Introduction
Lung cancer continues to be the leading cause of
cancer-related deaths worldwide due to its high incidence,
malignant behavior and lack of major advancements in treatment
strategy, accounting for ~26/100,000 patients in China (1,2).
Traditionally, lung cancer can be classified into two broad
categories, including small cell lung cancer and non-small cell
lung cancer (NSCLC) (3). Statistics
show that NSCLC accounts for 85–90% of all lung cancers and is
mainly comprised of squamous cell carcinoma, adenocarcinoma and
large-cell carcinoma (4,5). As a multifactorial cancer, NSCLC can
be triggered by many risk factors including tobacco smoking,
second-hand or passive smoking, diet, excess alcohol consumption,
air pollution, occupational exposure and genetic predisposition
(6,7). A combination of radiotherapy and
chemotherapy, along with surgical treatment, is the current
treatment standard to achieve local tumor control and prolonged
survival (8,9). Notably, previous evidence has shown
that the long-term survival rates of lung cancer patients remain
low and the 5-year overall survival rate of NSCLC is ~15% in the US
and <10% in China (1,10,11).
Therefore, there is a crucial need for finding prognostic markers
to improve the clinical management of NSCLC patients.
To the best of our knowledge, a dysregulated
inflammatory response is related to an increased risk of chronic
disease and cancers (12).
Chemokines, small pro-inflammatory chemotactic cytokines, play an
important role in many tumor-related processes including growth,
metastasis and angiogenesis (13).
Chemokine (C-C motif) ligand 22 (CCL22), a member of the CC
chemokine group, also known as macrophage-derived chemokine (MDC),
is expressed by monocytes and dendritic cells (14). CCL22 has been demonstrated to play a
homeostatic role in leukocyte trafficking in addition to its
well-documented effects in innate immune cell activation and Th2
immunopathology, acting through the combination of CC-chemokine
receptor 4 (CCR4) (15).
Interleukin-37 (IL-37), also known as IL-1F7, is a newly discovered
member of the interleukin family with anti-inflammatory and immune
inhibitory effects, and is mainly expressed in the brain, heart,
kidney, dendritic and peripheral blood of mononuclear cells
(16,17). Previous evidence revealed that IL-37
inhibits inflammation and immune reaction, and is found to be
related to many diseases, such as acute myocardial infarction,
sepsis and rheumatoid arthritis (18,19).
Furthermore, CCL22 also has functional effects on the
susceptibility to inflammation, immunological diseases and
malignancies (20,21). As far as we know, the occurrence and
development of lung cancer is closely related to inflammation
(22), and few studies have been
performed concerning the role and function of CCL22 and IL-37 in
NSCLC. Thus, we conducted the present study to investigate the
effects of CCL22 and IL-37 on the proliferation and the
epithelial-mesenchymal transition (EMT) of NSCLC A549 cells.
Materials and methods
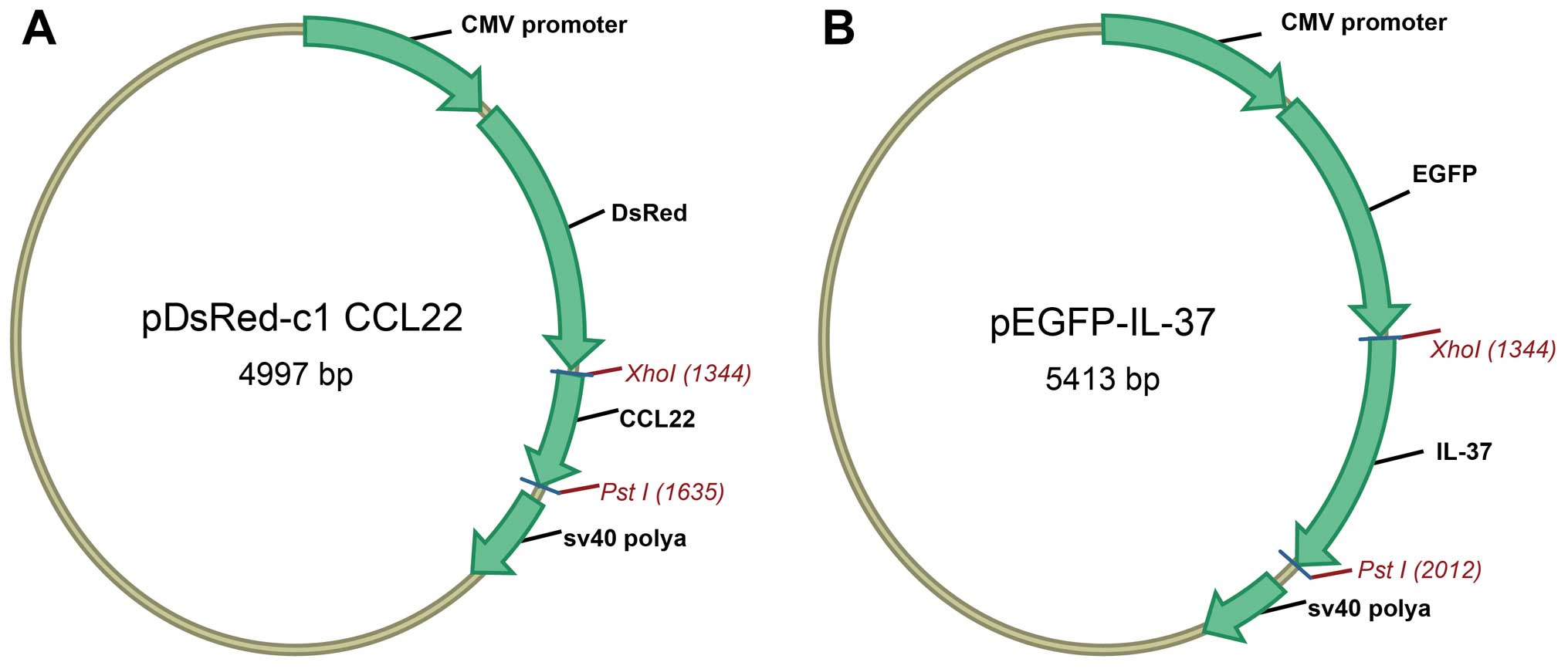
Plasmid construction
Polymerase chain reaction amplification was used for
obtaining cDNA sequences of CCL22 and IL-37. cDNA sequences were
recycled and inserted into pDsRed-N1 and pEGFP-C1 plasmids,
respectively, via enzyme digestion of XhoI and PstI,
to obtain pDsRed-CCL22 and pEGFP-IL-37 expression plasmids. PCR
conditions were: pre-denaturing at 95°C for 5 min, 95°C for 30 sec,
58°C for 30 sec, and 72°C for 30 sec with 25 cycles, and finally
elongation at 72°C for 5 min. The PCR primers were: CCL22 sense,
5′-cggctcgagatggatcgcctacagactgcactc and antisense,
5′-cggctgcagtcattggctcagcttattg; IL-37 sense,
5′-cggctcgagatgtcctttgtgggggag and antisense,
5′-cggctgcaggcgttcaatggggcagtttc. Enzyme digestion was conducted at
37°C for 6 h. Fig. 1 shows the
plasmid profiles.
