Introduction
Nasopharyngeal carcinoma (NPC), one of the most
common head and neck malignant tumors, is prevalent in Southern
China and Southeast Asia (1). For
patients with advanced NPC, first-line radiotherapy is far from
satisfactory. Chemotherapy is offered as the most efficacious
auxiliary treatment strategy (2).
However, similar to the obstacle of paclitaxel application in other
solid tumors, the emergence of resistant cancer cells to paclitaxel
limits its clinical efficiency in NPC, which eventually brings NPC
recurrence and metastasis (3).
Previous studies have focused on discovering diverse molecules
including protein, miRNAs and mRNAs to serve as biomarkers and
therapy targets of paclitaxel resistance (4,5).
However, paclitaxel resistance still remains unclear, making it
urgent to clarify paclitaxel resistance from a brand-new
perspective.
lncRNAs are a family of transcripts with >200
nucleotides (nt) in length with no protein-coding potential, which
are involved in numerous biological processes such as their
protein-coding counterparts (6).
This class of lncRNAs makes up the largest portion of the mammalian
non-coding transcriptomes. Most of the currently known lncRNAs show
their functional role by participating in the biological
progressions at the epigenetic, transcriptional and
post-transcriptional levels (7,8).
Moreover, dysregulated expression of lncRNAs in cancers involves in
a spectrum of disease progression, serving as an independent
predictor for patient outcomes and associates with therapeutic
efficacy (9,10). In vitro and in vivo
functional analyses also indicate that lncRNAs can take part in
multiple cancer malignant behaviors, such as unlimited
proliferation, metastasis, radioresistance and cancer stem cell
phenotype (11,12). In respect to the correlation between
lncRNA and chemoresistance, limited lncRNAs including MEG3,
MALAT-1, UCA1 and HOTAIR have been reported to regulate the
chemoresistance of cisplatin and doxorubicin in several solid human
tumor (13–15). Taken together, current research of
lncRNAs in tumors shows the need to illuminate the mechanism of
chemoresistance in NPC based on lncRNAs.
Paclitaxel, one of the taxane families, is widely
used in many solid human malignancies including NPC (16). The emergence of paclitaxel
resistance greatly restricts its clinical efficiency (3). Therefore, the present study
investigated lncRNAs association with paclitaxel resistance in NPC.
Considering the widespread application of next generation
sequencing (NGS) technology and its advantage of massively
discovering existing and novel molecule candidates, it is now
employed in our research to comprehensively and systematically
screen lncRNA candidates associated with paclitaxel resistance
(17).
Materials and methods
Cell culture and construction of
paclitaxel-resistant CNE-2 cells
Poorly differentiated NPC cell line CNE-2,5-8F and
6-10B were provided by the Cell Center of Central South University
and were cultured in RPMI-1640 medium (HyClone, Logan, UT, USA)
with 10% fetal bovine serum (FBS) and 1% antibiotics (both from
Gibco-BRL, Gaithersburg, MD, USA). Paclitaxel-resistant CNE-2 cell
line (termed CNE-2-Pr) was established by exposure to the cell
culture medium with gradually increased concentrations of
paclitaxel (Sigma-Aldrich, St. Louis, MO, USA). The initial and
final concentrations of Paclitaxel were 0.2 and 2 nM. The above
cells were propagated in an incubator at 37°C with saturated
humidity and 5% CO2. CNE-2-Pr was cultured in the
RPMI-1640 medium with 1 nM paclitaxel to keep its chemoresistance.
The CNE-2-Pr cells were sub-cultured <20 passages and used for
the following experiments.
Paclitaxel cytotoxicity assays by CCK-8
methods
Cells were suspended in essential medium, plated
into a 96-well plate (5×103 cells/well) and incubated
for 24 h at 37°C. Paclitaxel was separately added to each well at
different concentrations and the plate was incubated for another 48
h before further analyses. Triplicate wells were used for each drug
treatment. Cell Counting Kit-8 (CCK-8; Beyotime, Co., Guangzhou,
China) was used according to its instructions. The absorbance of
each well was then detected (18,19).
RNA extraction
Total RNA was extracted from paclitaxel resistant
CNE-2-Pr and parental CNE-2 cells using TRIzol (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's protocol. The
qualities and concentrations of RNA samples were monitored by gel
electrophoresis and at absorbance ratios of A260/A280 using a
NanoDrop ND-1000 spectrophotometer. RNAs of appropriate quality
were stored at −80°C for lncRNA detection and subsequent
experiments.
Construction of an RNA library for
sequencing
Total RNA was extracted from CNE-2-Pr and CNE-2
cells. After removal of rRNA, the remaining RNA was cut randomly
into short fragments. Using random hexamers, first-strand cDNA was
synthesized based on these fragments; second-strand cDNA was also
synthesized by mixing first-strand cDNA with buffer, dNTPs, RNase H
and DNA polymerase I. This cDNA was then purified by a QIAquick PCR
kit (Qiagen, Valencia, CA, USA) and subsequently degraded by
uracil-N-glycosylase. RNA fragments were separated by agarose gel
electrophoresis, and fragments were then expanded with polymerase
chain reaction (PCR). The PCR products were sequenced using an
Illumina HiSeq™ 2000 instrument (Illumina, Inc., San Diego, CA,
USA), and the original image data were converted into '.fq' files
by base calling software. The relative data were submitted to NCBI
under BioProject accession no. PRJNA 254709.
