Introduction
Metastasis is the most detrimental feature of cancer
that leads to patient mortality. It is a highly selective process
which is comprised of a number of complex and interrelated steps.
Cancer cells must escape from the primary tumor site, survive in
the circulation, invade multiple distant tissues or organs and
finally inhabit specific organ sites (1,2).
Cancer cells in primary and metastatic tumors exhibit differences
in biological heterogeneity that raise difficulty for the treatment
of metastatic tumors. Understanding the cellular and molecular
pathogenesis of metastasis is important to prevent metastasis and
to search for an effective therapy for metastatic cancers.
Cholangiocarcinoma (CCA) is a highly invasive tumor
that leads to an extremely poor prognosis. As CCA is difficult to
diagnose at the early stage, the vast majority of CCA patients are
diagnosed when the disease has progressed to the metastatic stage.
The common sites of metastasis are the liver, lymph nodes, lung and
bone (3–6). Metastasis is usually the cause of
death of CCA patients. Current treatments e.g., radiotherapy,
chemotherapy and surgery do not effectively ensure or prolong
patient survival (7).
At present, the understanding of the metastatic
process of CCA at the molecular level is limited. Experimental
systems that mimic endogenous contributing factors are required to
understand the complexity of metastasis. Most of the CCA cell lines
used rarely develop tumors that metastasize, making it difficult to
study these detrimental events in cancer progression. To understand
the molecular mechanism of metastasis in CCA, a highly metastatic
CCA cell line is needed. This cell line can then be used to
investigate the causes of metastasis while at the same time test
for new therapies. Moreover, the molecular networks and signaling
pathways between individual molecules and metastasis can be
elucidated.
In the present study, an in vivo selection
resulted in the establishment of a human CCA cell subline,
KKU-213L5, which possesses higher metastatic behaviors, i.e.,
growth rate, stem cell marker characteristics, migration and
invasion abilities, than those of the parental, KKU-213 cells. The
aggressiveness of KKU-213L5 compared to the parental cells was
demonstrated in the subcutaneous xenograft and tail vein injected
lung metastasis mouse models. Expression levels of various
molecules with regard to the metastatic processes were compared.
Finally, two genes, anterior gradient protein-2 (AGR2) and KiSS-1,
were identified in the KKU-213L5 cell line and their associations
with metastasis in CCA patients were validated. This newly
established subline should be of benefit not only to basic
investigations but also to translational research efforts on the
metastasis of CCA and of cancer as a whole.
Materials and methods
Cell lines and CCA patient tissues
The parental CCA cell line, KKU-213, was established
from the primary tumor of a Thai male with histologically confirmed
CCA as previously described (8).
The presence of liver fluke Opisthorchis viverrini eggs was
noted in this pathological record. The cell line KKU-213 (JCRB1557)
was purchased from the Japanese Collection of Research Bioresources
(JCBR) Cell Bank, Osaka, Japan.
Paraffin-embedded histologically confirmed CCA
tissues (n=32) were obtained from the specimen bank of the Liver
Fluke and Cholangiocarcinoma Research Center, Faculty of Medicine,
Khon Kaen University, Thailand. The protocol was reviewed and
approved by The Khon Kaen University Ethics Committee for Human
Research (HE571464) based on the Declaration of Helsinki and
ICH-Good Clinical Practice Guidelines.
Establishment of a highly metastatic
KKU-213L5 subline
A highly metastatic subline namely, KKU-213L5, was
established from the metastasized lung tissues of NOD/scid/Jak3
mice (NOJ mice) (9). Mice were
accommodated and monitored according to institutional guidelines.
The Institutional Animal Care and Use Committee of Kumamoto
University approved the experimental procedures.
The parental cells, KKU-213 (5×105
cells), were inoculated through tail veins of the 8- to 10-week old
male NOJ mice. Approximately 18 days after injection, the mice were
sacrificed and the pulmonary metastatic tumors were resected and
minced for in vitro culture. The cells were re-passaged and
cultured until the absence of fibroblasts occurred. Then, cultured
metastatic tumor cells were inoculated into the naïve mice in the
same manner. After repeating these in vivo selection
procedures 5 times, the cells which preferentially metastasized to
the lungs following intravenous injection were obtained and
designated as KKU-213L5 (lung metastatic variant 5).
KKU-213 and KKU-213L5 cells were cultured in Ham's
F-12 media supplemented with 10% heated inactivated fetal bovine
serum (FBS), 100 U/ml of penicillin G and 100 µg/ml
streptomycin. Cells were incubated at 37°C in a 5% CO2
incubator.
Cell proliferation and clonogenic
assays
Cell proliferation was determined using the dye
exclusion method. Briefly, exponentially growing cells
(5×103 cells/well) were plated into a 24-well plate.
Every other day, the cells were trypsinized and viable cells were
determined by trypan blue staining. Doubling times of each cell
line were calculated as described by Patterson (10).
Survival and proliferative activities were
determined using a clonogenic assay (11). Briefly, 200 cells in Ham's F-12 with
10% FBS were seeded in 6-well plates and incubated in a 5%
CO2 incubator at 37°C for 7 days. Colonies were fixed in
4% v/v paraformaldehyde, stained with 0.5% w/v crystal violet and
counted.
Cell surface marker determination
The expression levels of cell surface markers were
determined by flow cytometric analyses with phycoerythrin
(PE)-conjugated anti-CD29 (TS2/16; BioLegend, San Diego, CA, USA),
allophycocyanin (APC)-conjugated anti-CD34 (AC136; Miltenyi Biotec,
Teterow, Germany), fluoresecein isothiocyanate-conjugated (FITC)
anti-CD44 (IM7), Alexa Fluor® 647-conjugated anti-CD90
(F15-42-1), APC-conjugated anti-CD117 (104D2) (all from BioLegend)
and PE-conjugated anti-CD133 (AC133; Miltenyi Biotec). Single-cell
suspensions of ~1×106 cells were prepared in
phosphate-buffered saline (PBS) containing 3% FBS and 0.05% sodium
azide. After staining with each antibody and incubation for 30 min
on ice, the cells were washed twice with PBS and analyzed on an LSR
II flow cytometer (BD Biosciences, San Jose, CA, USA). The
expression levels of each surface marker were analyzed from at
least 20,000 cells using FlowJo software (Tree Star, San Carlos,
CA, USA) and presented as a percentage of positive cells.
Isotype-matched antibodies were used as controls.
Wound healing assay
Cells were seeded into a 12-well plate and incubated
overnight in complete media. A sterile 200-µl pipette tip
was used to create a scratch wound on the monolayer of cells and
the detached cells were removed. Cells were then incubated further
in 1% FBS media, and images were captured at 6, 12, 18 and 24 h.
The migrating distance was measured as the relative distance of
cell migration measured from 0 time as previously described
(12).
In vitro cell migration and invasion
assays
The migration and Matrigel cell invasion assays were
performed as previously described (12). Briefly, polycarbonate membranes with
8-µm pores were coated with 0.5 mg/ml Matrigel (BD
Biosciences) and allowed to stand overnight. The membranes were
rehydrated and 1.2×104 of CCA cells were placed onto the
upper chamber of a Transwell unit. Ham's F-12 media with 10% FBS
was used as a chemoattractant in the bottom chamber. After 18 h, 4%
paraformaldehyde was added to fix the migrated or invaded cells
before the cells were stained with 0.4% sulforhodamine B. The
membranes were dried and the numbers of migrated/invaded cells from
9 × low power fields (magnification, ×100) were counted. Mean
values were determined from triplicated assays. At least two
independent experiments were performed.
Tumor growth assay in xenografted
mice
KKU-213 or KKU-213L5 cells (1×106 cells)
were subcutaneously injected in each flank side of the 8- to
10-week old male BALB/c Rag-2−/−/Jak3−/− mice
(n=8). Tumor volumes were measured using a vernier every 3 days.
Tumors were removed and weighed at 12 days post-injection.
Pulmonary metastasis assay in vivo
NOJ mice aged 8–10 weeks were used to estimate the
in vivo metastatic potential to the lung. KKU-213 or
KKU-213L5 cells (5×105 cells/mouse) were intravenously
inoculated in mice via the tail vein. Lungs were removed on days 13
and 21 after inoculation, and fixed in 4% paraformladehyde
overnight for histochemical study. The total number of lung
micrometastases and macrometastases were microscopically evaluated.
Cell clusters with a diameter <100 µm were defined as
micrometastases and those of diameter >100 µm were
defined as macrometastases. For survival analysis, the survival of
mice was observed until 40 days.
Histological evaluations
The paraffin sections were prepared for histological
study according to the standard protocol. The sections were stained
with hematoxylin and eosin, or CK19 antibody for
immunohistochemistry. For CK19-immunohistochemistry, tissue
sections were incubated with anti-KRT19 (HPA002465; Sigma-Aldrich,
St. Louis, MO, USA) at 4°C overnight followed by biotinylated goat
anti-rabbit IgG (Vector Laboratories, Burlingame, CA, USA) at room
temperature for 30 min. Signals were enhanced using the Vectastain
Elite ABC standard kit (Vector Laboratories) and detection was
performed using the Histofine® DAB substrate kit
(Nichirei Biosciences, Inc., Tokyo, Japan).