Cell culture and grouping
The A549 cells (obtained from Shanghai Institute of
Cell Biology of the Chinese Academy of Sciences, Shanghai, China,
and retained in our laboratory) were cultured in 90% Dulbecco's
modified Eagle's medium (DMEM) + 10% fetal bovine serum (FBS) (both
from Thermo Fisher Scientific, Inc., Waltham, MA, USA). The A549
cells were divided into six groups in the present study: the
control, the pDsRed-N1 blank plasmid, the pEGFP-C1 blank plasmid,
the pDsRed-CCL22 plasmid, the pEGFP-IL-37 plasmid and the
pDsRed-CCL22 + pEGFP-IL-37 plasmid groups. After being cultured for
24 h, the cells were transfected with plasmids (total plasmid was 6
µg) via a Lipofectamine 3000 kit (Thermo Fisher
Scientific).
Confocal microscopy detection
A coverglass coated with 0.1% gelatin was placed
into a 6-well plate, and then the above six groups of the A549
cells were added into the 6-well plate for culturing, respectively.
When the cell density reached 80–90% confluency, the medium was
emptied and then the cells were washed three times with
phosphate-buffered saline (PBS). Next, the cells were fixed with 4%
paraformaldehyde for 15 min. Subsequently,
4′,6-diamidino-2-phenylindole (DAPI) was also added to mark the
nucleus. After being washed three times with PBS, the coverglass
was inverted on a glass slide for observation under confocal
microscopy (FV1000; Japan Olympus Corporation, Tokyo, Japan).
pDsRed-CCL22 and pEGFP-IL-37 were observed after excitation at 558
and 490-nm wavelengths.
Methyl thiazolyl-tetrazolium (MTT)
assay
A549 cells in six groups were cultured into a
96-well plate with a density of 5,000 cells in each well (each
group has four duplicated wells). After being cultured for 24 h,
cell proliferation was tested at four time points (24, 48, 72 and
96 h) via Vybrant® MTT Cell Proliferation Assay kit
(Thermo Fisher Scientific).
Real-time fluorescence quantitative
polymerase chain reaction (RT-qPCR)
After being cultured for 72 h, the cells were washed
three times with PBS, and then general RNA extraction Co., Ltd.,
Dalian, China) followed. PrimeScript RT reagent kit (Takara, Bao
Biological Engineering Co., Ltd.) and StepOnePlus (ABI) PCR system
(Thermo Fisher Scientific) were applied for reverse transcription
and RT-qPCR. The PCR system consisted of 1.6 µl cDNA, 5
µl 2X SYBR-Green Taq PCR Mix (Takara, Bao Biological
Engineering Co., Ltd.), 0.2 µl forward and 0.2 µl
reverse primers (10 µM) and 3 µl deuterium-depleted
water (DDW). The reaction system was: pre-denaturing at 95°C for 5
min, 95°C for 10 sec, 58°C for 10 sec, 72°C for 10 sec, with a
total of 60 cycles, and finally 72°C elongation for 10 min.
Vimentin (gene ID 7431), N-cadherin (gene ID 1000)
and E-cadherin (gene ID 999) were detected in the present
study, while β-actin (gene ID 2597) was regarded as the
internal reference. Data analyses were conducted with the
2−ΔΔCt method, 2−ΔΔCt representing the
multiple proportions between the test group and its control group.
The formula was: ΔΔCt = ΔCtcase group − ΔCtcontrol
group, and ΔCt = CtmiRNA −
Ctβ-actin. The primer sequences are listed in
Table I.
 | Table IPrimer sequences of vimentin,
E-cadherin, N-cadherin and β-actin by RT-qPCR. |
Table I
Primer sequences of vimentin,
E-cadherin, N-cadherin and β-actin by RT-qPCR.
| Sense | Antisense |
|---|
| Vimentin |
AATTTCACGCAGAGCAACAG |
CCACTAAGGCAGCACGTAAA |
| E-cadherin |
TTAACCTCACCAATCCTTGCT |
TAGCCCATTTCTTCCCAATC |
| N-cadherin |
AACCTAGCCTACTGGCCAAA |
AACATCGAGGTCGTAAACCC |
| β-actin |
GCAGGGGGGAGCCAAAAGGGT |
TGGGTGGCAGTGATGGCATGG |
Protein sample collection and western
blotting
After 72 h of culture, the cells were washed,
followed by the addition of RIPA lysis buffer (Thermo Fisher
Scientific) and protease inhibitor (Sigma-Aldrich, St. Louis, MO,
USA). After homogenization, the cells were centrifuged at 12,000 ×
g at 4°C for 10 min. The supernatant was gathered and used as the
protein sample, and the concentration of protein was tested by
bicinchoninic acid (BCA) assay. Then, the protein sample was stored
at −80°C. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) (10%) was used for western blotting. To each well 20
µg of protein sample was added and incubated overnight at
4°C with the use of rabbit polyclonal anti-vimentin, rabbit
polyclonal anti-N-cadherin, rabbit polyclonal anti-E-cadherin
(Abcam, Cambridge, UK) as primary antibodies. Horseradish
peroxidase (HRP)-conjugated-goat anti-rabbit IgG (1:25,000)
(Agrisera, Vännäs, Sweden) was regarded as the secondary antibody
added into the samples. Then, the samples were incubated in a
greenhouse for 30 min, followed by the addition of HRP (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) for coloration. ImageQuant
350 and ImageQuant TL-1 (GE Healthcare, Logan, UT, USA) were
applied for result analysis. β-actin was used as an internal
reference.
Statistical analyses
Data analyses were performed using SPSS version 20.0
software (SPSS, Inc., Chicago, IL, USA). Continuous data are
presented as the mean ± SD. Comparisons between the two groups were
conducted using a t-test, and comparisons among multiple groups by
repeated measurements were performed using a one-way analysis of
variance (ANOVA). P<0.05 provided evidence of significant
differences.