Transcript identification and
establishment of differential expression profiles
Sequence data (.fq files) was filtered to remove
rRNA sequences. We then used TopHat2 and Cufflinks strategies to
reconstruct transcripts in samples. The transcripts were BLASTed
against the NONCODE v3.0 database (http://www.noncode.org/NONCODERv3/) to identify
already known non-coding RNAs (identity >0.9 and coverage
>0.8 as the selection criteria). Transcripts without annotations
in the ncRNA library were then compared to protein databases, and
the mapped transcripts were considered as mRNA (identity >0.9
and coverage >0.8). The remaining transcripts, which were not
aligned with the protein library, were then entered into the Coding
Potential Calculator program to distinguish coding and non-coding
sequences (20). To obtain the
differential expression profiles, read counts and reads per
kilobase per million read (RPKM) values were calculated for each
gene, and a likelihood ratio test was used to assess the
significance of expression differences as described in our previous
study (21). lncRNAs were
considered to be differentially expressed when the fold-change was
>2 (|log2 ratio| ≥1) and the false discovery rate (FDR) was 0.01
or less.
Quantitative real-time reverse
transcription PCR (qRT-PCR)
Briefly, cDNA was synthesized from total RNA using a
PrimeScript RT reagent kit with gDNA Eraser (Takara, Shiga, Japan).
Primers for lncRNAs were designed and synthesized. Then, qPCR
assays were performed using a Bio-Rad iQ5 Multicolor Real-Time
qRT-PCR Detection system (Bio-Rad, Hercules, CA, USA). The
expression levels of lncRNAs were detected as previously described
(21). Human β-actin was used as a
housekeeping gene for normalization. The expression levels of
lncRNAs were measured in terms of the cycle threshold (CT) and were
then normalized to β-actin expression using the 2−ΔΔCt
method (21).
siRNA construction and transfection
To estimate inhibition of lncRNA n375709, the
specific lncRNA n375709 siRNA, universal negative control and the
transfection agent (riboFECT™ CP Buffer) were designed and
purchased from RiboBio Co. Ltd. (Guangzhou, China). NPC cells
(1×105) were grown in triplicate in 6-well plates.
