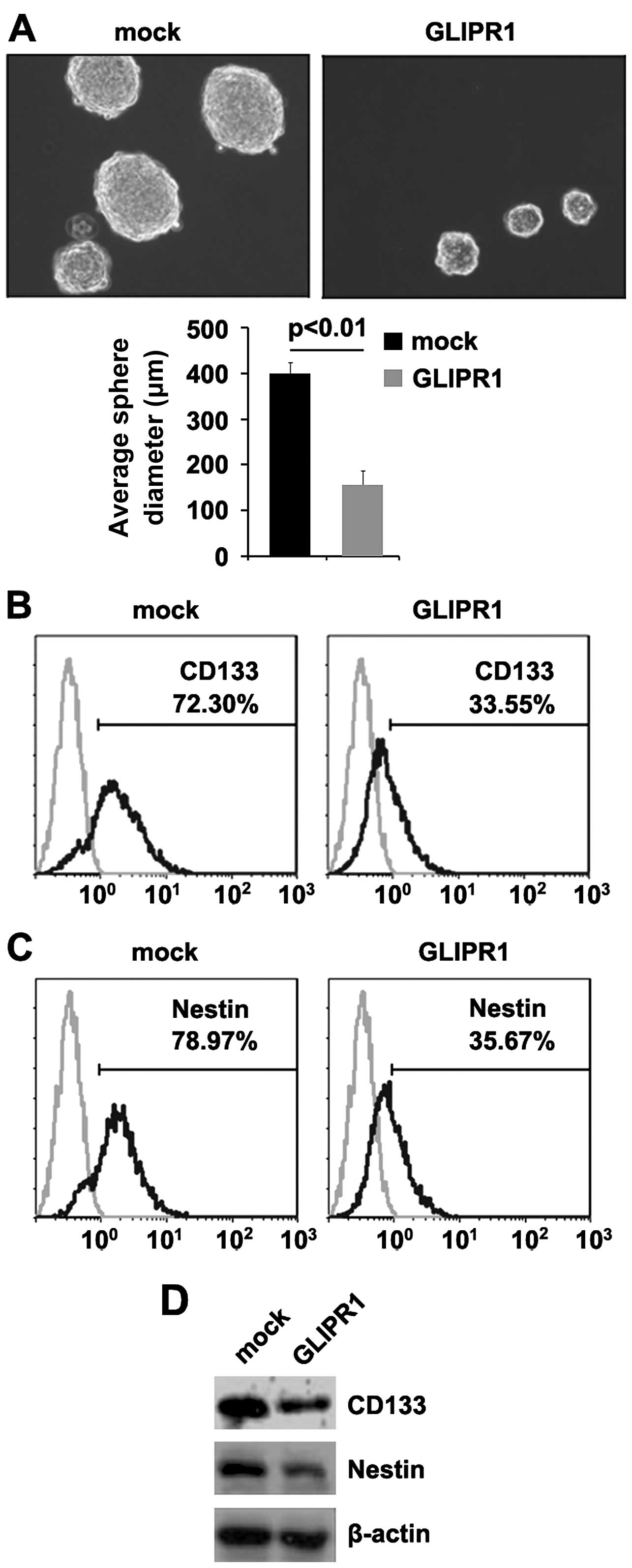
to the stem cell-like properties of a fraction of cells (25). Since GLIPR1 is expressed at
very low levels in osteosarcoma, but at high levels in
differentiated normal tissues, we aimed to determine the potential
role of GLIPR1 in the development and maintenance of the
stem-like property of osteosarcoma cells. A sphere forming assay
showed (Fig. 3A) that
GLIPR1-overexpressing cells formed much smaller spheres
after 7 days of culture when compared with the control cells
(~2-fold smaller in diameter), indicating markedly decreased
self-renewal ability by GLIPR1.
Consistent with these results, CD133- and
Nestin-positive cell proportions were significantly lower in the
GLIPR1-transfected cells than those in the control cells
(Fig. 3B and C). In further
experiments, we performed western blotting to detect CD133 and
Nestin protein in the MG63 cells following transfection with the
GLIPR1-expressing plasmids and empty vectors. We observed a
significantly faster decrease in CD133 and Nestin protein in the
MG63 cells transfected with GLIPR1 (Fig. 3D). Taken together, these data showed
that reintroduction of GLIPR1 in osteosarcoma cells reduced
the stem cell-like population and greatly attenuated the ability of
stem cell-like osteosarcoma cells to retain stemness.