Results
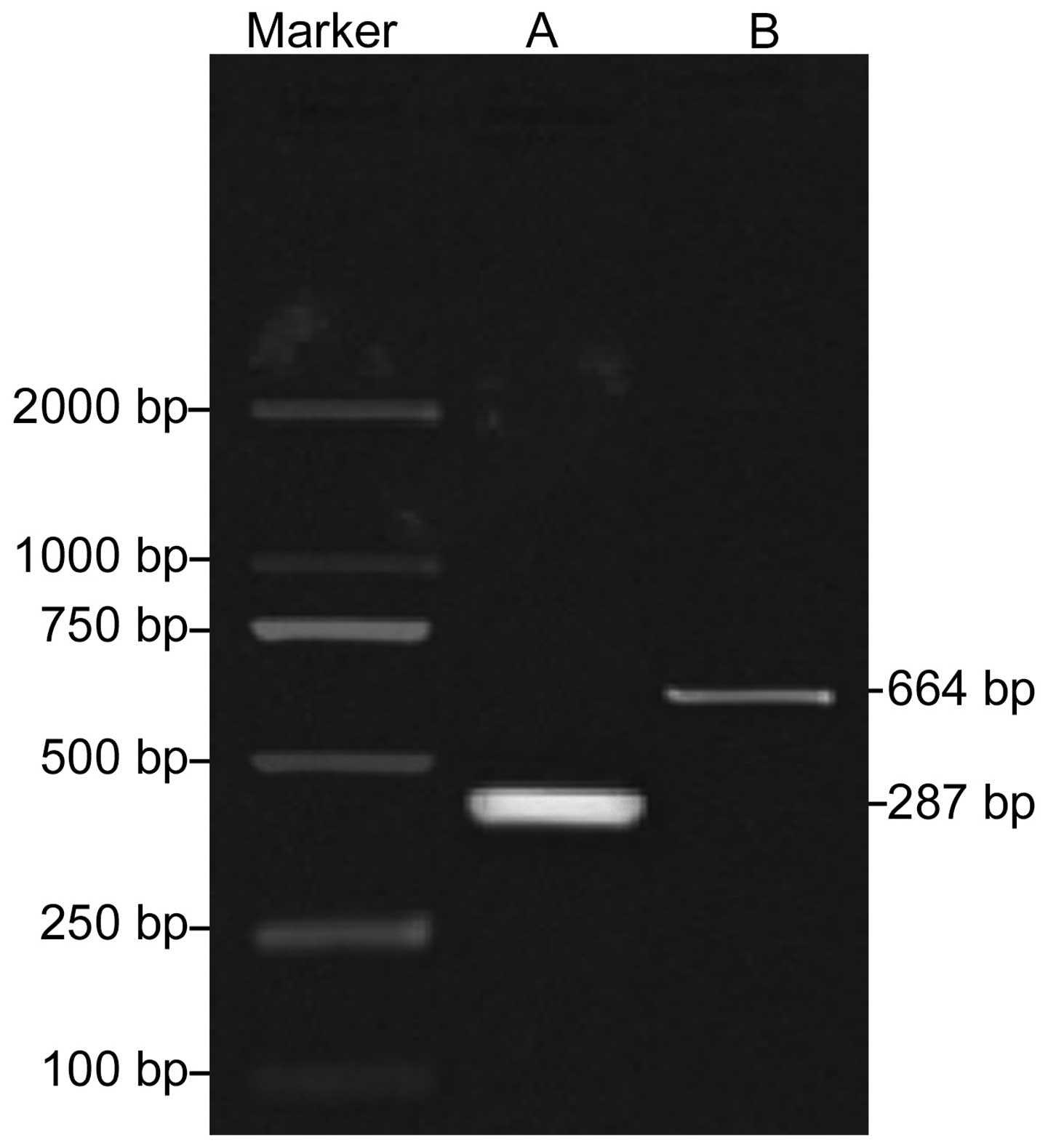
Construction and detection of the
pDsRed-CCL22 and pEGFP-IL-37 carrier
Fig. 2 shows the
enzyme digestion results of the pDsRed-CCL22 and pEGFP-IL-37
carrier. The fragment lengths of pDsRed-CCL22 and pEGFP-IL-37 were
287 and 664 bp, respectively, after the enzyme digestion by
XhoI and PstI, separately.
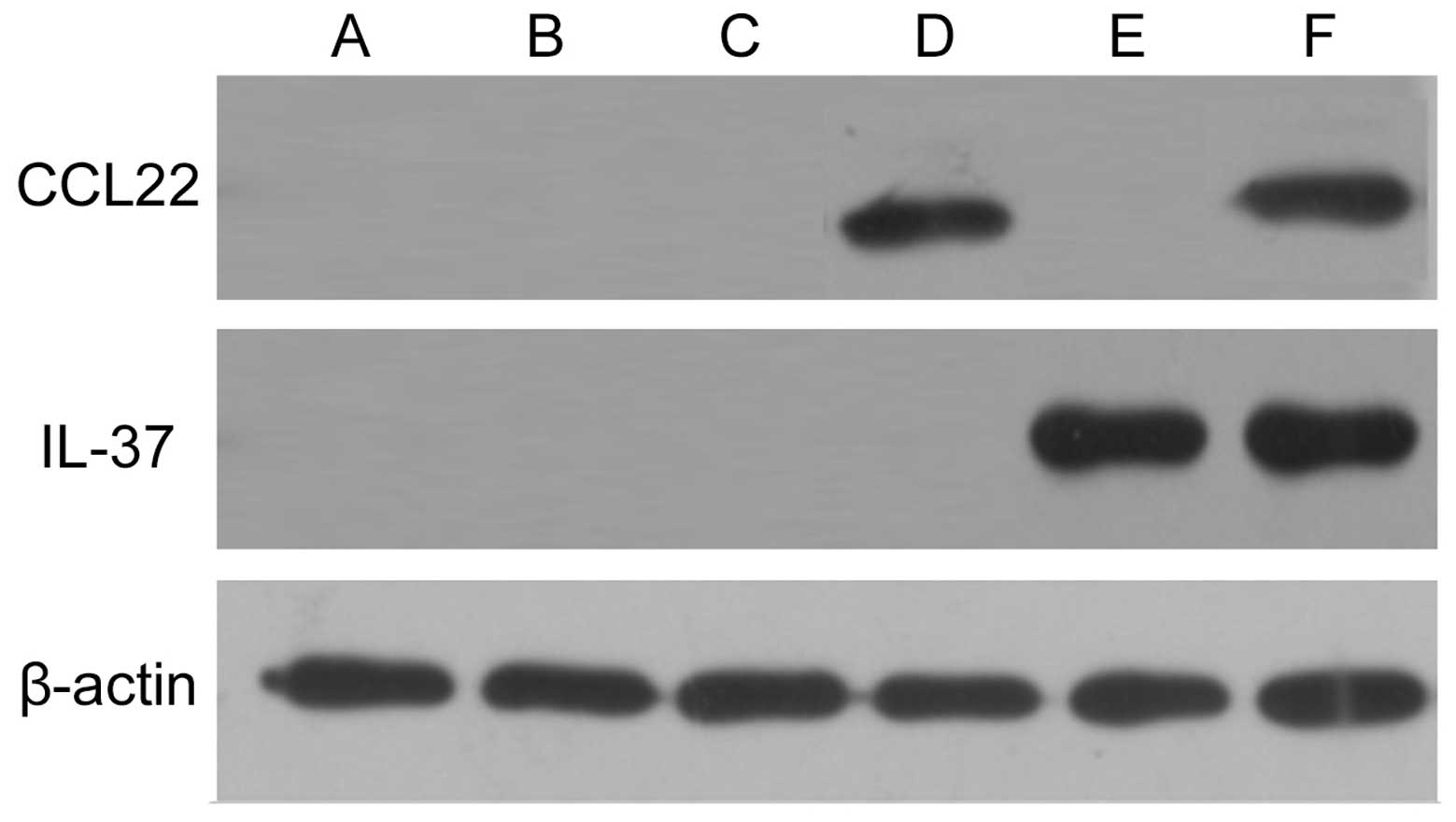
Expression levels of pDsRed-CCL22 and
pEGFP-IL-37 fusion proteins in the A549 cells
Expression levels of pDsRed-CCL22 and pEGFP-IL-37
fusion proteins in the A549 cells were detected by western
blotting. The results showed that no expression of CCL22 and IL-37
was found in the control, pDsRed-N1 blank plasmid and pEGFP-C1
blank plasmid groups; however, CCL22 was expressed in the
pDsRed-CCL22 plasmid group, IL-37 was expressed in the pEGFP-IL-37
plasmid group, and CCL22 and IL-37 were both found simultaneously
expressed in group 6 (Fig. 3).