Twenty-four hours later, 100 nM of lncRNA n375709 siRNA, the
transfection agent and negative control which were used as controls
were separately transfected into NPC cells according to the
manufacturer's instructions. Forty-eight hours later, the initial
transfection medium was changed for fresh medium and NPC cells were
harvested for following experiments.
Paclitaxel cytotoxicity assays
Cells (2×103) were separately cultured
into 96-well plates in triplicate and were treated with varying
concentrations of paclitaxel for 48 h. A CCK-8 was applied to
determine the growth curves for both cell lines. Inhibition
fraction was used to compare the sensitivity of paclitaxel in each
group. Each experiment was performed in triplicate. Inhibition
fraction = 1−A450 value (paclitaxel group)/A450 value (parental
group).
Statistical analysis
The results of the quantitative data in the present
study are expressed as the mean ± standard deviation. The
statistical significance of the differences between two groups were
analyzed using a two-sided, unpaired Student's t-tests (for equal
variances) or Welch's corrected t-tests (unequal variances)
performed in SPSS 18.0 software. Differences with P-values of
<0.05 were considered to indicate a statistically significant
result.
Results
Generation of paclitaxel-resistant NPC
cells
To obtain the lncRNAs correlated with paclitaxel
resistance, CNE-2 cells were exposed to gradually increased
concentration of paclitaxel in complete medium for 10 months, and
the remaining CNE-2 cells were abbreviated as CNE-2-Pr and its
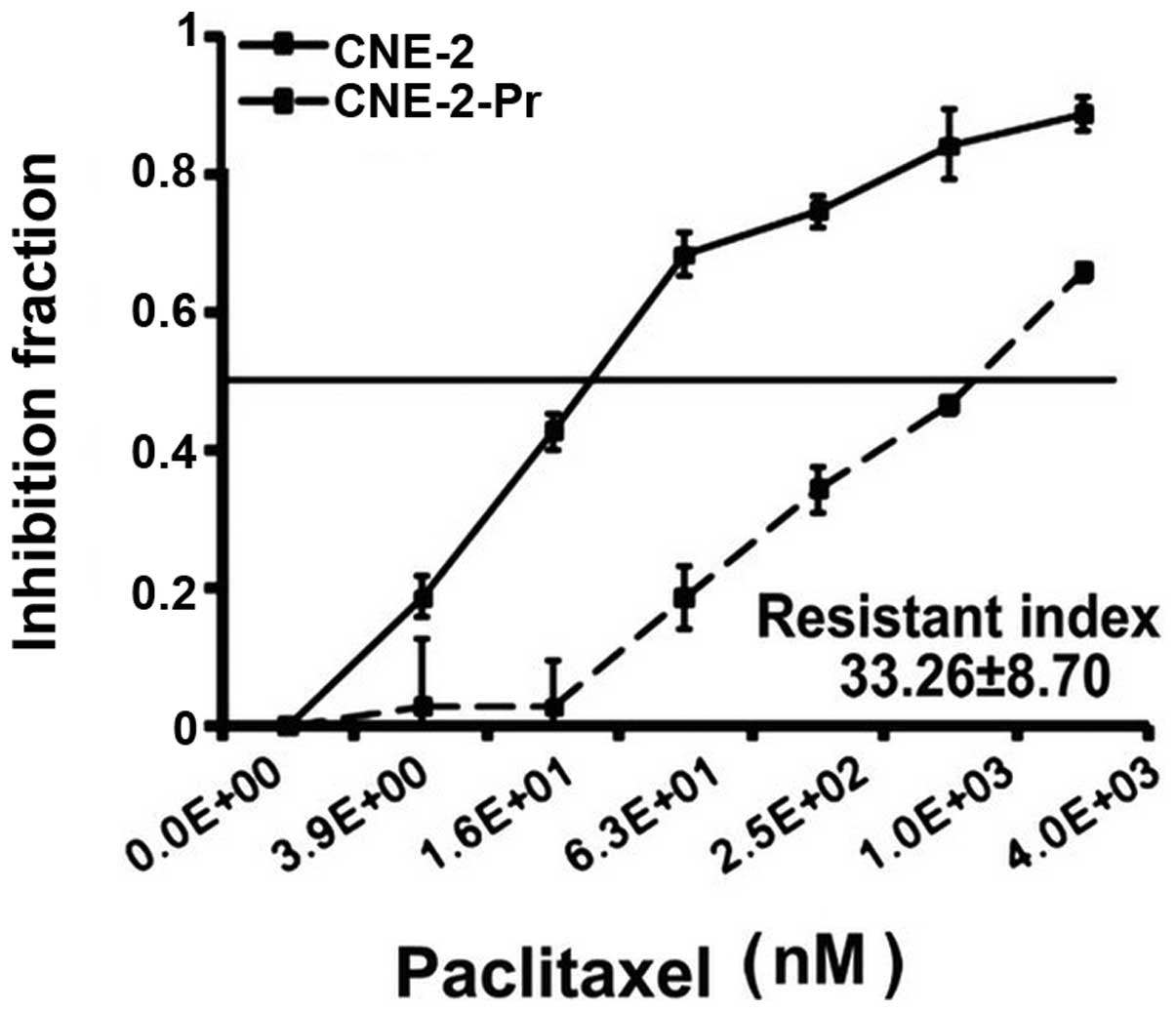
resistance to paclitaxel was then validated by CCK-8 assays. As
shown in Fig. 1, the half maximal
inhibitory concentration (IC50) of paclitaxel for
CNE-2-Pr and parental CNE-2 cells were 1098.66±31.95 and 32.76±1.67
nM, respectively, which indicates that paclitaxel resistance of
CNE-2-Pr cells was 33.26±8.70 times higher than that of its
parental CNE-2 cells. Therefore, we considered the remaining
CNE-2-Pr cells post-paclitaxel exposure as paclitaxel resistant
cells. CNE-2-Pr cells kept the paclitaxel resistant characteristics
after >20 passages under the continuous exposure to 1 nM
paclitaxel (data not shown), which were used for our further