AGR2 and KiSS-1 immunohistochemistry was performed
in 32 intrahepatic mass-forming CCA patient tissues using 1:500
rabbit polyclonal anti-AGR2 antibody (Abcam, Cambridge, MA, USA)
and 1:50 rabbit polyclonal anti-KiSS-1 antibody (Santa Cruz
Biotechnology, Dallas, TX, USA) at room temperature, overnight.
Anti-rabbit IgG EnVison (Dako, Carpinteria, CA, USA) was used as
the secondary antibody. The percentage of positive cells was scored
as follows: 0%, negative; 1–25%, +1; 26–50%, +2; and >50%, +3;
and the intensity of the staining as follows: weak, 1; moderate, 2;
and strong, 3. The expression levels of AGR2 and KiSS-1 were
semi-quantitatively determined using an immunohistochemistry (IHC)
index equal to the percentage of positive cells × intensity.
Real-time RT-PCR array
A real-time PCR array of 79 genes including 77
metastatic-associated genes and two internal control genes [β-actin
and β2 microglobuli (B2M)] was set (Table I). The metastatic-associated genes
were selected from the literature review, a commercial metastatic
PCR array, and the 'Epithelial mesenchymal transition gene
signatures in cancer drug discovery, diagnostics and treatment̓
(13). The primer sequences were
designed using Vector NTI (Thermo Fisher Scientific, Waltham, MA,
USA).
 | Table IList of 77 genes selected for the
metastatic PCR array. |
Table I
List of 77 genes selected for the
metastatic PCR array.
| No. | Gene symbol | Gene name | Accession no. |
|---|
| 1 | ACTN1 | Actinin, α1 | NM_001102.3 |
| 2 | AGR2 | Anterior gradient 2
homolog (Xenopus laevis) | NM_006408.3 |
| 3 | AKT1 | v-akt murine
thymoma viral oncogene homolog 1 | NM_001014431.1 |
| 4 | ANXA1 | Annexin A1 | NM_000700.1 |
| 5 | BSG | Basigin (OK blood
group) | NM_001728.3 |
| 6 | CAV1 | Caveolin 1 | NM_001172895.1 |
| 7 | CCND2 | Cyclin D2 | NM_001759.3 |
| 8 | CCNE2 | Cyclin E2 | NM_057749.2 |
| 9 | CD44 | CD44 molecule
(Indian blood group) | NM_000610.3 |
| 10 | CD82 | CD82 molecule | NM_001024844.1 |
| 11 | CDH1 | Cadherin 1, type 1,
E-cadherin (epithelial) | NM_004360.3 |
| 12 | CDH2 | Cadherin 2, type 1,
N-cadherin (neuronal) | NM_001792.3 |
| 13 | CFL1 | Cofilin 1
(non-muscle) | NM_005507.2 |
| 14 | CLDN4 | Claudin 4 | NM_001305.3 |
| 15 | CLDN7 | Claudin 7 | NM_001185022.1 |
| 16 | CST7 | Cystatin F
(leukocystatin) | NM_003650.3 |
| 17 | CTBP1 | C-terminal binding
protein 1 | NM_001012614.1 |
| 18 | CTNNB1 | Catenin
(cadherin-associated protein), β1 | NM_001904.3 |
| 19 | DSP | Desmoplakin | NM_001008844.1 |
| 20 | EGFR | Epidermal growth
factor receptor | NM_005228.3 |
| 21 | EPCAM | Epithelial cell
adhesion molecule | NM_002354.2 |
| 22 | ERBB3 | v-erb-b2
erythroblastic leukemia viral oncogene Homolog 3 (avian) | NM_001005915.1 |
| 23 | EREG | Epiregulin | NM_001432.2 |
| 24 | FLT4 | fms-related
tyrosine kinase 4 | NM_002020.4 |
| 25 | FN1 | Fibronectin 1 | NM_002026.2 |
| 26 | FOXC1 | Forkhead box
C1 | NM_001453.2 |
| 27 | GAPDH |
Glyceraldehyde-3-phosphate
dehydrogenase | NM_002046.4 |
| 28 | HOXB7 | Homeobox B7 | NM_004502.3 |
| 29 | HSPB2 | Heat shock 27 kDa
protein 2 | NM_001541.3 |
| 30 | ICAM1 | Intercellular
adhesion molecule 1 | NM_000201.2 |
| 31 | IFI27 | Interferon,
α-inducible protein 27 | NM_001130080.1 |
| 32 | ILK | Integrin-linked
kinase | NM_001014794.1 |
| 33 | ITGA5 | Integrin, α5 | NM_002205.2 |
| 34 | ITGB1 | Integrin, β1 | NM_002211.3 |
| 35 | JAG1 | Jagged 1 | NM_000214.2 |
| 36 | KISS1 | KiSS-1
metastasis-suppressor | NM_002256.3 |
| 37 | KRT13 | Keratin 13 | NM_002274.3 |
| 38 | LDHA | Lactate
dehydrogenase A | NM_005566.3 |
| 39 | METAP2 | Methionyl
aminopeptidase 2 | NM_006838.3 |
| 40 | MMP3 | Matrix
metallopeptidase 3 (stromelysin 1, progelatinase) | NM_002422.3 |
| 41 | MMP7 | Matrix
metallopeptidase 7 (matrilysin, uterine) | NM_002423.3 |
| 42 | MMP9 | Matrix
metallopeptidase 9 (gelatinase B, 92 kDa gelatinase, 92 kDa type IV
collagenase) | NM_004994.2 |
| 43 | MSN | Moesin | NM_002444.2 |
| 44 | NF2 | Neurofibromin 2
(merlin) | NM_000268.3 |
| 45 | NME1 | NME/NM23 nucleoside
diphosphate kinase 1 | NM_000269.2 |
| 46 | OCLN | Occludin | NM_001205254.1 |
| 47 | PGK1 | Phosphoglycerate
kinase 1 | NM_000291.3 |
| 48 | PLAUR | Plasminogen
activator, urokinase receptor | NM_001005376.2 |
| 49 | PTEN | Phosphatase and
tensin homolog | NM_000314.4 |
| 50 | PTK2 | Protein tyrosine
kinase 2 | NM_001199649.1 |
| 51 | PXN | Paxillin | NM_001080855.2 |
| 52 | RAC1 | Ras-related C3
botulinumtoxin substrate 1 (Rho family, small GTP binding protein
Rac1) | NM_006908.4 |
| 53 | RAC2 | Ras-related C3
botulinumtoxin substrate 2 (Rho family, small GTP binding protein
Rac2) | NM_002872.3 |
| 54 | REG1A | Regenerating
islet-derived 1α | NM_002909.4 |
| 55 | RHO | Rhodopsin | NM_000539.3 |
| 56 | RHOA | Ras homolog family
member A | NM_001664.2 |
| 57 | RHOB | Ras homolog family
member B | NM_004040.2 |
| 58 | RHOC | Ras homolog family
member C | NM_001042678.1 |
| 59 | ROCK1 | Rho-associated,
coiled-coil containing protein kinase 1 | NM_005406.2 |
| 60 | S100P | S100 calcium
binding protein P | NM_005980.2 |
| 61 | SERPINE1 | Serpin peptidase
inhibitor, clade E (nexin, plasminogen activator inhibitor type 1),
member 1 | NM_000602.4 |
| 62 | SNAI1 | Snail family zinc
finger 1 | NM_005985.3 |
| 63 | SNAI2 | Snail family zinc
finger 2 | NM_003068.4 |
| 64 | SPP1 | Secreted
phosphoprotein 1 | NM_001251830 |
| 65 | STAT3 | Signal transducer
and activator of transcription 3 (acute-phase response factor) | NM_003150.3 |
| 66 | TACSTD2 | Tumor-associated
calcium signal transducer 2 | NM_002353.2 |
| 67 | TGFB1 | Transforming growth
factor, β1 | NM_000660.4 |
| 68 | TIMP1 | TIMP
metallopeptidase inhibitor 1 | NM_003254.2 |
| 69 | TIMP2 | TIMP
metallopeptidase inhibitor 2 | NM_003255.4 |
| 70 | TM4SF1 | Transmembrane 4 L
six family member 1 | NM_014220.2 |
| 71 | TOB1 | Transducer of
ERBB2, 1 | NM_005749 |
| 72. | TWIST1 | Twist basic
helix-loop-helix transcription factor 1 | NM_000474.3 |
| 73 | VCAN | Versican | NM_004385 |
| 74 | VCL | Vinculin | NM_003373.3 |
| 75 | VEGFA | Vascular
endothelial growth factor A | NM_001025366.2 |
| 76 | VIM | Vimentin | NM_003380.3 |
| 77 | ZEB1 | Zinc finger E-box
binding homeobox 1 | NM_001128128.2 |
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was
used to extract total RNA from cells, and reverse transcription of
total RNA (2 µg) was performed using High Capacity cDNA
Reverse Transcription kits (Applied Biosystems, Foster City, CA,
USA). Quantitative real-time PCR, the PCR protocol and data
analysis were as described by Uthaisar et al (12). β2M was used as an
internal control.