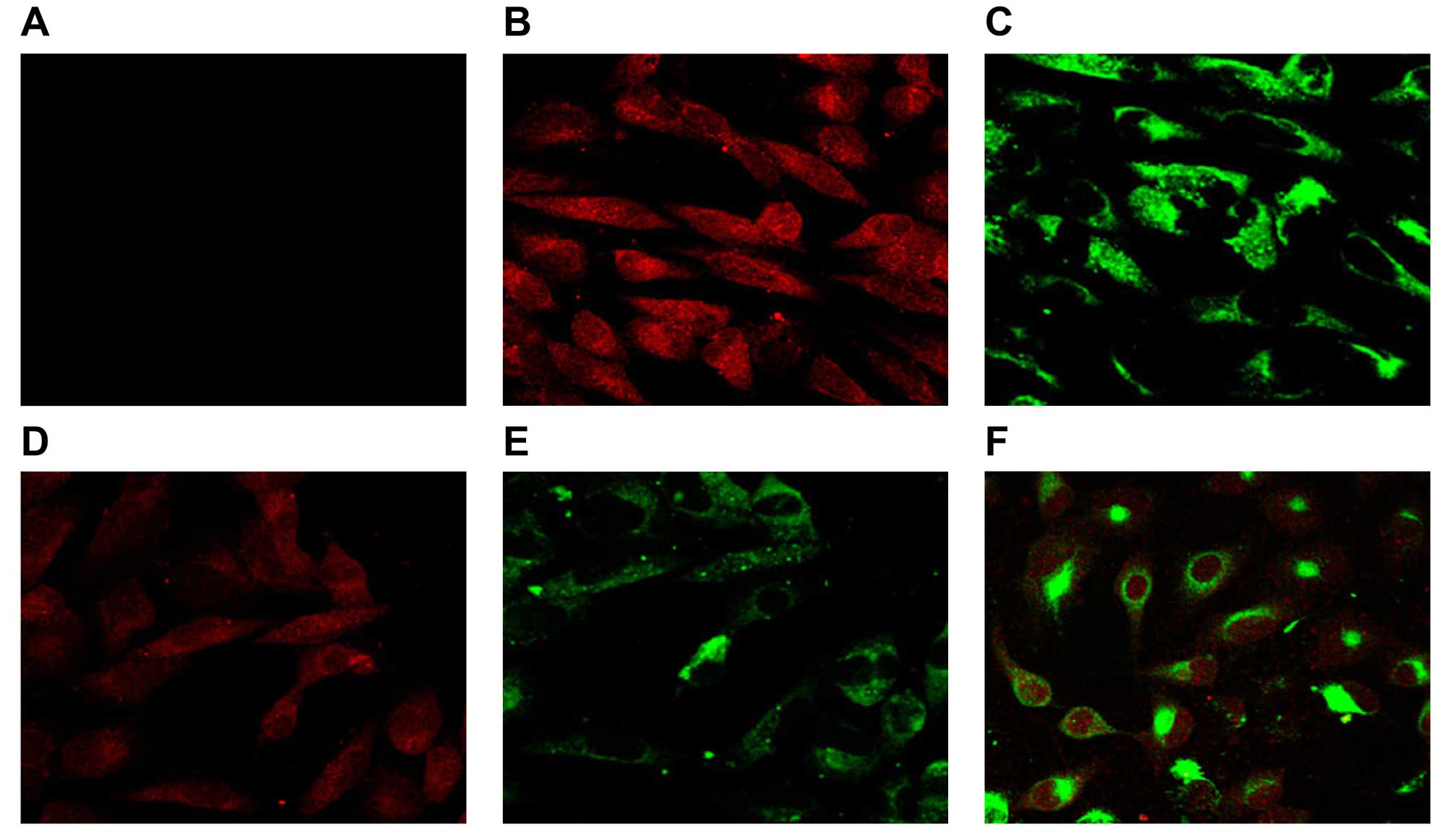
Result of confocal microscopy
detection
The results of confocal microscopy detection
(Fig. 4) showed that no
fluorescence was found in the control group, and fluorescence of
the pDsRed-N1 blank plasmid and pEGFP-C1 blank plasmid groups was
distributed in the cytoplasm. Expression of CCL22 and IL-37
proteins in the pDsRed-CCL22 plasmid and pEGFP-IL-37 plasmid
groups, respectively, were mainly distributed in the cytoplasm.
However, in the pDsRed-CCL22 + pEGFP-IL-37 plasmid group, CCL22 and
IL-37 proteins were both found in the cytoplasm. These results
revealed that CCL22 and IL-37 had a certain co-localization in the
A549 cells.