experiments to screen lncRNAs correlated with paclitaxel
resistance.
Establishment of lncRNA differential
expression profile associated with paclitaxel resistance in
NPC
To obtain lncRNAs associated with paclitaxel
resistance in NPC, cDNA libraries were initially constructed via
our previously established CNE-2-Pr and parental CNE-2 cells. As
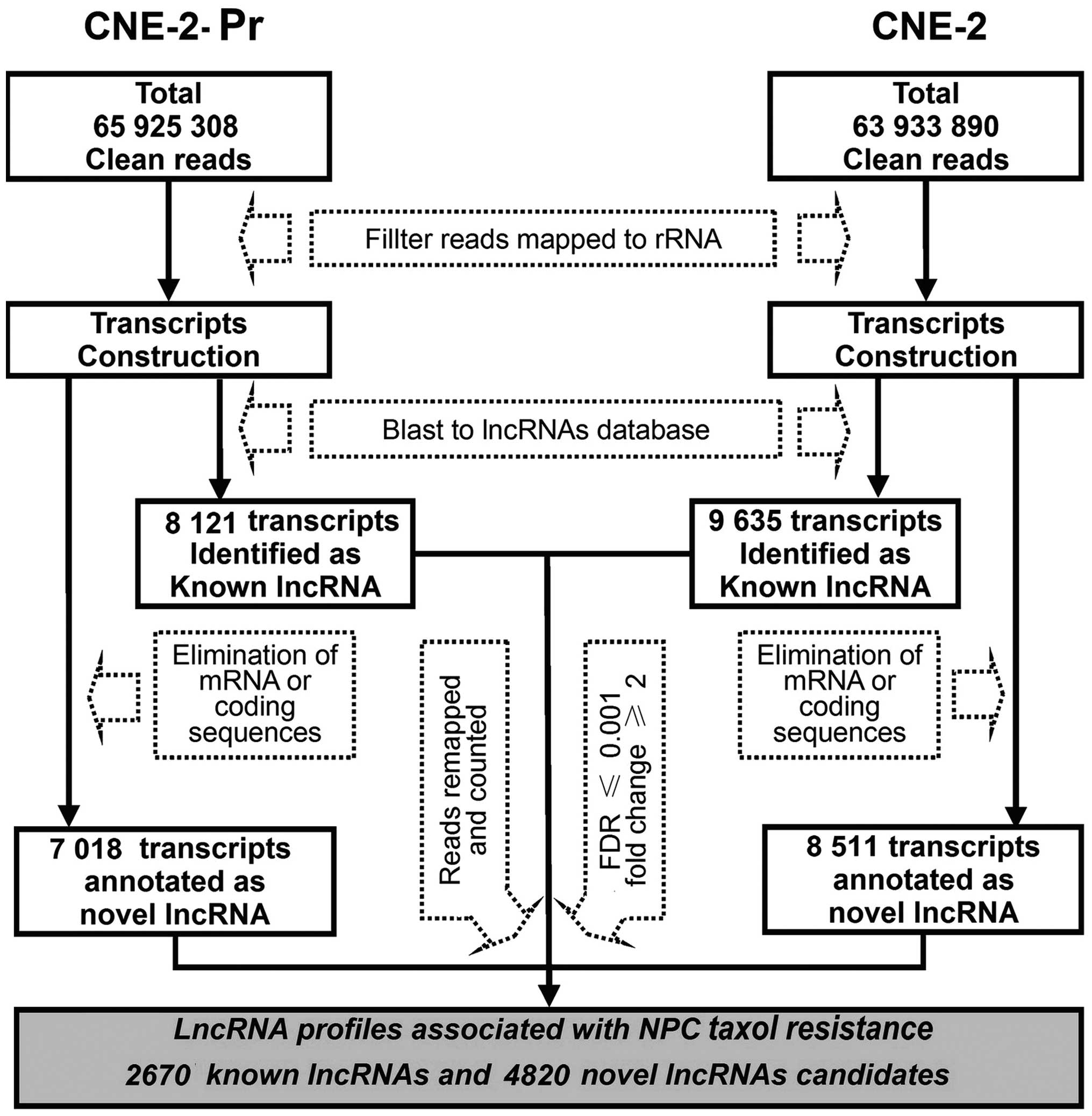
depicted in Fig. 2, a total of
65,925,308 and 63,933,890 clean reads were detected in CNE-2-Pr and
CNE-2 cells, respectively. After eliminating reads mapped to rRNA,
TopHat2 and Cufflinks were used to reconstruct transcripts in both
samples. Then, these reconstructed transcripts were BLASTed against
the NONCODE v3.0 database. Transcripts (8,121) in CNE-2-Pr and
9,635 transcripts in CNE-2 cells were annotated as known lncRNAs.
Additionally, following another elimination of transcripts mapped
to mRNA and coding sequences, 7,018 (CNE-2-Pr) and 8,511 (CNE-2)
transcripts were separately identified as novel lncRNAs. The unique
mapped reads for each lncRNA were then counted, and the RPKM value
for each lncRNA was calculated. Based on the criteria of an
absolute fold-change >2.0 and an FDR <0.001, 2,670 known
lncRNAs and 4,820 novel lncRNA candidates were finally obtained,
which constituted the differential lncRNA expression profiles
associated with NPC paclitaxel resistance.
Characteristics of known and novel
lncRNAs
Based on the above-mentioned lncRNA profiles, the
features of lncRNAs were further analyzed. Our data revealed that
the most known lncRNAs were abundant in 200 b-3 kb in length, and
novel candidates mainly distributed in the region of 200 b-2 kb.
Among these 2,670 known lncRNAs, 2,413 lncRNAs were expressed in
both cell lines, while 255 and 2 were present only in CNE-2-Pr or
CNE-2 cells, respectively. Compared with parental CNE-2 cells, 25
lncRNAs were upregulated and 2,645 lncRNAs were downregulated in
paclitaxel-resistant CNE-2-Pr cells. Similarly, in the 4,820 novel
lncRNA candidates, 3,518 lncRNAs were expressed in both cells,
while 1,264 and 38 existed only in CNE-2-Pr or CNE-2. Of the
lncRNAs, 193 were elevated and 4,627 were decreased in
paclitaxel-resistant CNE-2-Pr cells.