Statistical analysis
Data were analyzed using SPSS 16.0 Windows
Evaluation software (SPSS, Inc., Chicago, IL, USA). Quantitative
data are expressed as mean ± SD. Student's t-test was used for
comparison between two groups. A P-value of <0.05 was considered
to indicate a statistically significant result.
Results
Properties of the highly metastatic
subline KKU-213L5 in vitro
Morphology and growth properties of
the cultured cells
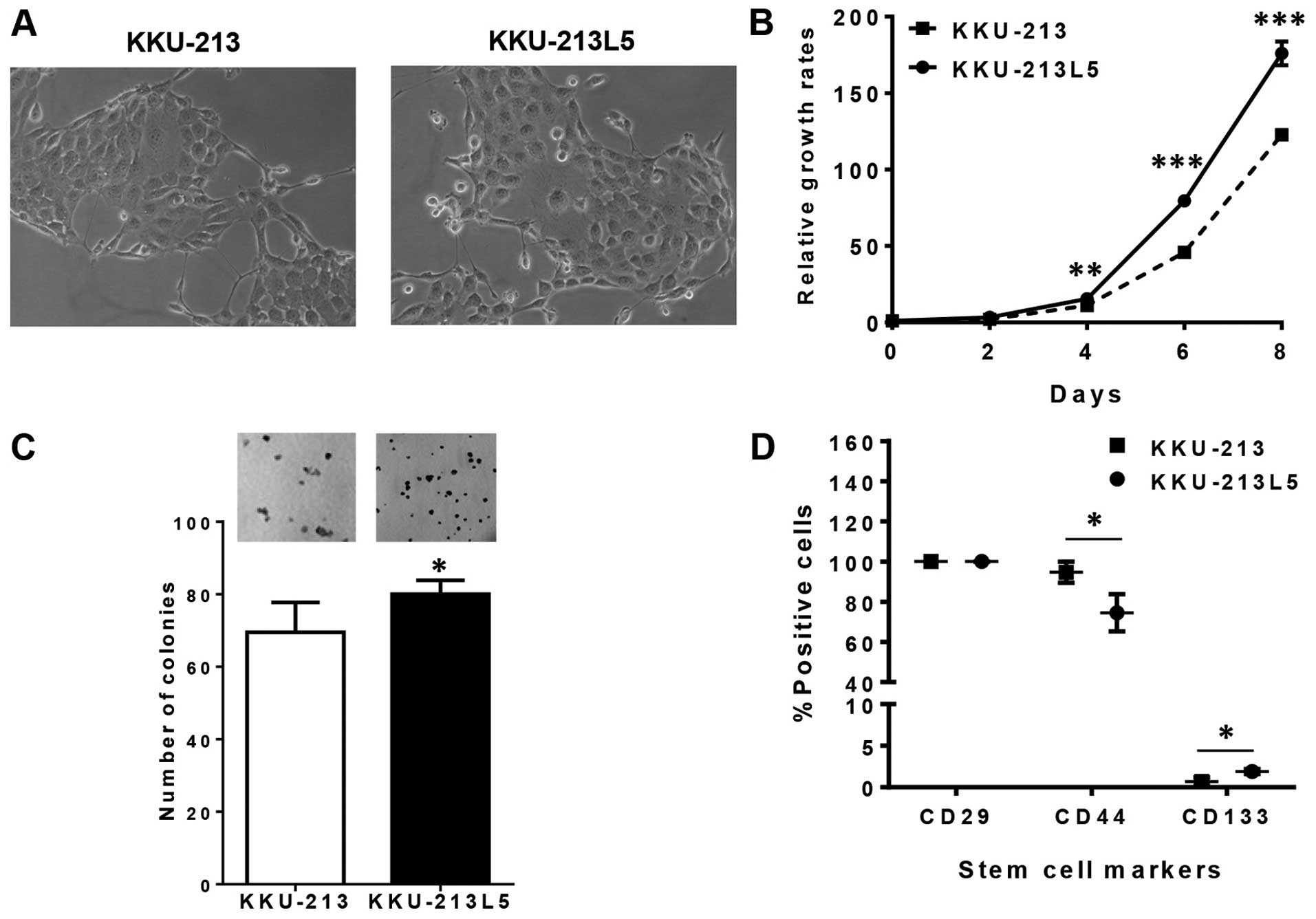
The culture appearance of the established KKU-213L5
cell subline and the parental KKU-213 cell line as determined by
phase contrast microscopy demonstrated considerably similar
epithelial morphology with a polygonal shape, and large nuclei
(Fig. 1A). The growth rate of the
KKU-213L5 subline exceeded that of the parental KKU-213 cells
(Fig. 1B). The average doubling
time for KKU-213 was 23.4 h and that for the metastatic KKU-213L5
was 20.3 h. The clonogenic assay that represents survival and
proliferative activities revealed that KKU-213L5 cells had a
significantly higher colony forming capability than that of the
KKU-213 cells (Fig. 1C).
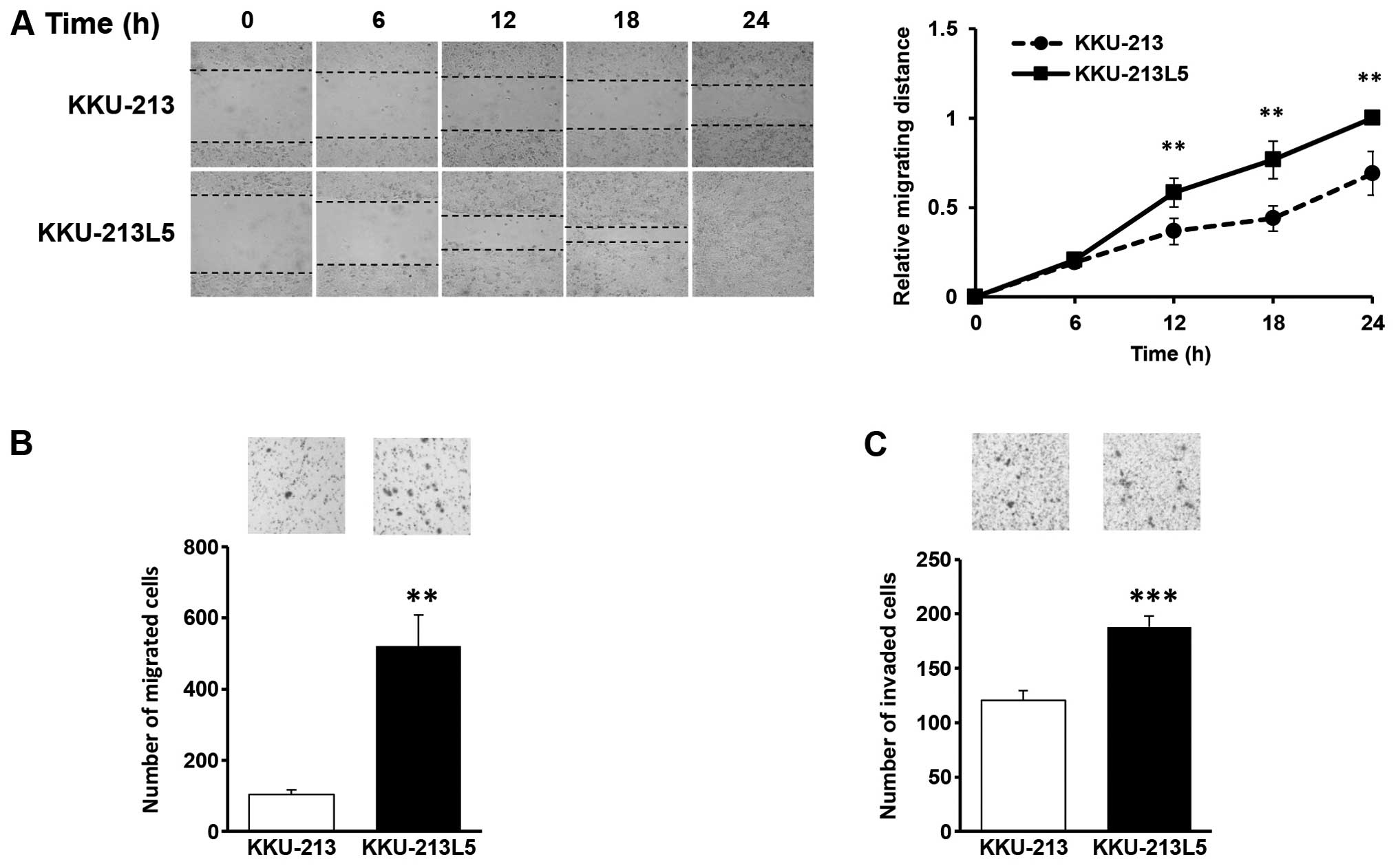
Migration and invasion activities
Migration and invasion are the most important
phenotypic characteristics of cancer metastasis. To examine the
ability of migration, cell scratching assays were performed and
compared between the KKU-213 and KKU-213L5 cells. Following
incubation of physically wounded cells at indicated time points,
the relative migrating distance of KKU-213L5 was significantly
longer than that of KKU-213 (P<0.001; Fig. 2A). The migration activities using
Boyden chamber assays demonstrated that KKU-213L5 cells had a
~5-fold higher migration ability than the parental line (P<0.01)
(Fig. 2B). The number of migrated
cells observed was 521±87 for KKU-213L5 and 104±11 for KKU-213.