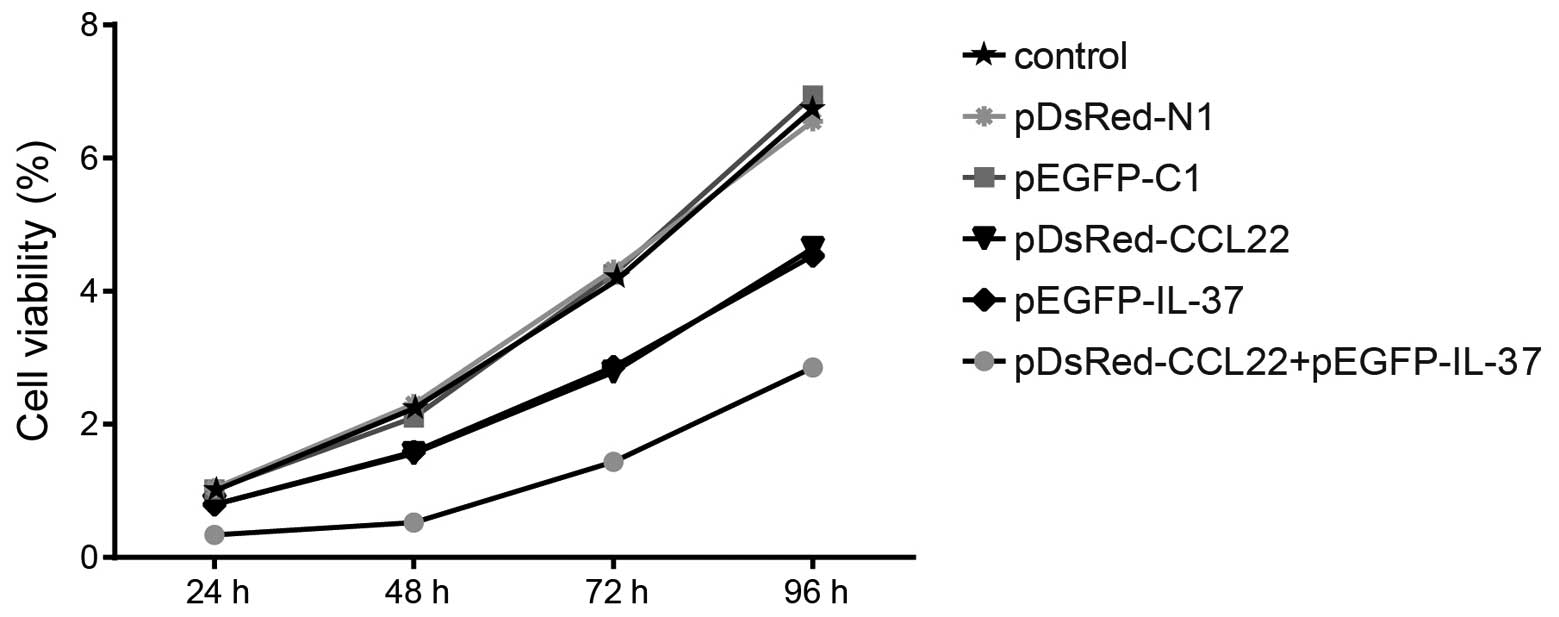
Proliferation of the six cell groups via
MTT assay
After being cultured for 24 h, the activity of the
A549 cells was detected every 24 h. The relative activity of the
cells in the control group was 1 at 24 h. After 24, 48, 72 and 96 h
of cell culture, no significant difference was found in the
control, pDsRed-N1 blank plasmid and pEGFP-C1 blank plasmid groups
(all P>0.05). When compared with the control, pDsRed-N1 blank
plasmid and pEGFP-C1 blank plasmid groups, lower cell proliferation
was found in the pDsRed-CCL22 plasmid, pEGFP-IL-37 plasmid and
pDsRed-CCL22 + pEGFP-IL-37 plasmid groups (all P<0.05). However,
the pDsRed-CCL22 + pEGFP-IL-37 plasmid group had lower cell
proliferation when compared with the pDsRed-CCL22 plasmid and
pEGFP-IL-37 plasmid groups alone (all P<0.05) (Fig. 5). These results confirmed that CCL22
and IL-37 inhibited the proliferation of the A549 cells.
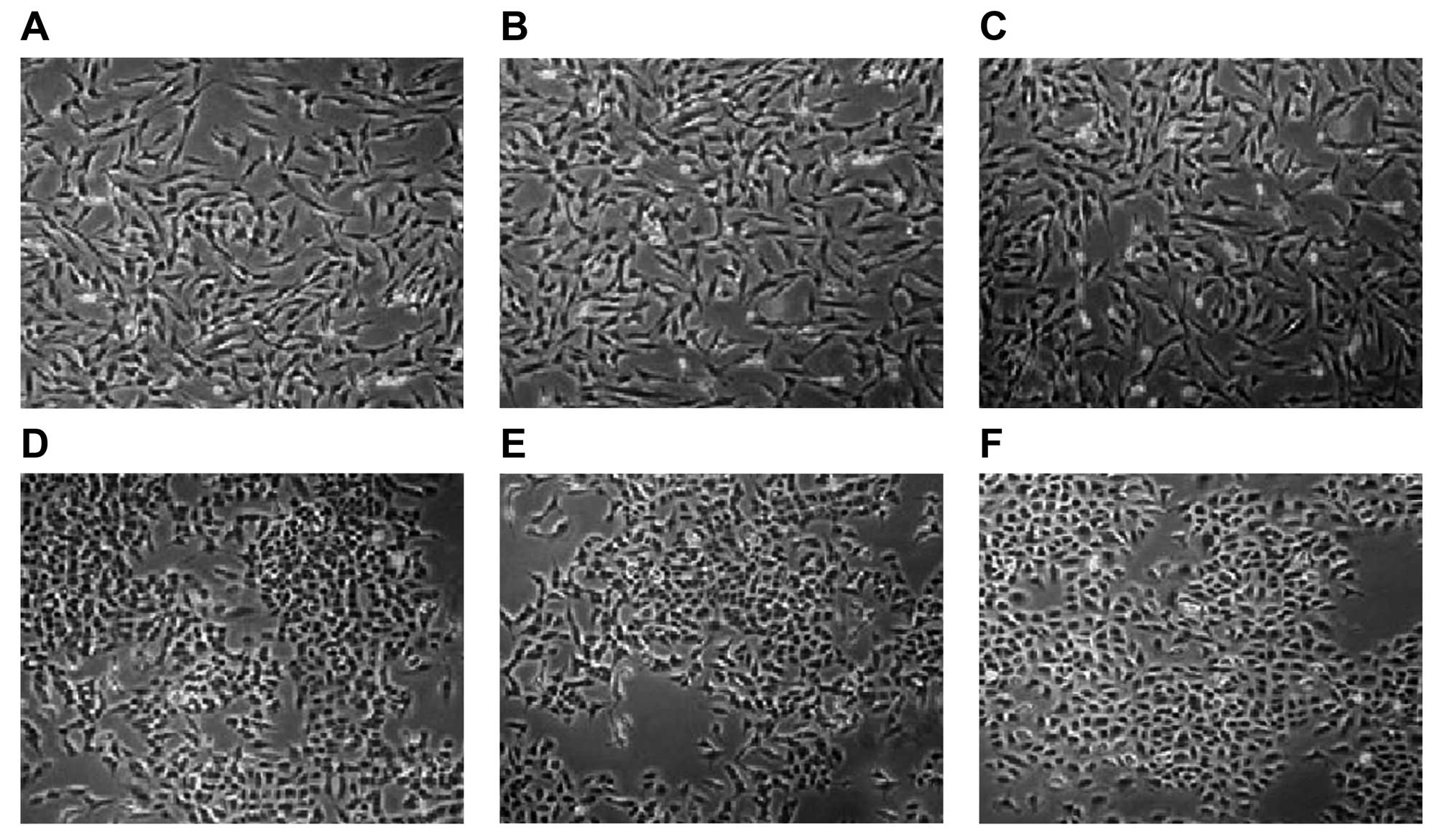
Cell morphologic observation under
EMT
After 72 h of cell culture, the EMT process was
observed under a microscope (Fig.
6). The results showed that obvious EMT was found in the
control, pDsRed-N1 blank plasmid, and pEGFP-C1 blank plasmid
groups. The cell morphologies were altered from pebble shaped cells
into fusiform cells with elongated cell appearance. In addition,
the cells were separated from peripheral cells, and the cells
exhibited loose connections with larger intercellular space
(Fig. 6A–C). EMT was also found in
a few cells in the pDsRed-CCL22 plasmid and pEGFP-IL-37 plasmid
groups. Cells showed fusiform shape and were loosely connected, but
most of the cell morphology did not change significantly (still
pebble shaped cells) (Fig. 6D and
E). While, in the pDsRed-CCL22 plasmid + pEGFP-IL-37 plasmid
group, epithelia maintained their pebble shape, tight junctions
between cells and no EMT was noted (Fig. 6F). These results revealed that CCL22
and IL-37 inhibited the EMT process of the A549 cells and the
inhibitory function was enhanced after the combination of CCL22 and
IL-37.