GLIPR1 upregulates miR-16 expression and
miR-16 suppresses the proliferation, migration and invasion in
osteosarcoma
Tumor-suppressor genes can exert their functions by
regulating miRNA expression in cancer (26) and in regards to the miRNA
involvement in lung cancer pathogenesis, some of them function as
tumor-suppressor genes or oncogenes (27,28).
Thus, we reasoned that GLIPR1 functions as a tumor-suppressor gene
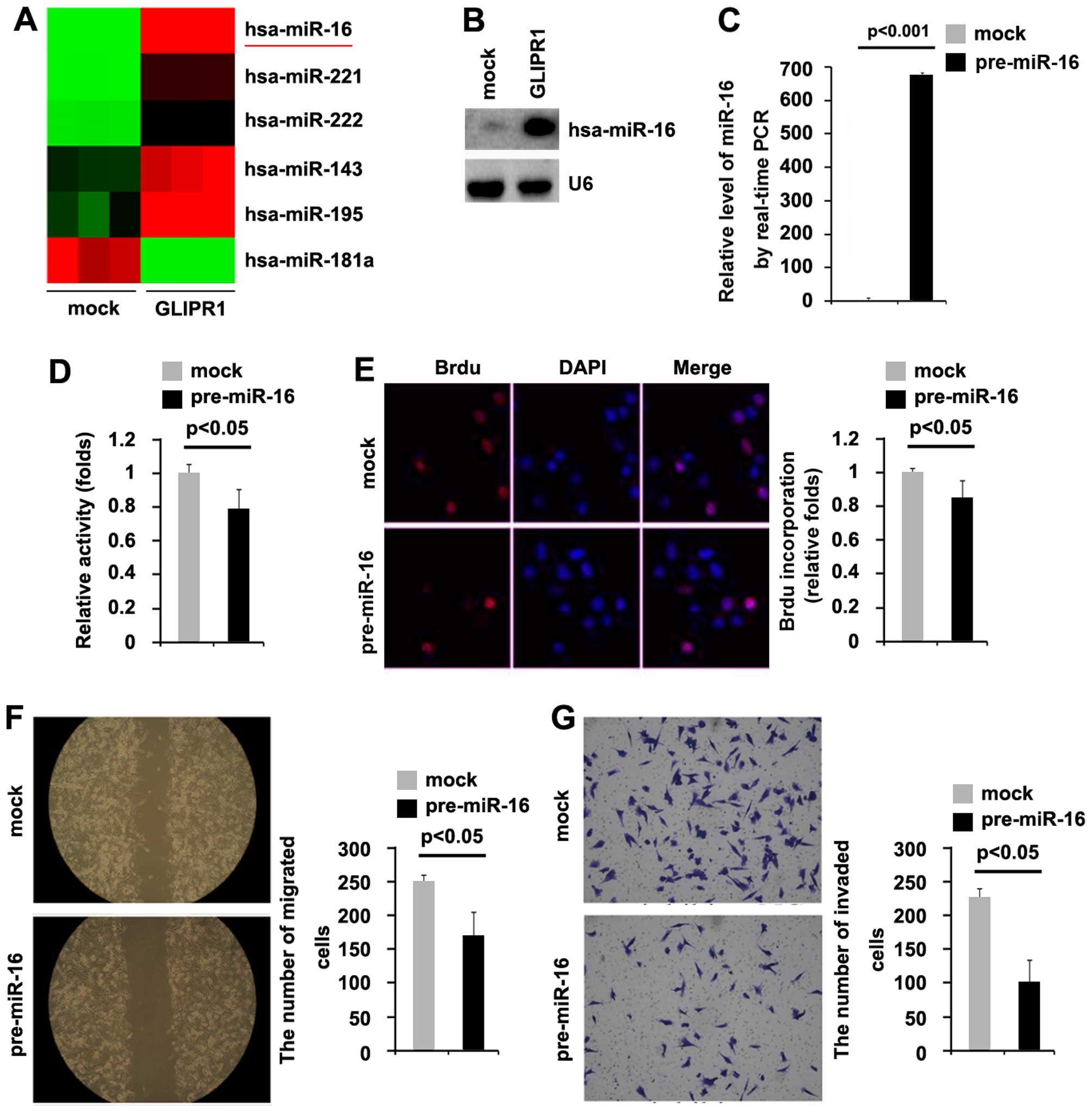
by regulating relevant miRNAs. miRNA microarray was performed. RNAs
isolated from the MG63 cells transfected with GLIPR1 or empty
vectors were hybridized to a custom miRNA microarray platform.