Validation of the lncRNA differential
expression profile via qRT-PCR
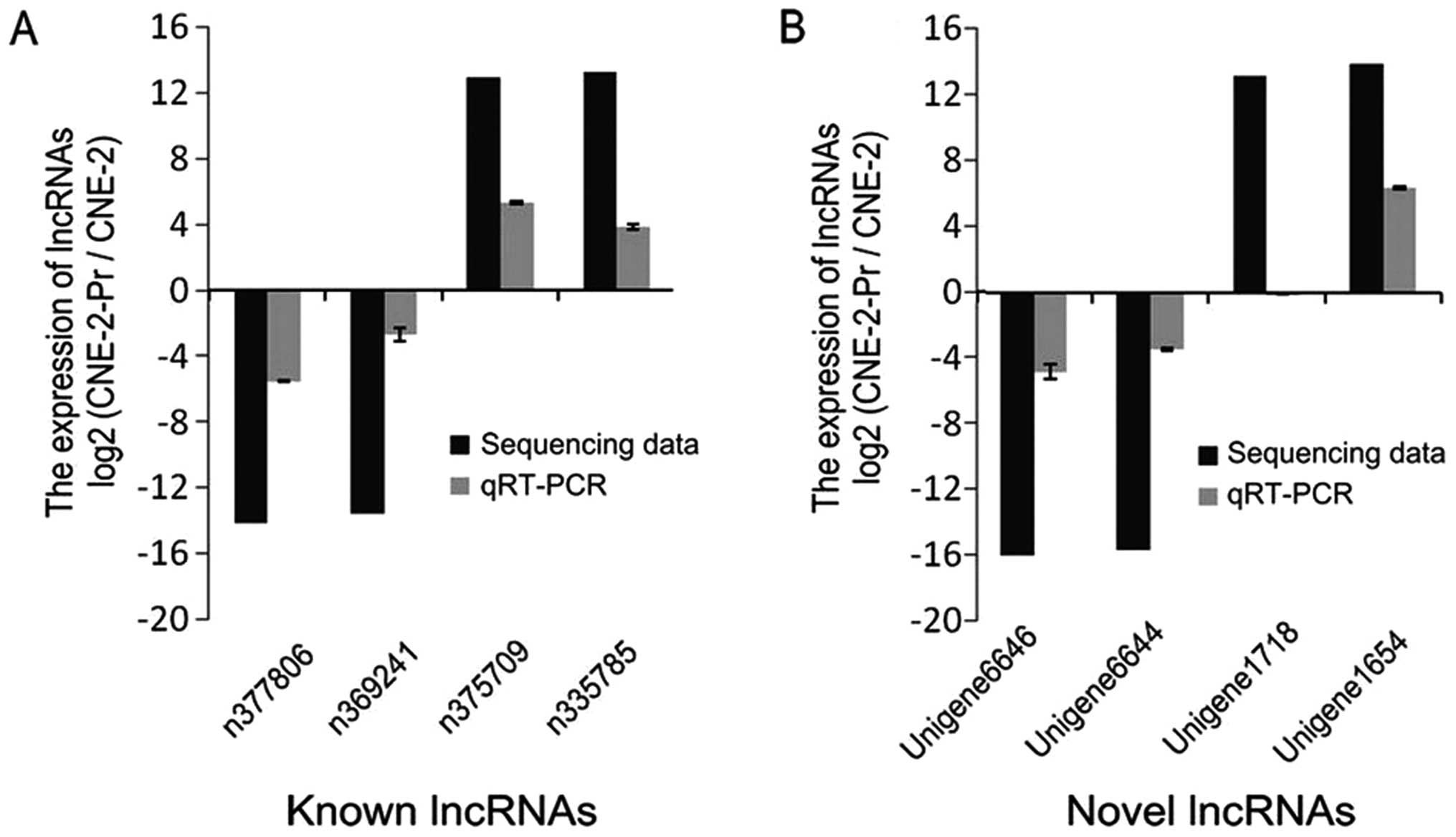
To confirm the lncRNA expression profile findings,
the expression level of top 8 upregulated and downregulated in both
known and novel lncRNAs were detected by qRT-PCR. Compared with the
deep sequencing prediction findings, qRT-PCR verified that the same
significant differential expression trends were presented in 7 of 8
lncRNAs, which included all the known lncRNAs (n375709, n377806,
n369241 and n335785) (Fig. 3A) and
3 novel lncRNAs (Unigene6646, Unigene6644 and Unigene1654)
(Fig. 3B). Our validation showed a
high consistency with the lncRNAs predicted by the expression
profile established via next generation deep sequencing.
Inhibition of lncRNA n375709 in NPC cells
increases their sensitivity to paclitaxel
The above abundant lncRNAs provide us a solid basis
to investigate paclitaxel resistance of NPC. Our group initially
focused on lncRNA n375709, which was the most significantly
overexpressed lncRNA in known lncRNAs. Thus, the role of lncRNA
n375709 in paclitaxel resistance of NPC was further explored in
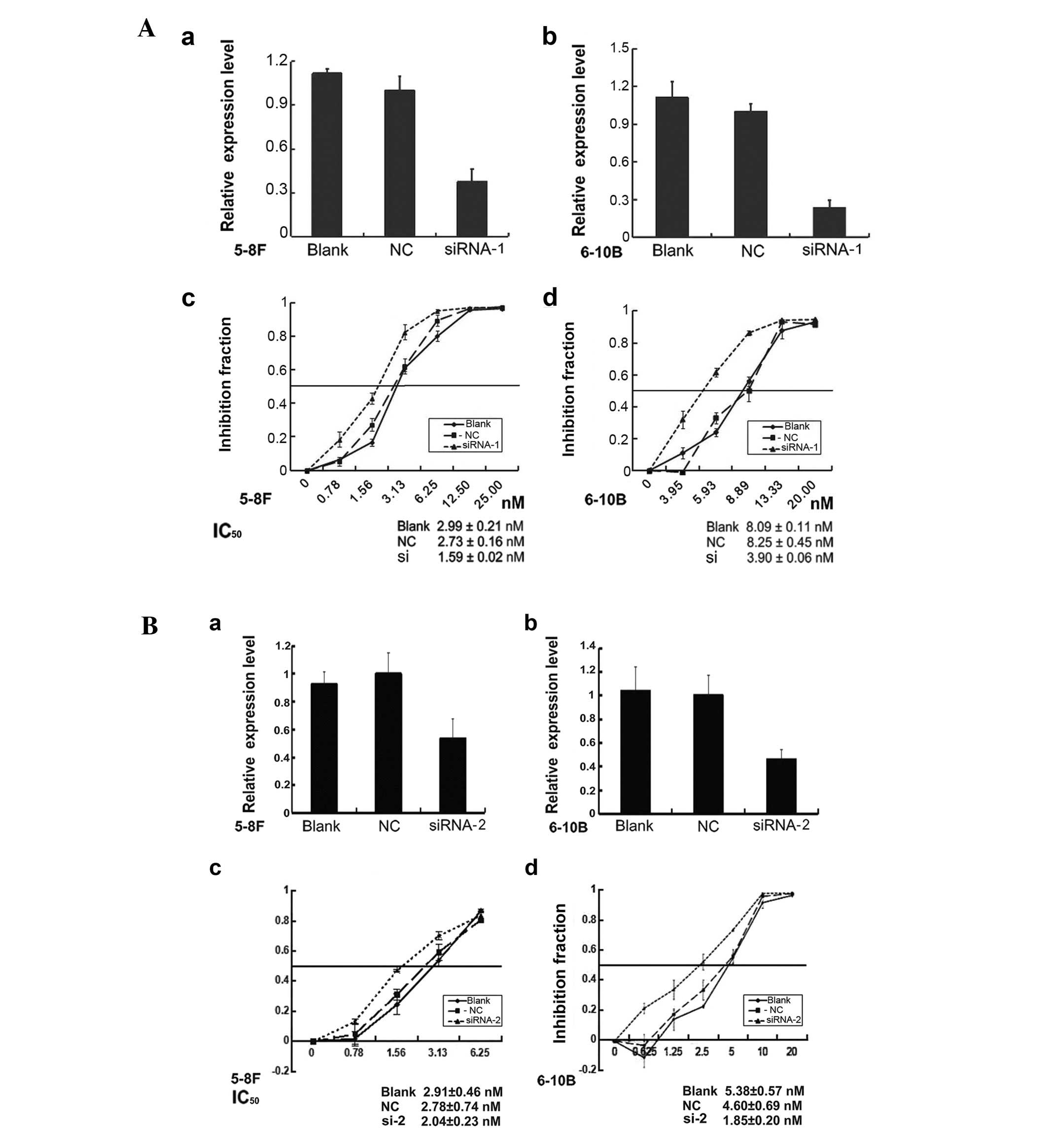
vitro. siRNA interference was employed to inhibit the
expression of lncRNA n375709 in both 5-8F and 6-10B cells. To make
the results more credible, two different siRNAs targeting the
different sites of lncRNA n375709 were used. qRT-PCR assays
revealed that lncRNA n375709 was successfully downregulated by two
siRNA in NPC 5-8F and 6-10B cell (Fig.
4A-a and -b, and B-a and -b). Further paclitaxel cytotoxic
assays demonstrated that inhibition of lncRNA n375709 increased the
paclitaxel sensitivity of NPC cells in vitro, which was
indicated by the increased inhibition fraction of both cell lines
(Fig. 4A-c and -d, and B-c and -d;
p<0.05). Together, these data clearly showed that lncRNA n375709
could regulate the paclitaxel resistance of NPC cells.
Discussion
Paclitaxel resistance restricts the clinical