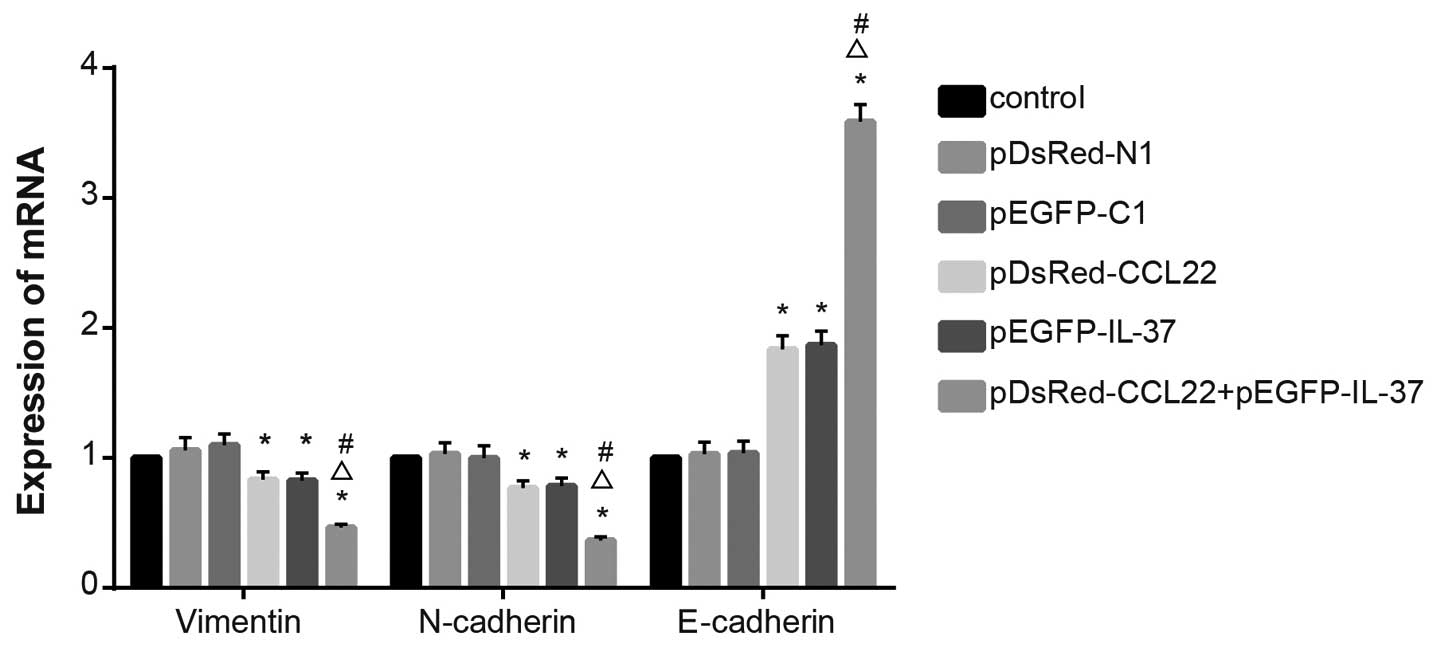
mRNA levels of EMT-related genes
During EMT, the expression levels of vimentin
and N-cadherin are increased, while the expression of
E-cadherin is decreased. The results of RT-qPCR revealed
that no significant difference was found when comparing
vimentin, N-cadherin and E-cadherin among the
control, the pDsRed-N1 blank plasmid, and the pEGFP-C1 blank
plasmid groups (P>0.05). Decreased vimentin and
N-cadherin mRNA expression levels, and increased
E-cadherin mRNA expression were found in the pDsRed-CCL22
plasmid, the pEGFP-IL-37 plasmid and the pDsRed-CCL22 plasmid +
pEGFP-IL-37 plasmid groups when compared with the control, the
pDsRed-N1 blank plasmid and the pEGFP-C1 blank plasmid groups (all
P<0.05). Decreased vimentin and N-cadherin mRNA
expression levels and increased E-cadherin mRNA expression
were found in the pDsRed-CCL22 plasmid + pEGFP-IL-37 plasmid groups
when compared with the pDsRed-CCL22 plasmid and the pEGFP-IL-37
plasmid group (all P<0.05) (Fig.
7). These results showed that CCL22 and IL-37 inhibited the EMT
process in the A549 cells.
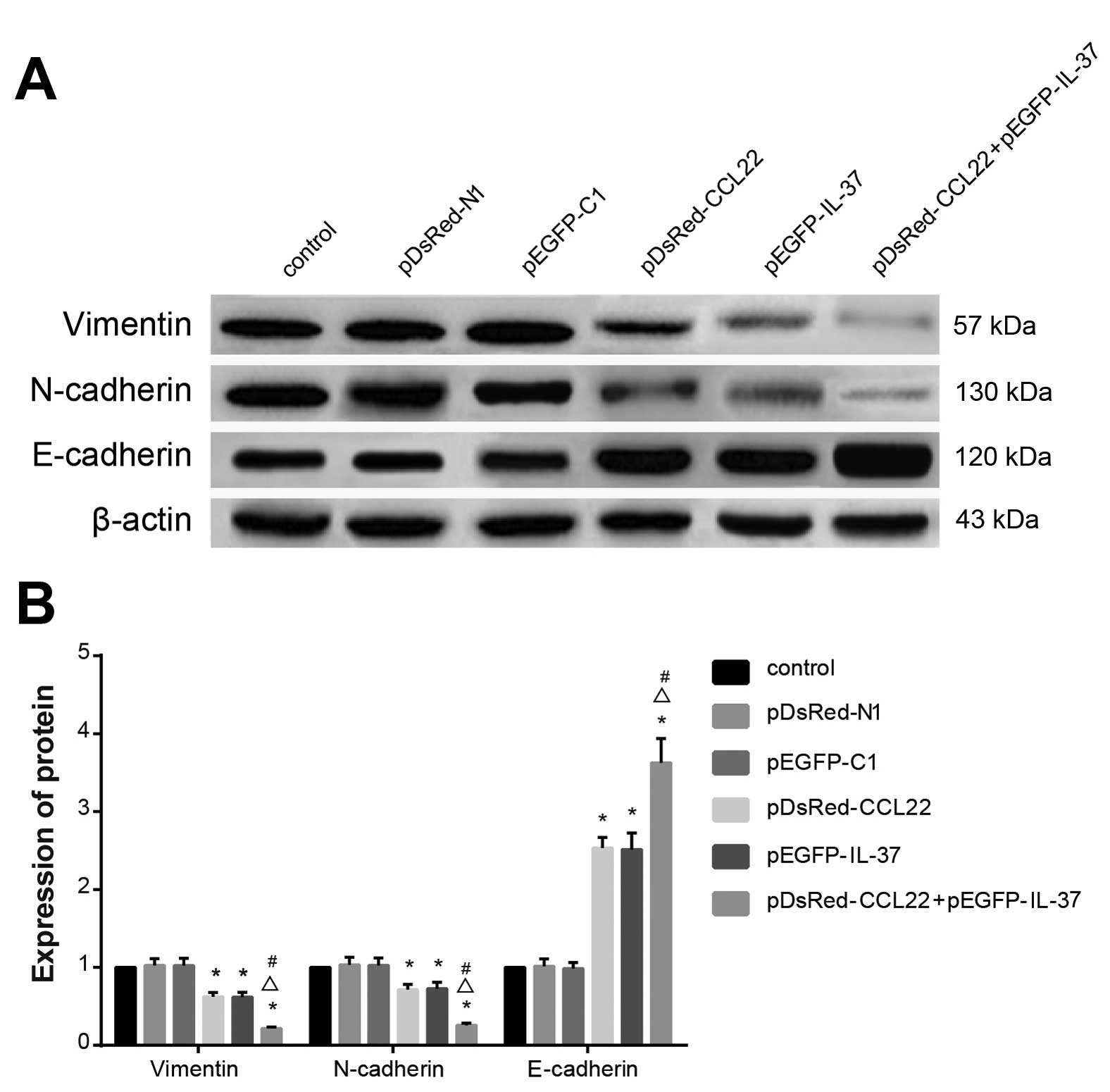
Protein expression levels of EMT-related
genes
The results from western blotting showed that no
significant differences were found when comparing vimentin,
N-cadherin and E-cadherin protein expression levels among the
control, pDsRed-N1 blank plasmid and pEGFP-C1 blank plasmid groups
(all P>0.05). Vimentin and N-cadherin expression levels were
decreased, and E-cadherin expression was increased in the
pDsRed-CCL22 plasmid, pEGFP-IL-37 plasmid and pDsRed-CCL22 plasmid
+ pEGFP-IL-37 plasmid groups when compared with the control,
pDsRed-N1 blank plasmid and pEGFP-C1 blank plasmid groups (all
P<0.05), and decreased vimentin and N-cadherin expression levels
and increased E-cadherin expression were found in the pDsRed-CCL22
plasmid + pEGFP-IL-37 plasmid groups when compared with the
pDsRed-CCL22 plasmid and the pEGFP-IL-37 plasmid group (all
P<0.05) (Fig. 8). These results
further confirmed that CCL22 and IL-37 inhibited the EMT process in
the A549 cells.
Discussion
The present study represents a comprehensive
analysis of CCL22 and IL-37 in regards to the proliferation and EMT
of A549 cells. Our in-depth analyses concluded that CCL22 and IL-37
with co-localization in the A549 cells inhibited the proliferation
and EMT process. The antitumor effects of CCL22 and IL37 provide a
strategy for the treatment of NSCLC.
In the present study, we found that when compared
with the control group, the pDsRed-N1 plasmid and the pEGFP-C1
plasmid group, lower cell proliferation was found in the
pDsRed-CCL22 plasmid, the pEGFP-IL-37 plasmid and pDsRed-CCL22
plasmid + pEGFP-IL-37 plasmid groups; moreover, the pDsRed-CCL22
plasmid + pEGFP-IL-37 plasmid group had lower cell proliferation
when compared with the pDsRed-CCL22 plasmid and the pEGFP-IL-37
plasmid group alone, suggesting that CCL22 and IL-37 can inhibit
the proliferation of the A549 cells. Lung cancer is one of the
typical inflammation-associated cancers, since smoking can lead to
chronic inflammation (such as chronic obstructive pulmonary
disease), and bronchioles and lungs are open organs which are more
receptive to suffer the stimulation from a variety of physical
factors that lead to inflammation (23). Chemokines represent a family of
endogenous mediators of inflammation attracting various leukocyte
subtypes to sites of infection and have been reported to affect
malignancy in various ways, either by promoting or inhibiting tumor
growth and metastasis formation via the activation of seven