After three times of hybridization, quantification and
normalization, 200 miRNAs were upregulated >5-fold in the cells.
We were interested in miR-16 (Fig.
4A), since it is downregulated in osteosarcoma indicating that
it may be a tumor-suppressor gene.
To further identify whether GLIPR1 upregulates the
miR-16 level in MG63 cells, northern blotting was performed. Our
results demonstrated that miR-16 was upregulated by GLIPR1 in the
MG63 cells (Fig. 4B).
To investigate whether miR-16 can affect the
proliferation of osteosarcoma cells, firstly using real-time PCR,
we tested whether pre-miR-16 stably expressed miR-16 in the MG63
cells. The results showed that miR-16 was significantly increased
by pre-miR-16 in the cells (Fig.
4C). In addition, we performed MTT assay to detect the
proliferation of MG63 cells following transfection with pre-miR-16.
The results showed that miR-16 inhibited the proliferation in MG63
cells after 48 h of transfection (Fig.
4D). To further show the effects of miR-16 on proliferation, we
performed BrdU incorporation assay to detect DNA synthesis in the
cells. The results confirmed that miR-16 significantly inhibited
DNA synthesis in the cells (Fig.
4E). In an attempt to identify the role of miR-16 in regulating
migration and invasion of MG63 cells, we performed wound healing
and invasion assays to detect the migration and invasion of MG63
cells following transfection with pre-miR-16 and control miR.
Ectopic miR-16 inhibited the motility and invasion by ~2-fold in
the cells (Fig. 2F and G).
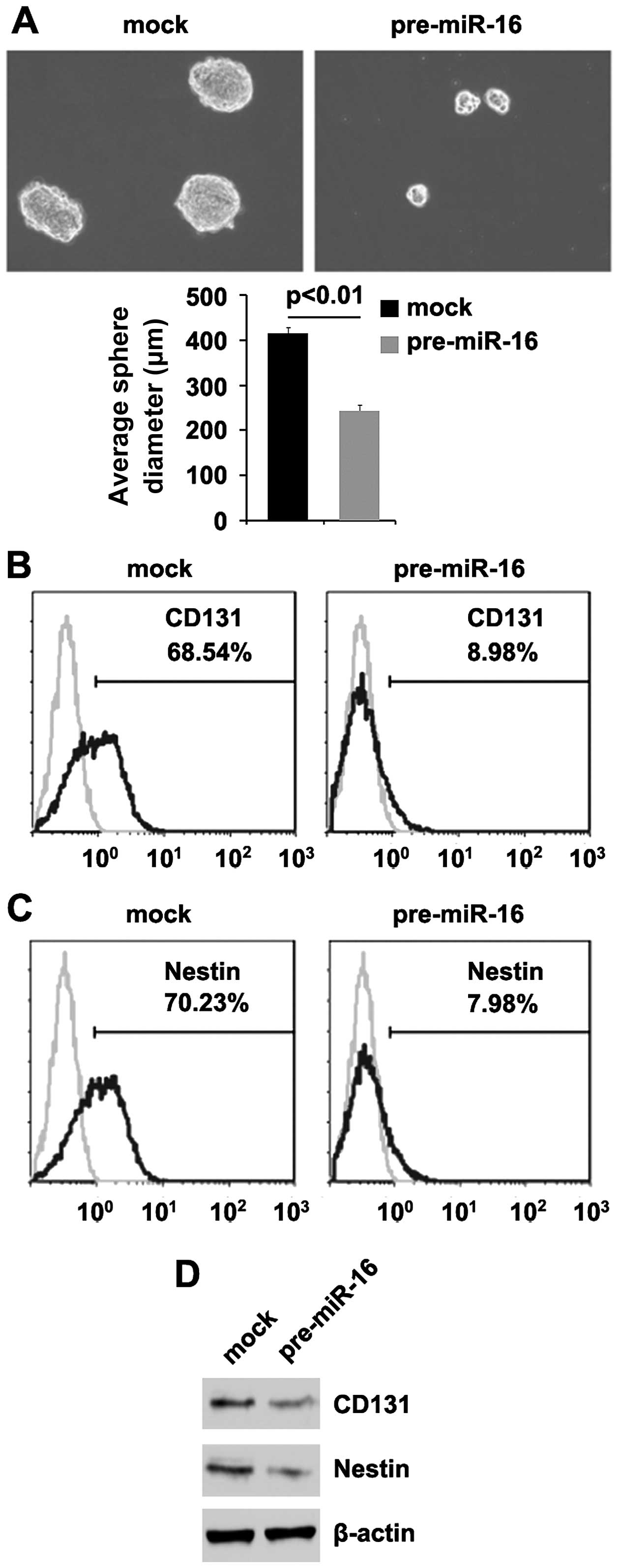
miR-16 induces the differentiation of
CICs in osteosarcoma
Since GLIPR1 induces the differentiation of CICs and
it upregulates miR-16 expression, we aimed to determine the
potential role of miR-16 in the development and maintenance
of the stem-like properties of osteosarcoma cells. Sphere-forming
assay showed (Fig. 5A) that
miR-16-overexpressing cells formed much smaller spheres
after 7 days of culture as compared with the control cells (~2-fold