Invasion is a crucial step in metastasis; the
invasion ability of the KKU-213L5 cells was compared to that of
KKU-213 cells using the Matrigel invasion assay. As shown in
Fig. 2C, KKU-213L5 had an
aggressive capacity to invade the matrix compared to the parental
cells (P<0.001). The number of invaded cells for KKU-213L5 was
188±9 and for KKU-213 the number was 120±8.
Differential expression of stem cell
markers
Emerging roles of cancer stem cells (CSCs) in
tumorigenesis, growth and metastasis have been recognized (14). The high plasticity and
stress-resistant phenotypes of CSCs make them potential candidates
for metastatic cells. It was speculated that the in vivo
selection of KKU-213 cells may preferentially select CSCs in
KKU-213L5. Six CSC markers (CD29, CD34, CD44, CD90, CD117 and
CD133) from common liver stem cell markers (15) were investigated in the parental
KKU-213 and highly metastatic KKU-213L5 cells. Only 3 markers;
CD29, CD44 and CD133, were expressed on the surface of these CCA
cells and only CD44 and CD133 were differentially expressed
(Fig. 1D). As compared to KKU-213,
the expression of CD133 was increased (1.9±0.3 vs. 0.7±0.5;
P<0.05) whereas that of CD44 was decreased (74.5±9.3 vs.
94.7±5.2; P<0.05) in the KKU-213L5 cells.
Properties of the highly metastatic
subline KKU-213L5 in vivo
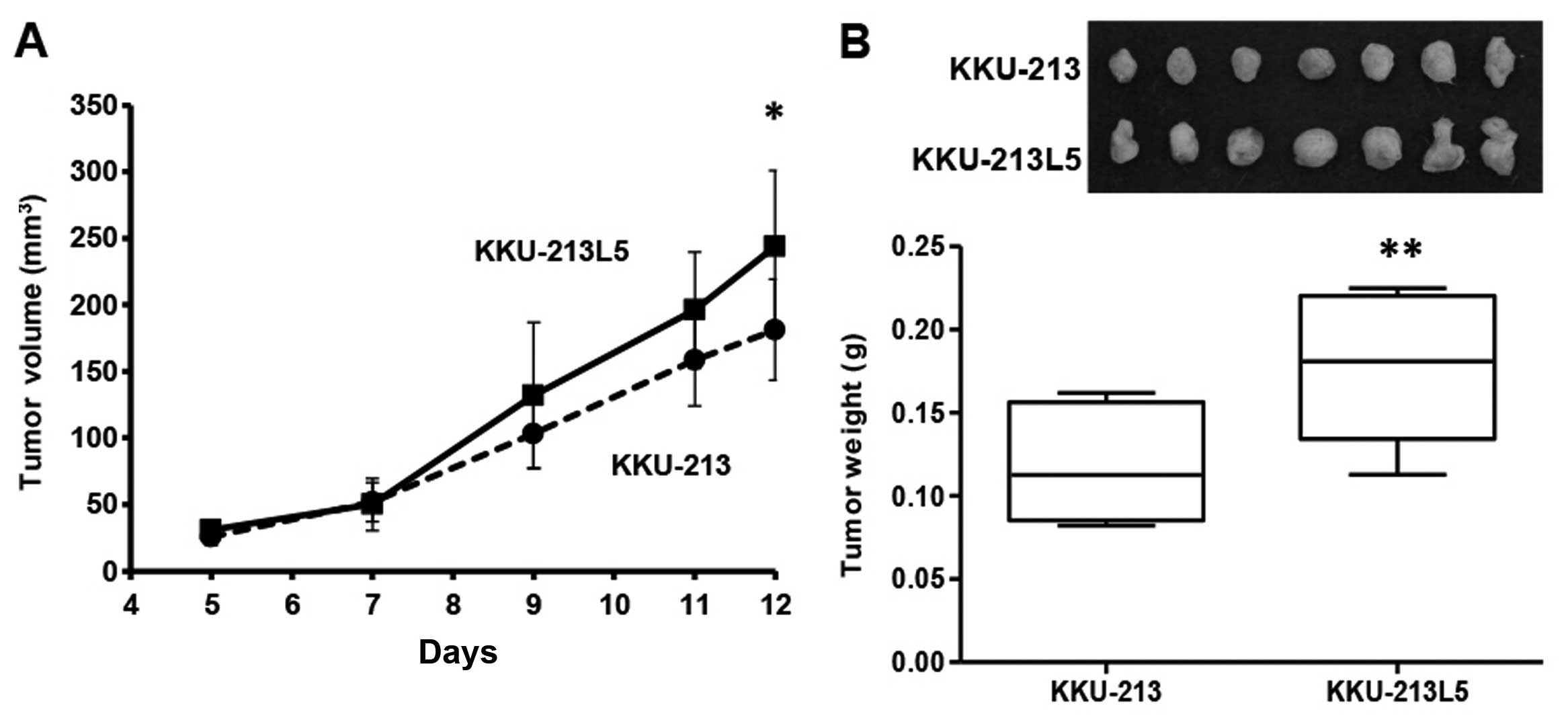
Tumorigenicity
Both KKU-213 and KKU-213L5 cells were tumorigenic as
the sizes of inoculums were progressively increased and reached
sizes of 0.1–0.25 g within 12 days. The tumor growths of the
KKU-213L5 subline, however, exceeded those of the parental KKU-213
cells both in tumor volume (Fig.
3A) and tumor weight (Fig. 3B).
At 12 days post-inoculation, the average tumor weight of the
KKU-213L5 cells (0.18±0.04 g) was significantly higher than that of
the parental KKU-213 cells (0.11±0.03 g; P<0.01). Distant organ
metastases, e.g., lung and liver were not observed in this period
of time.
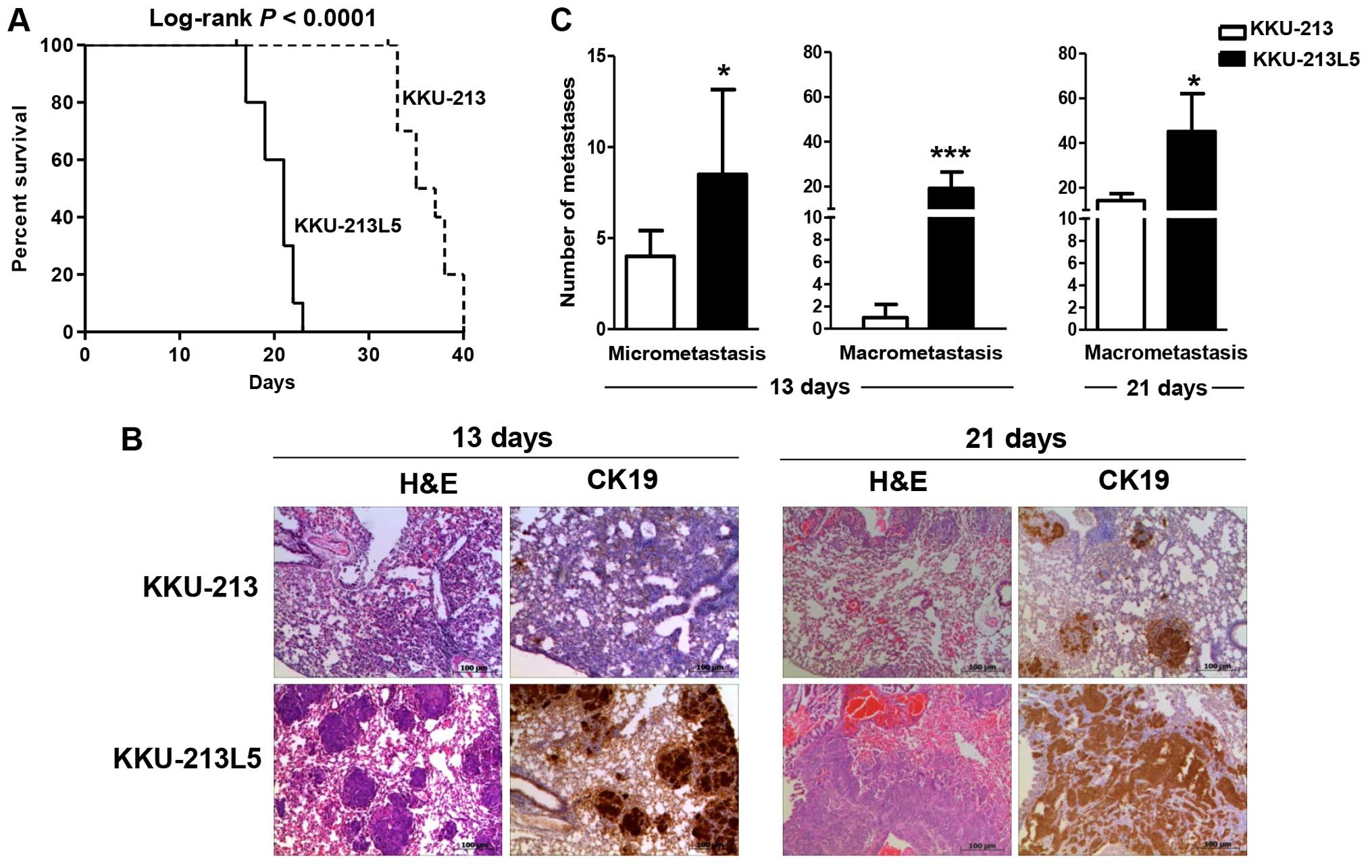
Metastatic activity
The aggressiveness of the metastatic KKU-213L5 cells
over the parental cells was determined by the percent survival of
the mice after tail-vein injection (Fig. 4A). Mice injected with KKU-213L5 had
shorter survival with a median survival of 21 days compared to 36
days of the parental cell group (log-rank; P<0.0001). The
KKU-213L5 cells developed lung metastases with high efficiency in
the animal model. After 13 and 21 days post intravenous injection,
the CCA cells that were colonized in the lungs were examined using
cytokeratin-19 (CK19) immunohistochemistry (Fig. 4B). KKU-213L5-injected mice
significantly developed numerous lung colonizations with
micrometastatic and macrometastatic foci within 13 days of
injection, whereas the metastatic foci were hardly detected in the
KKU-213 cell group (Fig. 4C;
P<0.001). Macrometastases were observed from the KKU-213 cells
at 21 days post-injection but the number was considerably fewer
than those observed for the KKU-213L5 cells (P<0.05).