transmembrane domain receptors coupled to heterotrimeric G proteins
(12,24). Previous evidence revealed that the
binding of chemokines and their receptors can induce aggregation of
actin, and regulate cell movement and migration (25). As far as we know, the main source of
CCL22 is dendritic cells, which are the most powerful
antigen-presenting cells in vivo, and capable of stimulating
T cells into Th1 and Th2; and Th1 can secret interferon-γ and
inhibit CCL22, thereby inhibiting the proliferation of tumor cells
(26). IL-37 is a cytokine which
plays an important role in inflammation, autoimmunity, and other
immunological disorders, and is synthesized as a protein which is
processed to its mature form after stimulation (27). It has also been reported that IL-37
has the ability to kill microorganisms and increase production of
several cytokines for the expansion of the acquired immune function
(28). One study also demonstrated
that IL-37 downregulated inflammation in a mouse model (29). Mature IL-37 can bind to Smad3 to
regulate the key enzymes of multiple signaling pathways, including
focal adhesion kinase (FAK), proline-rich tyrosine kinase (Pyk2),
MAP kinase p38α, signal transducer and activator of transcription
(STAT) p53 and mTOR. The expression of these key enzymes is
downregulated and these pathways have been confirmed to be related
to cell proliferation and migration (17).
Furthermore, we also found that vimentin and
N-cadherin expression levels were decreased, and E-cadherin
expression was increased in the groups transfected with CCL22,
IL-37 or both CCL22 and IL-37, suggesting that CCL22 and IL-37 can
inhibit the EMT process in A549 cells. EMT was first described in
the 1980's, and it was reported that EMT is not only a key
biological process during embryonic morphogenesis, but also one of
the earliest steps in solid tumor progression. EMT is related to
tumor growth, metastasis as well as invasion and results in the
conversion of tumors from low- to high-grade malignancy (30). E-cadherin, a crucial mediator of
EMT, has been proven to result in rapid cell growth and metastasis
in most cell types by loss of cell adhesion when E-cadherin loses
its function (31). Li and Liu
demonstrated that macrophages and activated fibroblasts can release
a large number of growth factors, chemokines and matrix proteases,
and then activate multiple signaling pathways such as the Smads,
the ERK-MAPK and the PI3K-AKT signaling pathways to initiate EMT
(32). Thus, we suspect that CCL22
and IL-37 inhibit the EMT process via multiple signaling pathways,
including the Smads, the ERK-MAPK and the PI3K-AKT signaling
pathways. However, the underlying mechanism of CCL22 and IL-37
inhibition in the EMT process warrants additional confirmation by
further study.
Collectively, the present study demonstrated that
CCL22 and IL-37 with co-localization in the A549 cells inhibited
the proliferation and the EMT process in the A549 cells. The
antitumor effects of CCL22 and IL37 provide a strategy for the
treatment of NSCLC. The data from the present study can be explored
by studying the effects of cytokines on the invasion and metastasis
of NSCLC and by considering different cell lines for further
substantiation of the aforementioned results.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (No. 30972779,
81273237 and 81102850), the Science and Technology Project of
Zhanjiang (2016B01036) and the Guangdong Medical College Research
Fund (No. M2014026). We would like to acknowledge the reviewers for
their helpful comments on this study.