smaller in diameter), indicating the markedly decreased
self-renewal ability by miR-16.
Consistent with these results, CD133- and
Nestin-positive proportions were significantly lower in the
miR-16-expressing cells than levels in the control cells
(Fig. 5B and C). In further
experiments, we performed western blotting to detect CD133 and
Nestin protein in the MG63 cells following transfection with
pre-miR-16 and miR control. We also observed a significantly faster
decrease in CD133 and Nestin protein in the MG63 cells transfected
with pre-miR-16 (Fig. 5D). Taken
together, these data showed that reintroduction of miR-16 in
the osteosarcoma cells reduced the stem cell-like population and
greatly attenuated the ability of stem cell-like osteosarcoma cells
to retain stemness.
Discussion
Recent studies have indicated that solid tumors,
including osteosarcoma, are driven by a population of tumor stem
cells (TSCs) or tumor-initiating cells (TICs) (29,30).
It is believed that TSCs can fuel tumor growth and seed metastasis.
Although conventional chemotherapy can target proliferating tumor
cells and it is common to find tumor regression, some osteosarcoma
patients develop tumor relapse or metastasis after chemotherapy and
surgery. This clinical clue allows us to presume that current
approaches are not efficient to target TSCs, according to the
concept of TSCs (31,32).
Thus, finding novel targets for eliminating TSCs is
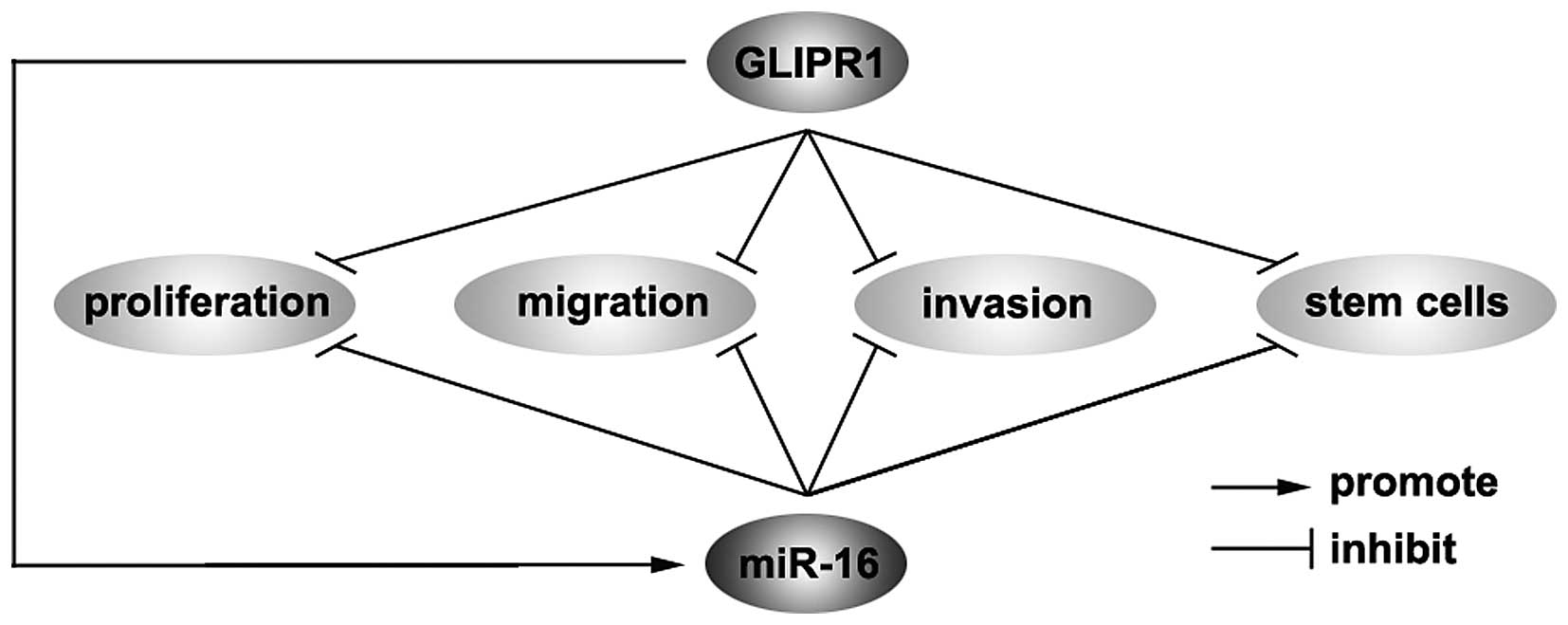
vital. The present study showed that GLIPR1 protein is
downregulated in osteosarcoma and GLIPR1 inhibits proliferation,
migration and invasion in osteosarcoma, meanwhile inducing
differentiation of cancer-initiating cells (CICs) in osteosarcoma
(Fig. 6). CSCs have been shown to
be more resistant to standard chemotherapeutic agents and
radiotherapy. Traditional treatment may lead to tumor shrinkage,
however most tumors can recur after treatment, likely since CSCs
survive and regenerate tumor growth (33,34).
Treatment strategies to restore GLIPR1 protein has the potential to
be an effective therapy for osteosarcoma.