Identification of metastasis-related
genes in KKU-213L5
Cancer metastasis is a complicated process supported
by abnormal expression of metastasis-associated genes. To
understand the molecular mechanisms supporting the highly
metastatic activities of KKU-213L5, differential expression levels
of 77 metastatic-associated genes regarding cell proliferation,
cell migration and cell invasion (Table
I) were determined in the KKU-213L5 cells in comparison with
the KKU-213 cells using real-time PCR.
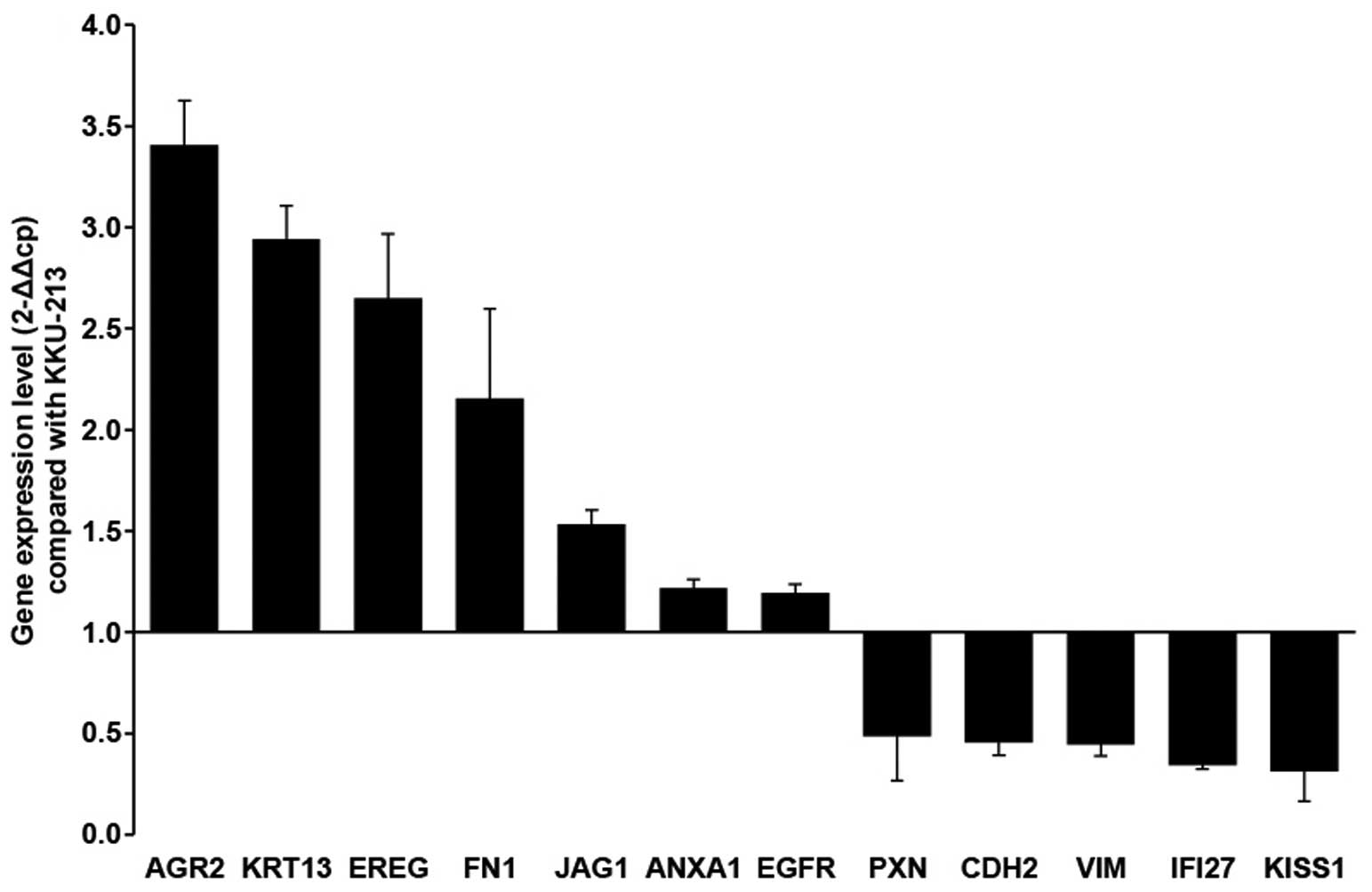
To increase the chance for uncovering the
metastatic-associated genes, the expression levels of genes that
were >1.2-fold different were designated as upregulated and
those <0.5-fold were designated as downregulated. Cluster
analysis (dChip) of the metastatic-associated genes revealed 21
differentially expressed genes in KKU-213L5 with 7 upregulated and
14 downregulated. AGR2, KRT13, EREG and FN1 were upregulated with a
>2-fold difference; whereas JAG1, ANXA1 and EGFR3 had a
>1.2-fold difference. In contrast, KiSS-1, IFI27, VIM, CDH2 and
PXN were downregulated with a <0.5-fold difference (Fig. 5; Table
II). Using STRING9.1 (http://string-db.org), these differentially expressed
genes were suggested to be involved in wound healing and cellular
component movement.
 | Table IIUpregulated and downregulated genes
in the KKU-213L5 cells compared with the KKU-213 cells. |
Table II
Upregulated and downregulated genes
in the KKU-213L5 cells compared with the KKU-213 cells.