response and prognosis of diverse human cancers including NPC
(3), therefore it is urgent to find
biomarkers and therapeutic targets associated with paclitaxel
resistance. In the present study, next generation sequencing
technology was used to establish differential expression profile of
lncRNAs associated with paclitaxel resistance in NPC, which
embraced both known and novel lncRNAs. This profile provided us
numerous potential lncRNAs that may function in NPC paclitaxel
resistance, furthermore indicating that much more effort should be
made to validate the exact function of these known and novel
lncRNAs in paclitaxel resistance.
Currently, it is undeniable that available
chemoresistant tissues are the best choice for screening biomarkers
and targets correlated with paclitaxel resistance, particularly
under the application of NGS technology (22). However, NPC is not suitable for
surgery and shows high response to radiotherapy, which is the
reason why these specific NPC tissues are hard to obtain in
clinical settings. Therefore, we established paclitaxel resistant
NPC CNE-2 cells named CNE-2-Pr, which were then used in NGS and
bioinformatics analyses. The construction of paclitaxel resistant
NPC cells was by exposure of NPC cells to gradually increasing
doses of paclitaxel. Compared with methods reported by other
groups, such as high-dose paclitaxel impulse, high-dose paclitaxel
impulse followed with low-dose paclitaxel maintenance, our method
avoided lethal cell damage and unrepeatable dosage used in
high-dose paclitaxel impulse (23–25).
lncRNAs have been demonstrated to be involved in the
development of different human diseases, such as diabetes and
obesity (26), cardiovascular
diseases (27), neurodegenerative
disorders (28) and muscular
dystrophies (29). Indeed, strong
links between lncRNAs and cancers are also abundant. The most
studied lncRNAs, such as HOTAIR and MALAT1, are abnormally
expressed in a wide range of solid tumors and function in cancer
malignant behavior (10,30-32).