A large body of evidence indicates that miRNAs are
frequently deregulated in a variety of human malignancies (35). Studies have shown a direct link
between miRNA function and oncogenesis which is supported by
examining the expression of miRNAs in clinical samples (36,37).
The profiling of miRNA expression showed that most of them are
downregulated in tumors compared to normal tissues (38), such as let-7 in lung cancers
(39) and miR-127 in human bladder
cancers (40). However, there are
other miRNAs which are upregulated in tumors, such as miR-150 in
gastric cancer (41), miR-21 in
prostate cancer (42) and the
miR-17-92 cluster in renal cell carcinoma (43). We focused on miR-16 since previous
studies demonstrated that the expression of miR-16 was
significantly decreased in primary osteosarcoma samples as compared
with adjacent normal tissues (22).
We showed that miR-16 inhibited the proliferation, motility and
invasion in osteosarcoma, meanwhile inducing differentiation of
CICs in osteosarcoma cells (Fig.
6).
In addition, we demonstrated that GLIPR1 can
upregulate miR-16 expression in ostesosarcoma cells (Fig. 6). In the future, we may continue to
analyze whether the GLIPR1 protein level is positively associated
with miR-16 expression in osteosarcoma tissues and to detect
whether GLIPR1 upregulates the miR-16 level by activating its
promoter.
In the present study, GLIPR1-mediated miR-16
regulation in osteosarcoma has potential basic and clinical
implications (Fig. 6). On the one
hand, GLIPR1 could be a powerful tumor-suppressor gene by promoting
proliferation, motility and invasion as well as regulating
osteosarcoma stem cells and pharmacological inhibition of GLIPR1
may represent a promising therapeutic strategy. On the other hand,
miR-16 is a tumor-suppressor gene and its expression is promoted by
GLIPR1 in osteosarcoma. Yet, the role of GLIPR1-mediated miR-16
regulation in osteosarcoma warrants further investigation. We will
continue to identify downstream target genes of miR-16 in
osteosarcoma.