| No. | Gene symbol | Gene name | Fold-change | P-value |
|---|
| Upregulated
genes |
| 1 | AGR2 | Anterior gradient 2
homolog (Xenopus laevis) | 3.41 | 1.00E-08 |
| 2 | KRT13 | Keratin 13 | 2.94 | 1.60E-07 |
| 3 | EREG | Epiregulin | 2.65 | 2.75E-06 |
| 4 | FN1 | Fibronectin 1 | 2.15 | 3.72E-02 |
| 5 | JAG1 | Jagged 1 | 1.53 | 4.59E-03 |
| 6 | ANXA1 | Annexin A1 | 1.22 | 3.34E-05 |
| 7 | EGFR | Epidermal growth
factor receptor | 1.20 | 1.92E-03 |
| Downregulated
genes |
| 1 | KISS1 | KiSS-1
metastasis-suppressor | 0.31 | 3.30E-02 |
| 2 | IFI27 | Interferon,
α-inducible protein 27 | 0.34 | 9.21E-06 |
| 3 | VIM | Vimentin | 0.45 | 4.85E-06 |
| 4 | CDH2 | Cadherin 2, type 1,
N-cadherin (neuronal) | 0.46 | 5.45E-03 |
| 5 | PXN | Paxillin | 0.49 | 7.97E-03 |
Expression levels of AGR2 and KiSS-1
are associated with metastatic potential of KKU-213L5 cells
As shown in Fig. 5,
AGR2 was in the first rank of upregulated genes and KiSS-1, a
metastasis suppressor was in the first rank of downregulated genes
found in the KKU-213L5 cells when compared to those of the parental
KKU-213 cells. To validate the association of these two genes in
the metastasis of CCA, the expression levels of AGR2 and KiSS-1
were confirmed in clinical specimens from CCA patients with
different metastatic statuses. Paraffin-embedded tumor tissues from
histologically confirmed intrahepatic CCA patients with different
tumor staging (n=32) were examined for AGR2 and KiSS-1
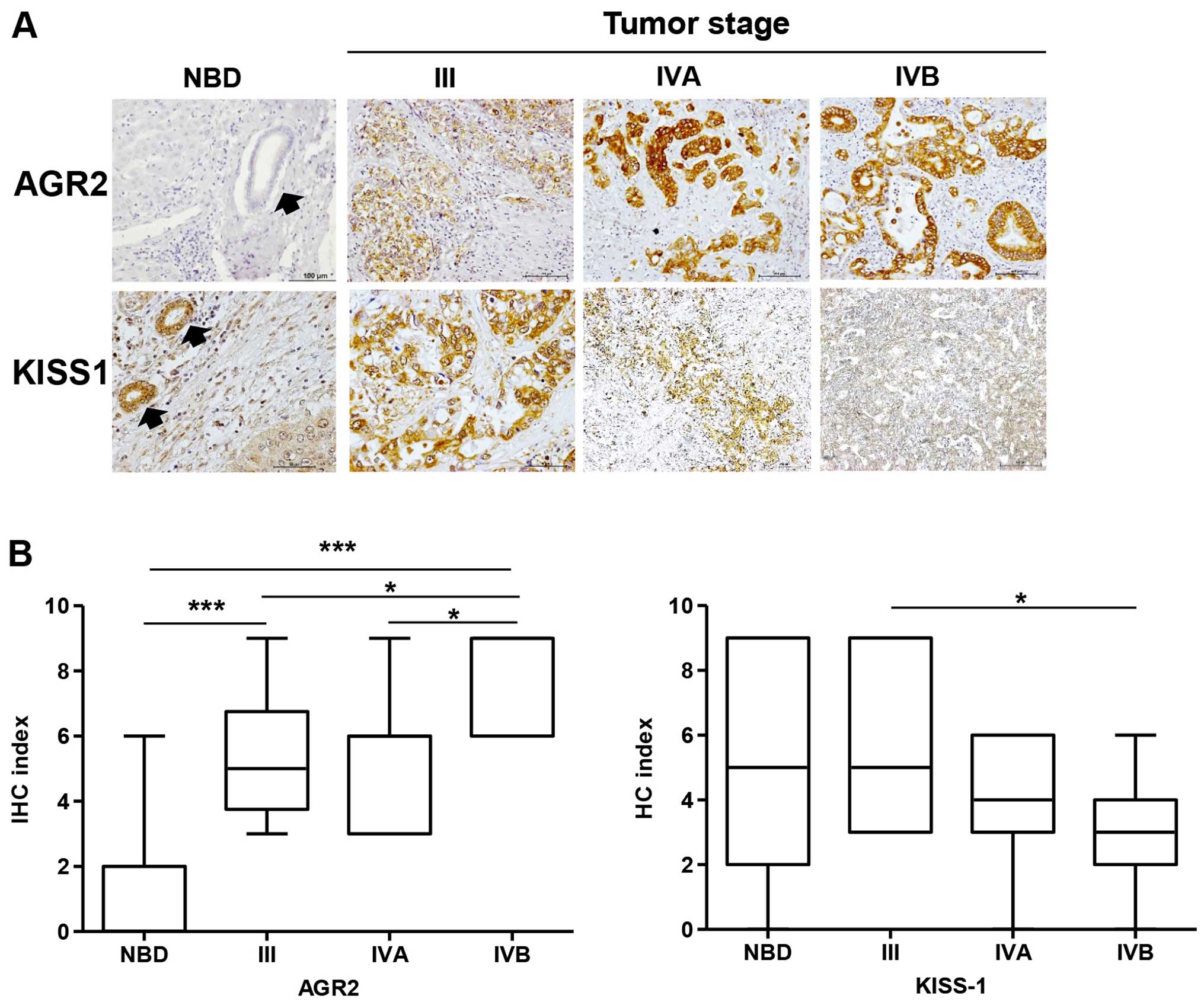
immunohistochemical analysis. Normal bile duct epithelia from the
adjacent non-tumorous tissues were assessed. Normal bile ducts
occasionally expressed AGR2, however, it was frequently found in
CCA tissues with strong intensity. The expression of AGR2 was
significantly higher in the CCA tissues than that in the normal
bile ducts (Fig. 6A and B)
(P<0.001). Moreover, AGR2 expression was gradually increased
with higher stages of tumor development. Expression of AGR2 in the
CCA tissues of stage IVB was significantly higher than these levels
in stages IVA and III (P<0.05). In contrast to AGR2, KiSS-1 was
strongly expressed in normal bile ducts and gradually decreased
with increasing tumor stages (Fig. 6A
and B) (P<0.05).
Discussion
Metastasis is the major cause of cancer morbidity
and mortality. The complex nature of metastasis dictates the
necessity for experimental systems that more closely simulate
endogenous contributing factors. In the present study, we report
the establishment and comparative characterization of a novel
highly metastatic subline, designated KKU-213L5 and its parental,
KKU-213 cell line. The cell lines were compared in vitro for
cell proliferation, stem cell markers, migratory and invasive
abilities. The xenografted and tail vein metastatic mouse models
were used to compare tumorigenicity and lung metastatic capability.
Furthermore, AGR2 and KiSS-1 were first identified and shown to be
associated with the metastatic status of the CCA patients.
The present search from 1985–2016 revealed that
there were 21 CCA established cell lines from 16 reports, 3 of
which were established from Thai CCA patients which were assumed to
be associated with liver fluke infections: HuCCA-1 (16), KKU-100 (8) and RMCCA-1 (17). Similar to other solid tumors, CCA is
characterized as a highly metastatic cancer, and the lung is the
common site for metastasis (18). A
greater understanding of the biology of pulmonary metastases is
needed to improve treatment outcomes. There are several models of
highly metastatic cell lines established in the mouse with varied
advantages and disadvantages to each. The present study used the
tail-vein metastasis model which involves the injection of tumor
cells via tail vein and analysis of their ability to form tumors
and/or to colonize at distant sites e.g. lung via hematogenous
spread. This model generates lung colonization of CCA without other
distant colonization sites. The pitfall of this assay is the lack
of measurement of the earlier invasive and angiogenic stages of
malignant progression, and therefore may be an incomplete measure
of the metastatic process.
Cell morphology of the KKU-213L5 cells was not
obviously different from that of the parental cells, however, the
KKU-213L5 cells demonstrated a significant increased rate of
proliferation, migration and invasion compared to the KKU-213
cells. LM8, a lung metastatic osteosarcoma cell line, obtained from
murine Dunn osteosarcoma using a similar mouse protocol also had a
higher growth rate than its parental cell line (19). Higher growth rate, migration and
invasion than the parental cells seem to be distinct
characteristics of highly metastatic cells as shown in the present
study and previous studies (19–22).
As it is well-known, cancer stem cells which represent a relatively
tiny portion of the tumor are responsible for building a cancer
mass and enhancing metastatic potential (14). In the present study, cells that
possessed the surface stem cell markers were enriched in the
KKU-213L5 cells containing considerably more of an apparent
CD133-positive subpopulation than the KKU-213 cells. The
association of a subgroup of hepatocellular carcinoma cells with
CD133+CD44+ and hematogenous metastasis was
reported (23). In addition, the
correlation of high CD133 expression in CCA patient tissues with
poor prognosis, with the tubular subtype, a higher tumor stage
(T4), and shorter survival of these Thai CCA patients was
previously reported (24). It is
worth noting that the expression of CD44 appeared to be lower in
the KKU-213L5 cells. CD44 is a pleomorphic protein and it has been
shown that only specific variants of CD44 promote tumorigenesis
(25). Altogether, these data
support the hypothesis that the CSC population may confer higher
metastatic activities to KKU-213L5.
The aggressiveness of the KKU-213L5 and KKU-213
cells was also demonstrated in the in vivo mouse models. The
tumorigenicity and metastatic potential of KKU-213L5 were
significantly higher than those of the KKU-213 cells as shown in
the subcutaneous xenografted and tail-vein metastatic mouse models.
The greater aggressiveness of KKU-213L5 was more obvious in the
metastatic mouse model, by the fact that spontaneous pulmonary
metastasis was significantly observed by determining KKU-213L5 >
KKU-213. A few micrometastases were detected in the KKU-213 group
13 days post-injection, whereas at the same time period, a greater
number of micrometastasis and macrometastasis (×20-fold) were found
in lung tissues of the KKU-213L5-injected mice. The greater
pulmonary metastasis of the KKU-213L5 group may be the cause of the
significantly shorter survival in all KKU-213L5-injected mice when
compared with the KKU-213-injected mice.