As to NPC, lncRNA profiles associated with metastasis and
recurrence except to paclitaxel resistance has been recently
reported (33,34). These groups used the microarray to
screen differential lncRNAs, which is different to the NGS
technology we used in the present study. NGS not only allowed
massive parallel analyses of genome-wide expression as microarrays,
but also had the advantages of calculating the absolute abundance
of the transcripts, identifying variations in lncRNA sequences and
discovering novel lncRNAs. According to the CPC website and the
cited literature (20), a true
protein-coding transcript is more likely to have a long and
high-quality open reading frame (ORF) compared with a non-coding
transcript. In the present study, we considered the following 6
features, such as log-odds score, coverage of the predicted ORF,
integrity of the predicted ORT, number of HITs, hit score and frame
score. When the transcripts matched the above criterion, we
identified the transcripts as a candidate of protein-coding
transcript. Accordingly, in addition to 2,670 known lncRNAs, we
also obtained 4,820 novel lncRNA candidates that were associated
with paclitaxel resistance. These lncRNAs may greatly enrich the
human lncRNA pool.
Aberrant expression and dysfunction of lncRNAs in
cancers, along with its tissue specific expression pattern,
indicates specific-lncRNAs may be used as promising targets for the
development of novel anticancer therapy (30–32).
So far, multiple strategies have been proposed to restore the
homeostatic levels of lncRNAs, in which siRNA is one of the most
frequently used approaches to inhibit the upregulation of oncogenic
lncRNAs (35,36). These siRNAs are complementary to
target lncRNAs and induce degradation in RISC (RNA-induced
silencing) complex, and consequently decline the levels of lncRNA
transcripts (37). Although
numerous lncRNAs were found in the present study, we initially
focused on the most increased lncRNA n375709. As expected, siRNA
inhibition of lncRNA n375709 led to enhanced paclitaxel sensitivity
in two NPC cell lines. These data indicate that we successfully
identified lncRNA n375709 as a potential target to regulate
paclitaxel resistance in this differential expression profile. As a
novel biomarker, there is scarce research of lncRNA n375709 in the
field of tumor biology. lncRNA n375709 is a ~0.5 kb long non-coding
RNA which non-coded gene ID is NONHSAG021653. However, we have to
strengthen two points: firstly, we just confirmed the phenotype of
lncRNA n375709 in regulating paclitaxel resistance. Many
experiments are necessary to illuminate how it functions in the
process; secondly, apart from lncRNA n375709, the remaining lncRNAs
are also waiting to be evaluated and functionally analyzed.
In summary, we established an expression profile of
lncRNAs associated with NPC paclitaxel resistance in human NPC
cells and found that lncRNA expression differed in the
chemoresistant cells, suggesting that these unique non-coding
transcripts may contribute to the acquisition of chemoresistance in
NPC. Although additional in vivo studies and clinical trials
are needed to verify the actual value of the lncRNAs mentioned
above, the present study provides important insights into novel
potential treatment strategies or prognostic indicators for
patients with NPC.
Acknowledgments
Grants were provided by the National Natural Science
Foundation of China (nos. 81202128, 81372426 and 81172558), the
Research Fund for the Doctoral Program of Higher Education of China
(no. 20120162120049) and the Natural Science Foundation of Hunan
Province (nos. 2015JJ3137 and 14JJ2018).
References
|
1
|
Yu MC and Yuan JM: Epidemiology of
nasopharyngeal carcinoma. Semin Cancer Biol. 12:421–429. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chan AT, Grégoire V, Lefebvre JL, Licitra
L, Hui EP, Leung SF and Felip E; EHNS-ESMO-ESTRO Guidelines Working
Group: Nasopharyngeal cancer: EHNS-ESMO-ESTRO Clinical Practice
Guidelines for diagnosis, treatment and follow-up. Ann Oncol.
23(Suppl 7): vii83–vii85. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yusuf RZ, Duan Z, Lamendola DE, Penson RT
and Seiden MV: Paclitaxel resistance: Molecular mechanisms and
pharmacologic manipulation. Curr Cancer Drug Targets. 3:1–19. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tao WY, Liang XS, Liu Y, Wang CY and Pang
D: Decrease of let-7f in low-dose metronomic paclitaxel
chemotherapy contributed to upregulation of thrombospondin-1 in
breast cancer. Int J Biol Sci. 11:48–58. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Choi JH, Sheu JJ, Guan B, Jinawath N,
Markowski P, Wang TL and Shih IeM: Functional analysis of 11q13.5
amplicon identifies Rsf-1 (HBXAP) as a gene involved in paclitaxel
resistance in ovarian cancer. Cancer Res. 69:1407–1415. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nagano T, Mitchell JA, Sanz LA, Pauler FM,
Ferguson-Smith AC, Feil R and Fraser P: The Air noncoding RNA
epigenetically silences transcription by targeting G9a to
chromatin. Science. 322:1717–1720. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Harries LW: Long non-coding RNAs and human
disease. Biochem Soc Trans. 40:902–906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun M, Gadad SS, Kim DS and Kraus WL:
Discovery, annotation, and functional analysis of long noncoding
RNAs controlling cell-cycle gene expression and proliferation in
breast cancer cells. Mol Cell. 59:698–711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jin C, Yan B, Lu Q, Lin Y and Ma L: The
role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal
carcinoma. Tumour Biol. 37:4025–4033. 2016. View Article : Google Scholar
|
|
13
|
Jiao F, Hu H, Han T, Yuan C and Wang L,
Jin Z, Guo Z and Wang L: Long noncoding RNA MALAT-1 enhances stem
cell-like phenotypes in pancreatic cancer cells. Int J Mol Sci.
16:6677–6693. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: Long non-coding RNA UCA1 increases chemoresistance of
bladder cancer cells by regulating Wnt signaling. FEBS J.