References
|
1
|
Ottaviani G and Jaffe N: The etiology of
osteosarcoma. Cancer Treat Res. 152:15–32. 2009. View Article : Google Scholar
|
|
2
|
Wang SW, Wu HH, Liu SC, Wang PC, Ou WC,
Chou WY, Shen YS and Tang CH: CCL5 and CCR5 interaction promotes
cell motility in human osteosarcoma. PLoS One. 7:e351012012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gill J, Ahluwalia MK, Geller D and Gorlick
R: New targets and approaches in osteosarcoma. Pharmacol Ther.
137:89–99. 2013. View Article : Google Scholar
|
|
4
|
Bjørnland K, Flatmark K, Pettersen S,
Aaasen AO, Fodstad O and Maelandsmo GM: Matrix metalloproteinases
participate in osteosarcoma invasion. J Surg Res. 127:151–156.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ren C, Li L, Goltsov AA, Timme TL, Tahir
SA, Wang J, Garza L, Chinault AC and Thompson TC: mRTVP-1, a novel
p53 target gene with proapoptotic activities. Mol Cell Biol.
22:3345–3357. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ren C, Li L, Yang G, Timme TL, Goltsov A,
Ren C, Ji X, Addai J, Luo H, Ittmann MM, et al: RTVP-1, a tumor
suppressor inactivated by methylation in prostate cancer. Cancer
Res. 64:969–976. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiao YH, Li XH, Tan T, Liang T, Yi H, Li
MY, Zeng GQ, Wan XX, Qu JQ, He QY, et al: Identification of GLIPR1
tumor suppressor as methylation-silenced gene in acute myeloid
leukemia by microarray analysis. J Cancer Res Clin Oncol.
137:1831–1840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chilukamarri L, Hancock AL, Malik S,
Zabkiewicz J, Baker JA, Greenhough A, Dallosso AR, Huang TH,
Royer-Pokora B, Brown KW, et al: Hypomethylation and aberrant
expression of the glioma pathogenesis-related 1 gene in Wilms
tumors. Neoplasia. 9:970–978. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li L, Abdel Fattah E, Cao G, Ren C, Yang
G, Goltsov AA, Chinault AC, Cai WW, Timme TL and Thompson TC:
Glioma pathogenesis-related protein 1 exerts tumor suppressor
activities through proapoptotic reactive oxygen
species-c-Jun-NH2 kinase signaling. Cancer Res.
68:434–443. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Satoh T, Timme TL, Saika T, Ebara S, Yang
G, Wang J, Ren C, Kusaka N, Mouraviev V and Thompson TC: Adenoviral
vector-mediated mRTVP-1 gene therapy for prostate cancer. Hum Gene
Ther. 14:91–101. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sonpavde G, Thompson TC, Jain RK, Ayala
GE, Kurosaka S, Edamura K, Tabata K, Ren C, Goltsov AA, Mims MP, et
al: GLIPR1 tumor suppressor gene expressed by adenoviral vector as
neoadjuvant intraprostatic injection for localized intermediate or
high-risk prostate cancer preceding radical prostatectomy. Clin
Cancer Res. 17:7174–7182. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Naruishi K, Timme TL, Kusaka N, Fujita T,
Yang G, Goltsov A, Satoh T, Ji X, Tian W, Abdelfattah E, et al:
Adenoviral vector-mediated RTVP-1 gene-modified tumor cell-based
vaccine suppresses the development of experimental prostate cancer.
Cancer Gene Ther. 13:658–663. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karantanos T, Tanimoto R, Edamura K,
Hirayama T, Yang G, Golstov AA, Wang J, Kurosaka S, Park S and
Thompson TC: Systemic GLIPR1-ΔTM protein as a novel therapeutic
approach for prostate cancer. Int J Cancer. 134:2003–2013. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pasquinelli AE, Reinhart BJ, Slack F,
Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B,
Müller P, et al: Conservation of the sequence and temporal
expression of let-7 heterochronic regulatory RNA. Nature.
408:86–89. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Farh KK, Grimson A, Jan C, Lewis BP,
Johnston WK, Lim LP, Burge CB and Bartel DP: The widespread impact
of mammalian microRNAs on mRNA repression and evolution. Science.