Metastasis requires functionally distinct sets of
genes that provide metastatic initiation and progression. Cell
migration and invasion are essential for early events in the
metastatic process (26,27); therefore, attention was focused on a
small group of potentially important genes related to cell motility
and invasion. The differential expression of 77 selected genes
analyzed between KKU-213L5 and KKU-213 revealed significant
upregulation of AGR2 and downregulation of KiSS-1. These two genes
exhibited the highest differential expression levels between the
two cell lines and have never been previously described in CCA
metastasis. Since the differences in the expression may or may not
account for the highly metastatic potential of KKU-213L5 cells,
AGR2 and KiSS-1 were further investigated for their association
with metastasis in CCA patient tissues.
The metastasis-associated genes retrieved from the
KKU-213L5 cells can reflect the metastasis involvement of these
genes in CCA patients. The expression levels of AGR2 and KiSS-1
were verified in CCA patient tissues using immunohistochemistry.
The potential relevance of AGR2 and KiSS-1 in metastasis of
intrahepatic mass-forming CCA is suggested. AGR2 was rarely
expressed in normal bile duct epithelia, but was increased
gradually according to the progression of the metastatic stage. The
significant increase in AGR2 expression and concomitant tumor stage
progression may imply the association of AGR2 in the metastasis of
CCA.
AGR2 belongs to a family of protein disulfide
isomerases (PDIs). The secreted form of AGR2 has been shown to
function at the cell surface and the extracellular matrix (28,29).
Overexpression of AGR2 has been reported in various types of
cancers and has been intensively studied in breast cancer.
Overexpression of AGR2 was found to be related to the increase in
cell survival and proliferation, whereas loss of AGR2 leads to a
decrease in cell cycle progression and cell death (30–32).
In addition, AGR2 was shown to promote metastasis of breast
epithelial cells in an in vivo metastasis assay (33) and the secreted AGR2 plays roles in
cellular adhesion and dissemination of metastatic tumor cells
(34). Therefore, inhibition of
AGR2 was suggested to be useful in the targeted therapy of breast
cancer.
There are only two previous studies on AGR2 in CCA.
The expression of AGR2 in normal bile duct epithelia and CCA from
these studies is still controversial. Lepreux et al
(35) observed AGR2 only in the
peribiliary glands and the tall epithelial cells of hilar large
bile ducts, while Kim et al (36) found general expression of AGR2 in
the epithelia of intrahepatic and extrahepatic biliary tracts.
Moreover, from Lepreux's study, only 21% (3/14) of intrahepatic CCA
expressed AGR2, whereas those from Kim's study, 82% (9/11) were
positive for AGR2. In the present study, upregulation of AGR2
expression in highly metastatic CCA cells and tissues was noted.
Expression of AGR2 in advanced stage CCA was higher than that of
earlier stage CCA. The discrepancy of AGR2 expression in CCA is
varied among different study cohorts. Determination of AGR2
expression in association with clinicopathological information of
patients in a larger sample size should be carefully conducted.
KiSS-1 is a member of the metastatic suppressors
which can block metastasis without preventing primary tumor
development. KiSS-1 effectively inhibits colonization of tumor
cells, the last step of the metastatic cascade. Until the present
search, there is no report on KiSS-1 in CCA. The role of KiSS-1 as
a metastatic suppressor in CCA is emphasized in the present study
by the fact that KiSS-1 expression was downregulated in the highly
metastatic KKU-213L5 cells and was also decreased in the advanced
stage CCA patient tissues. This observation agreed with clinical
observations from various cancer patients. Immunohistochemistry
studies of KiSS-1 in ovarian cancer revealed that the positivity of
KiSS-1 was individually correlated with favorable prognosis and
superior overall survival (37,38).
Low KiSS-1 expression was found to be correlated with metastases
and poor survival of patients with gastrointestinal (39,40)
and pancreatic cancers (41,42).
In contrast, the antimetastatic role of KiSS-1 in breast cancer is
still controversial. Loss of KiSS-1 expression was found to be
related to positive lymph nodes in breast adenocarcinomas (43) and brain metastases (44). Moreover, patients with high KiSS-1
expression exhibited shorter disease-free survival (45).
In conclusion, understanding the molecular
mechanisms of CCA progression and metastasis is essential for
improving the prevention and ensuring the effective control of the
metastasis of CCA. To the best of our knowledge, KKU-213L5 is the
first CCA cell line with relatively high metastatic activity, not
only in vitro but also in vivo. The
metastasis-associated genes retrieved from KKU-213L5 can reflect
the metastasis involvement of these genes in CCA patients. It is
likely that differential expression of genes between the parental
KKU-213 and the highly metastatic KKU-213L5 cells may be valuable
to discover novel genes that control metastasis. This cell line
will be an excellent tool for further dissection of the molecular
mechanisms underlying the metastasis of CCA as well as an
appropriate model for studying inhibitory agents against metastasis
and the treatment of CCA.
Acknowledgments
The present study was co-supported by Joint Research
Grants from Khon Kaen University, the TRF Senior Research Scholar
Grant, Thailand Research Fund to S.W. (RTA5780012), the Higher
Education Research Promotion and National Research University
Project of Thailand, Office of the Higher Education Commission,
through the Health Cluster to S.W. (NRU58-2014), the Khon Kaen
University Research Fund to K.V. (KKU59-2403), the Joint Funding
through Royal Golden Jubilee Ph.D. Program and Khon Kaen University
to K.U. and S.W. (PHD/0211/2550), and Japan Student Services
Organization to K.U. and S.O. We would like to acknowledge
Professor James A. Will for editing the manuscript via Publication
Clinic KKU, Thailand.
References
|
1
|
Fidler IJ: The pathogenesis of cancer
metastasis: The 'seed and soil̓ hypothesis revisited. Nat Rev
Cancer. 3:453–458. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paget S: The distribution of secondary
growths in cancer of the breast. 1889. Cancer Metastasis Rev.
8:98–101. 1989.PubMed/NCBI
|
|
3
|
Wakahara T, Tsukamoto T, Kitamura S,
Watanabe A, Tsujimura T, Nakamura Y, Toyokawa A, Onishi N, Hamabe
Y, Mukai H, et al: Metastatic colon cancer from intrahepatic
cholangiocarcinoma. J Hepatobiliary Pancreat Surg. 12:415–418.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yamamoto M, Takasaki K and Yoshikawa T:
Lymph node metastasis in intrahepatic cholangiocarcinoma. Jpn J
Clin Oncol. 29:147–150. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kuroki T, Fukuda K, Tajima Y, Matsuzaki S,
Kitajima T, Furui J and Kanematsu T: Parapapillary
choledochoduodenal fistula associated with cholangiocarcinoma. J
Hepatobiliary Pancreat Surg. 12:143–146. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takahashi I, Nakamura Y, Suzuki Y,
Mitsuhashi N, Niibe H and Osada J: Case report: Bone metastasis
from cholangiocarcinoma showing unusual accumulation on bone
scintigraphy and 67Ga scintigraphy. Br J Radiol. 67:303–305. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li SQ, Liang LJ, Hua YP, Peng BG, He Q, Lu
MD and Chen D: Long-term outcome and prognostic factors of
intrahepatic cholangiocarcinoma. Chin Med J. 122:2286–2291.
2009.
|
|
8
|
Sripa B, Leungwattanawanit S, Nitta T,
Wongkham C, Bhudhisawasdi V, Puapairoj A, Sripa C and Miwa M:
Establishment and characterization of an opisthorchiasis-associated
cholangiocarcinoma cell line (KKU-100). World J Gastroenterol.
11:3392–3397. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Okada S, Harada H, Ito T, Saito T and Suzu
S: Early development of human hematopoietic and acquired immune
systems in new born NOD/Scid/Jak3null mice intrahepatic
engrafted with cord blood-derived CD34+ cells. Int J
Hematol. 88:476–482. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Patterson MK Jr: Measurement of growth and
viability of cells in culture. Methods Enzymol. 58:141–152. 1979.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Grenman R, Burk D, Virolainen E, Buick RN,
Church J, Schwartz DR and Carey TE: Clonogenic cell assay for
anchorage-dependent squamous carcinoma cell lines using limiting
dilution. Int J Cancer. 44:131–136. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Uthaisar K, Seubwai W, Srikoon P,
Vaeteewoottacharn K, Sawanyawisuth K, Okada S and Wongkham S:
Cepharanthine suppresses metastatic potential of human
cholangiocarcinoma cell lines. Asian Pac J Cancer Prev. 13(Suppl):
S149–S154. 2012.
|
|
13
|
Kan J, Thomson S, Argast GM, O'Connor ME,
Robinson M, Feng B, Heyer J, Chiu MI and Nicoletti R: Use of EMT
gene signatures in cancer drug discovery, diagnostics and
treatment. (US Patent Application Publication. Pub. No.:
US2012/0302572 A1). 2012
|
|
14
|
Li F, Tiede B, Massagué J and Kang Y:
Beyond tumorigenesis: Cancer stem cells in metastasis. Cell Res.