281:1750–1758. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W,
De W, Wang Z and Wang R: The long noncoding RNA HOTAIR contributes
to cisplatin resistance of human lung adenocarcinoma cells via
downregualtion of p21WAF1/CIP1 expression. PLoS One.
8:e772932013. View Article : Google Scholar
|
|
16
|
van der Burg ME, Vergote I, Onstenk W,
Boere IA, Leunen K, van Montfort CA and van Doorn HC: Long-term
results of weekly paclitaxel carboplatin induction therapy: An
effective and well-tolerated treatment in patients with
platinum-resistant ovarian cancer. Eur J Cancer. 49:1254–1263.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoshida K, Sanada M and Ogawa S: Deep
sequencing in cancer research. Jpn J Clin Oncol. 43:110–115. 2013.
View Article : Google Scholar
|
|
18
|
Li G, Liu Y, Su Z, Ren S, Zhu G, Tian Y
and Qiu Y: MicroRNA-324-3p regulates nasopharyngeal carcinoma
radioresistance by directly targeting WNT2B. Eur J Cancer.
49:2596–2607. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li G, Wang Y, Liu Y, Su Z, Liu C, Ren S,
Deng T, Huang D, Tian Y and Qiu Y: miR-185-3p regulates
nasopharyngeal carcinoma radioresistance by targeting WNT2B in
vitro. Cancer Sci. 105:1560–1568. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ,
Wei L and Gao G: CPC: Assess the protein-coding potential of
transcripts using sequence features and support vector machine.
Nucleic Acids Res. 35:W345–W349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li G, Qiu Y, Su Z, Ren S, Liu C, Tian Y
and Liu Y: Genome-wide analyses of radioresistance-associated miRNA
expression profile in nasopharyngeal carcinoma using next
generation deep sequencing. PLoS One. 8:e844862013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Moulder S, Helgason T, Janku F, Wheler J,
Moroney J, Booser D, Albarracin C, Morrow PK, Atkins J, Koenig K,
et al: Inhibition of the phosphoinositide 3-kinase pathway for the
treatment of patients with metastatic metaplastic breast cancer.
Ann Oncol. 26:1346–1352. 2015.PubMed/NCBI
|
|
23
|
Cai J, Chen S, Zhang W, Zheng X, Hu S,
Pang C, Lu J, Xing J and Dong Y: Salvianolic acid A reverses
paclitaxel resistance in human breast cancer MCF-7 cells via
targeting the expression of transgelin 2 and attenuating PI3 K/Akt
pathway. Phytomedicine. 21:1725–1732. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang YY, Xie KM, Yang GQ, Mu HJ, Yin Y,
Zhang B and Xie P: The effect of glucosylceramide synthase on
P-glycoprotein function in K562/AO2 leukemia drug-resistance cell
line. Int J Hematol. 93:361–367. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li W, Tan GL, Ma YH, Peng XW and He GX:
Role of folate receptor 1 in paclitaxel-resistance of
nasopharyngeal carcinoma cells. Zhonghua Er Bi Yan Hou Tou Jing Wai
Ke Za Zhi. 45:1035–1040. 2010.In Chinese.
|
|
26
|
Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li
YJ, Yan B and Jiang Q: Pathogenic role of lncRNA-MALAT1 in
endothelial cell dysfunction in diabetes mellitus. Cell Death Dis.
5:e15062014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fiedler J, Breckwoldt K, Remmele CW,
Hartmann D, Dittrich M, Pfanne A, Just A, Xiao K, Kunz M, Müller T,
et al: Development of long noncoding RNA-based strategies to
modulate tissue vascularization. J Am Coll Cardiol. 66:2005–2015.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou X and Xu J: Identification of
Alzheimer's disease-associated long noncoding RNAs. Neurobiol
Aging. 36:2925–2931. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Neguembor MV, Jothi M and Gabellini D:
Long noncoding RNAs, emerging players in muscle differentiation and
disease. Skelet Muscle. 4:82014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xue X, Yang YA, Zhang A, Fong KW, Kim J,
Song B, Li S, Zhao JC and Yu J: LncRNA HOTAIR enhances ER signaling
and confers tamoxifen resistance in breast cancer. Oncogene. Sep
14–2015.Epub ahead of print. PubMed/NCBI
|
|
31
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
interacts with miR-205. Cancer Res. 75:1322–1331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar
|
|
33
|
Zhang W, Wang L, Zheng F, Zou R, Xie C,
Guo Q, Hu Q, Chen J, Yang X, Yao H, et al: Long noncoding RNA
expression signatures of metastatic nasopharyngeal carcinoma and
their prognostic value. Biomed Res Int. 2015:6189242015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gao W, Chan JY and Wong TS: Differential
expression of long noncoding RNA in primary and recurrent
nasopharyngeal carcinoma. Biomed Res Int. 2014:4045672014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou Z, Liu H, Wang C, Lu Q, Huang Q,
Zheng C and Lei Y: Long non-coding RNAs as novel expression
signatures modulate DNA damage and repair in cadmium toxicology.
Sci Rep. 5:152932015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cheng Z, Guo J, Chen L, Luo N, Yang W and
Qu X: A long noncoding RNA AB073614 promotes tumorigenesis and
predicts poor prognosis in ovarian cancer. Oncotarget.
6:25381–25389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|