310:1817–1821. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Calin GA and Croce CM: MicroRNA-cancer
connection: The beginning of a new tale. Cancer Res. 66:7390–7394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen L, Wang Q, Wang GD, Wang HS, Huang Y,
Liu XM and Cai XH: miR-16 inhibits cell proliferation by targeting
IGF1R and the Raf1-MEK1/2-ERK1/2 pathway in osteosarcoma. FEBS
Lett. 587:1366–1372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu H, Wu Y, Zheng W and Lu S: CO-029 is
overexpressed in gastric cancer and mediates the effects of EGF on
gastric cancer cell proliferation and invasion. Int J Mol Med.
35:798–802. 2015.PubMed/NCBI
|
|
24
|
Xu WG, Shang YL, Cong XR, Bian X and Yuan
Z: MicroRNA-135b promotes proliferation, invasion and migration of
osteosarcoma cells by degrading myocardin. Int J Oncol.
45:2024–2032. 2014.PubMed/NCBI
|
|
25
|
Sanai N, Alvarez-Buylla A and Berger MS:
Neural stem cells and the origin of gliomas. N Engl J Med.
353:811–822. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Seike M, Goto A, Okano T, Bowman ED,
Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, et
al: MiR-21 is an EGFR-regulated anti-apoptotic factor in lung
cancer in never-smokers. Proc Natl Acad Sci USA. 106:12085–12090.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lal A, Navarro F, Maher CA, Maliszewski
LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O,
et al: miR-24 Inhibits cell proliferation by targeting E2F2, MYC,
and other cell-cycle genes via binding to 'seedless' 3′UTR microRNA
recognition elements. Mol Cell. 35:610–625. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gupta PB, Chaffer CL and Weinberg RA:
Cancer stem cells: Mirage or reality? Nat Med. 15:1010–1012. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: Accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Siclari VA and Qin L: Targeting the
osteosarcoma cancer stem cell. J Orthop Surg. 5:782010. View Article : Google Scholar
|
|
32
|
Chumsri S and Burger AM: Cancer stem cell
targeted agents: Therapeutic approaches and consequences. Curr Opin
Mol Ther. 10:323–333. 2008.PubMed/NCBI
|
|
33
|
Costello RT, Mallet F, Gaugler B, Sainty
D, Arnoulet C, Gastaut JA and Olive D: Human acute myeloid leukemia
CD34+/CD38− progenitor cells have decreased
sensitivity to chemotherapy and Fas-induced apoptosis, reduced
immunogenicity, and impaired dendritic cell transformation
capacities. Cancer Res. 60:4403–4411. 2000.PubMed/NCBI
|
|
34
|
Dean M, Fojo T and Bates S: Tumour stem
cells and drug resistance. Nat Rev Cancer. 5:275–284. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Deng S, Calin GA, Croce CM, Coukos G and
Zhang L: Mechanisms of microRNA deregulation in human cancer. Cell
Cycle. 7:2643–2646. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and downregulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar
|
|
37
|
Calin GA, Ferracin M, Cimmino A, Di Leva
G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et
al: A MicroRNA signature associated with prognosis and progression
in chronic lymphocytic leukemia. N Engl J Med. 353:1793–1801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Saito Y, Liang G, Egger G, Friedman JM,
Chuang JC, Coetzee GA and Jones PA: Specific activation of
microRNA-127 with downregulation of the proto-oncogene BCL6 by
chromatin-modifying drugs in human cancer cells. Cancer Cell.
9:435–443. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K,
Ren G, Su T, Pan Y, Feng B, et al: MiR-150 promotes gastric cancer
proliferation by negatively regulating the pro-apoptotic gene EGR2.
Biochem Biophys Res Commun. 392:340–345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ribas J and Lupold SE: The transcriptional
regulation of miR-21, its multiple transcripts, and their
implication in prostate cancer. Cell Cycle. 9:923–929. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chow TF, Mankaruos M, Scorilas A, Youssef
Y, Girgis A, Mossad S, Metias S, Rofael Y, Honey RJ, Stewart R, et
al: The miR-17-92 cluster is over expressed in and has an oncogenic
effect on renal cell carcinoma. J Urol. 183:743–751. 2010.
View Article : Google Scholar
|