17:3–14. 2007. View Article : Google Scholar
|
|
15
|
Mishra L, Banker T, Murray J, Byers S,
Thenappan A, He AR, Shetty K, Johnson L and Reddy EP: Liver stem
cells and hepatocellular carcinoma. Hepatology. 49:318–329. 2009.
View Article : Google Scholar :
|
|
16
|
Sirisinha S, Tengchaisri T, Boonpucknavig
S, Prempracha N, Ratanarapee S and Pausawasdi A: Establishment and
characterization of a cholangiocarcinoma cell line from a Thai
patient with intrahepatic bile duct cancer. Asian Pac J Allergy
Immunol. 9:153–157. 1991.PubMed/NCBI
|
|
17
|
Rattanasinganchan P, Leelawat K,
Treepongkaruna SA, Tocharoentanaphol C, Subwongcharoen S,
Suthiphongchai T and Tohtong R: Establishment and characterization
of a cholangiocarcinoma cell line (RMCCA-1) from a Thai patient.
World J Gastroenterol. 12:6500–6506. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goodman ZD, Terracciano LM and Wee A: 14 -
Tumours and tumour-like lesions of the liver. MacSween's Pathology
of the Liver. Burt AD, Portmann BC and Ferrell LD: 6th edition.
Churchill Livingstone; Edinburgh: pp. 761–851. 2012, View Article : Google Scholar
|
|
19
|
Asai T, Ueda T, Itoh K, Yoshioka K, Aoki
Y, Mori S and Yoshikawa H: Establishment and characterization of a
murine osteosarcoma cell line (LM8) with high metastatic potential
to the lung. Int J Cancer. 76:418–422. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Tang ZY, Ye SL, Liu YK, Chen J, Xue
Q, Chen J, Gao DM and Bao WH: Establishment of cell clones with
different metastatic potential from the metastatic hepatocellular
carcinoma cell line MHCC97. World J Gastroenterol. 7:630–636.
2001.
|
|
21
|
Barroga EF, Kadosawa T, Okumura M and
Fujinaga T: Establishment and characterization of the growth and
pulmonary metastasis of a highly lung metastasizing cell line from
canine osteosarcoma in nude mice. J Vet Med Sci. 61:361–367. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Su Y, Luo X, He BC, Wang Y, Chen L, Zuo
GW, Liu B, Bi Y, Huang J, Zhu GH, et al: Establishment and
characterization of a new highly metastatic human osteosarcoma cell
line. Clin Exp Metastasis. 26:599–610. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hou Y, Zou Q, Ge R, Shen F and Wang Y: The
critical role of CD133+CD44+/high tumor cells
in hematogenous metastasis of liver cancers. Cell Res. 22:259–272.
2012. View Article : Google Scholar
|
|
24
|
Thanan R, Pairojkul C, Pinlaor S,
Khuntikeo N, Wongkham C, Sripa B, Ma N, Vaeteewoottacharn K,
Furukawa A, Kobayashi H, et al: Inflammation-related DNA damage and
expression of CD133 and Oct3/4 in cholangiocarcinoma patients with
poor prognosis. Free Radic Biol Med. 65:1464–1472. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo W and Frenette PS: Alternative CD44
splicing in intestinal stem cells and tumorigenesis. Oncogene.
33:537–538. 2014. View Article : Google Scholar
|
|
26
|
Hajra KM and Fearon ER: Cadherin and
catenin alterations in human cancer. Genes Chromosomes. Cancer.
34:255–268. 2002.
|
|
27
|
Skubitz AP: Adhesion molecules. Cancer
Treat Res. 107:305–329. 2002.PubMed/NCBI
|
|
28
|
Adam PJ, Boyd R, Tyson KL, Fletcher GC,
Stamps A, Hudson L, Poyser HR, Redpath N, Griffiths M, Steers G, et
al: Comprehensive proteomic analysis of breast cancer cell
membranes reveals unique proteins with potential roles in clinical
cancer. J Biol Chem. 278:6482–6489. 2003. View Article : Google Scholar
|
|
29
|
Dumartin L, Whiteman HJ, Weeks ME,
Hariharan D, Dmitrovic B, Iacobuzio-Donahue CA, Brentnall TA,
Bronner MP, Feakins RM, Timms JF, et al: AGR2 is a novel surface
antigen that promotes the dissemination of pancreatic cancer cells
through regulation of cathepsins B and D. Cancer Res. 71:7091–7102.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vanderlaag KE, Hudak S, Bald L,
Fayadat-Dilman L, Sathe M, Grein J and Janatpour MJ: Anterior
gradient-2 plays a critical role in breast cancer cell growth and
survival by modulating cyclin D1, estrogen receptor-α and survivin.
Breast Cancer Res. 12:R322010. View Article : Google Scholar
|
|
31
|
Park K, Chung YJ, So H, Kim K, Park J, Oh
M, Jo M, Choi K, Lee EJ, Choi YL, et al: AGR2, a mucinous ovarian
cancer marker, promotes cell proliferation and migration. Exp Mol
Med. 43:91–100. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Z, Hao Y and Lowe AW: The
adenocarcinoma-associated antigen, AGR2, promotes tumor growth,
cell migration, and cellular transformation. Cancer Res.
68:492–497. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu D, Rudland PS, Sibson DR,
Platt-Higgins A and Barraclough R: Human homologue of cement gland
protein, a novel metastasis inducer associated with breast
carcinomas. Cancer Res. 65:3796–3805. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chanda D, Lee JH, Sawant A, Hensel JA,
Isayeva T, Reilly SD, Siegal GP, Smith C, Grizzle W, Singh R, et
al: Anterior gradient protein-2 is a regulator of cellular adhesion
in prostate cancer. PLoS One. 9:e899402014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lepreux S, Bioulac-Sage P and Chevet E:
Differential expression of the anterior gradient protein-2 is a
conserved feature during morphogenesis and carcinogenesis of the
biliary tree. Liver Int. 31:322–328. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim SJ, Kim DH, Kang D and Kim JH:
Expression of anterior gradient 2 is decreased with the progression
of human biliary tract cancer. Tohoku J Exp Med. 234:83–88. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hata K, Dhar DK, Watanabe Y, Nakai H and
Hoshiai H: Expression of metastin and a G-protein-coupled receptor
(AXOR12) in epithelial ovarian cancer. Eur J Cancer. 43:1452–1459.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Prentice LM, Klausen C, Kalloger S, Köbel
M, McKinney S, Santos JL, Kenney C, Mehl E, Gilks CB, Leung P, et
al: Kisspeptin and GPR54 immunoreactivity in a cohort of 518
patients defines favourable prognosis and clear cell subtype in
ovarian carcinoma. BMC Med. 5:332007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dhar DK, Naora H, Kubota H, Maruyama R,
Yoshimura H, Tonomoto Y, Tachibana M, Ono T, Otani H and Nagasue N:
Downregulation of KiSS-1 expression is responsible for tumor
invasion and worse prognosis in gastric carcinoma. Int J Cancer.
111:868–872. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guan-Zhen Y, Ying C, Can-Rong N, Guo-Dong
W, Jian-Xin Q and Jie-Jun W: Reduced protein expression of
metastasis-related genes (nm23, KISS1, KAI1 and p53) in lymph node
and liver metastases of gastric cancer. Int J Exp Pathol.
88:175–183. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Masui T, Doi R, Mori T, Toyoda E, Koizumi
M, Kami K, Ito D, Peiper SC, Broach JR, Oishi S, et al: Metastin
and its variant forms suppress migration of pancreatic cancer
cells. Biochem Biophys Res Commun. 315:85–92. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nagai K, Doi R, Katagiri F, Ito T, Kida A,
Koizumi M, Masui T, Kawaguchi Y, Tomita K, Oishi S, et al:
Prognostic value of metastin expression in human pancreatic cancer.
J Exp Clin Cancer Res. 28:92009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kostadima L, Pentheroudakis G and Pavlidis
N: The missing kiss of life: Transcriptional activity of the
metastasis suppressor gene KiSS1 in early breast cancer. Anticancer
Res. 27:2499–2504. 2007.PubMed/NCBI
|
|
44
|
Stark AM, Tongers K, Maass N, Mehdorn HM
and Held-Feindt J: Reduced metastasis-suppressor gene
mRNA-expression in breast cancer brain metastases. J Cancer Res
Clin Oncol. 131:191–198. 2005. View Article : Google Scholar
|
|
45
|
Marot D, Bieche I, Aumas C, Esselin S,
Bouquet C, Vacher S, Lazennec G, Perricaudet M, Kuttenn F, Lidereau
R, et al: High tumoral levels of Kiss1 and G-protein-coupled
receptor 54 expression are correlated with poor prognosis of
estrogen receptor-positive breast tumors. Endocr Relat Cancer.
14:691–702. 2007. View Article : Google Scholar : PubMed/NCBI
|