References
|
1
|
Aberle DR, Adams AM, Berg CD, Black WC,
Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM and Sicks
JD; National Lung Screening Trial Research Team: Reduced
lung-cancer mortality with low-dose computed tomographic screening.
N Engl J Med. 365:395–409. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu H, Qiao N, Wang Y, Jiang M, Wang S,
Wang C and Hu L: Association between the telomerase reverse
transcriptase (TERT) rs2736098 polymorphism and cancer risk:
Evidence from a case-control study of non-small-cell lung cancer
and a meta-analysis. PLoS One. 8:e763722013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu J, Yin Z, Gao W, Liu L, Yin Y, Liu P
and Shu Y: Genetic variation in a microRNA-502 minding site in SET8
gene confers clinical outcome of non-small cell lung cancer in a
Chinese population. PLoS One. 8:e770242013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pao W and Girard N: New driver mutations
in non-small-cell lung cancer. Lancet Oncol. 12:175–180. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dela Cruz CS, Tanoue LT and Matthay RA:
Lung cancer: Epidemiology, etiology, and prevention. Clin Chest
Med. 32:605–644. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Raz DJ, Gomez SL, Chang ET, Kim JY, Keegan
TH, Pham J, Kukreja J, Hiatt RA and Jablons DM: Epidemiology of
non-small cell lung cancer in Asian Americans: Incidence patterns
among six subgroups by nativity. J Thorac Oncol. 3:1391–1397. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Albain KS, Swann RS, Rusch VW, Turrisi AT
III, Shepherd FA, Smith C, Chen Y, Livingston RB, Feins RH, Gandara
DR, et al: Radiotherapy plus chemotherapy with or without surgical
resection for stage III non-small-cell lung cancer: A phase III
randomised controlled trial. Lancet. 374:379–386. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kitano H, Chung JY, Ylaya K, Conway C,
Takikita M, Fukuoka J, Doki Y, Hanaoka J and Hewitt SM: Profiling
of phospho-AKT, phospho-mTOR, phospho-MAPK and EGFR in non-small
cell lung cancer. J Histochem Cytochem. 62:335–346. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee SH, Suh IB, Lee EJ, Hur GY, Lee SY,
Lee SY, Shin C, Shim JJ, In KH, Kang KH, et al: Relationships of
coagulation factor XIII activity with cell-type and stage of
non-small cell lung cancer. Yonsei Med J. 54:1394–1399. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song G, Qin T, Liu H, Xu GB, Pan YY, Xiong
FX, Gu KS, Sun GP and Chen ZD: Quantitative breath analysis of
volatile organic compounds of lung cancer patients. Lung Cancer.
67:227–231. 2010. View Article : Google Scholar
|
|
12
|
Ma H, Shu Y, Pan S, Chen J, Dai J, Jin G,
Hu Z and Shen H: Polymorphisms of key chemokine genes and survival
of non-small cell lung cancer in Chinese. Lung Cancer. 74:164–169.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Singh S, Sadanandam A and Singh RK:
Chemokines in tumor angiogenesis and metastasis. Cancer Metastasis
Rev. 26:453–467. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Niens M, Visser L, Nolte IM, van der
Steege G, Diepstra A, Cordano P, Jarrett RF, Te Meerman GJ, Poppema
S and van den Berg A: Serum chemokine levels in Hodgkin lymphoma
patients: Highly increased levels of CCL17 and CCL22. Br J
Haematol. 140:527–536. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Freier CP, Kuhn C, Rapp M, Endres S, Mayr
D, Friese K, Anz D and Jeschke U: Expression of CCL22 and
infiltration by regulatory T cells are increased in the decidua of
human miscarriage placentas. Am J Reprod Immunol. 74:216–227. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tete S, Tripodi D, Rosati M, Conti F,
Maccauro G, Saggini A, Cianchetti E, Caraffa A, Antinolfi P,
Toniato E, et al: IL-37 (IL-1F7) the newest anti-inflammatory
cytokine which suppresses immune responses and inflammation. Int J
Immunopathol Pharmacol. 25:31–38. 2012.PubMed/NCBI
|
|
17
|
Nold MF, Nold-Petry CA, Zepp JA, Palmer
BE, Bufler P and Dinarello CA: IL-37 is a fundamental inhibitor of
innate immunity. Nat Immunol. 11:1014–1022. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu D, Wang A, Jiang F, Hu J and Zhang X:
Effects of interleukin-37 on cardiac function after myocardial
infarction in mice. Int J Clin Exp Pathol. 8:5247–5251.
2015.PubMed/NCBI
|
|
19
|
Xia L, Shen H and Lu J: Elevated serum and
synovial fluid levels of interleukin-37 in patients with rheumatoid
arthritis: Attenuated the production of inflammatory cytokines.
Cytokine. 76:553–557. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang G, Yu D, Tan W, Zhao D, Wu C and Lin
D: Genetic polymorphism in chemokine CCL22 and susceptibility to
Helicobacter pylori infection-related gastric carcinoma. Cancer.
115:2430–2437. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Erfani N, Nedaei Ahmadi AS, Ghayumi MA and
Mojtahedi Z: Genetic polymorphisms of CCL22 and CCR4 in patients
with lung cancer. Iran J Med Sci. 39:367–373. 2014.PubMed/NCBI
|
|
22
|
Dougan M, Li D, Neuberg D, Mihm M, Googe
P, Wong KK and Dranoff G: A dual role for the immune response in a
mouse model of inflammation-associated lung cancer. J Clin Invest.
121:2436–2446. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jin Y, Xiong X, Su Y, Hu J and Tao X:
Serum vascular endothelial growth factor levels in patients with
non-small cell lung cancer and its relations to the micrometastasis
in peripheral blood. J Huazhong Univ Sci Technolog Med Sci.
29:462–465. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thelen M and Stein JV: How chemokines
invite leukocytes to dance. Nat Immunol. 9:953–959. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Speyer CL and Ward PA: Role of endothelial
chemokines and their receptors during inflammation. J Invest Surg.
24:18–27. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xia S, Wei J, Wang J, Sun H, Zheng W, Li
Y, Sun Y, Zhao H, Zhang S, Wen T, et al: A requirement of dendritic
cell-derived interleukin-27 for the tumor infiltration of
regulatory T cells. J Leukoc Biol. 95:733–742. 2014. View Article : Google Scholar
|
|
27
|
Boraschi D, Lucchesi D, Hainzl S, Leitner
M, Maier E, Mangelberger D, Oostingh GJ, Pfaller T, Pixner C,
Posselt G, et al: IL-37: A new anti-inflammatory cytokine of the
IL-1 family. Eur Cytokine Netw. 22:127–147. 2011.PubMed/NCBI
|
|
28
|
Ma P, Gu B, Xiong W, Tan B, Geng W, Li J
and Liu H: Glimepiride promotes osteogenic differentiation in rat
osteoblasts via the PI3K/Akt/eNOS pathway in a high glucose
microenvironment. PLoS One. 9:e1122432014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maloy KJ and Powrie F: Intestinal
homeostasis and its breakdown in inflammatory bowel disease.
Nature. 474:298–306. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lindsey S and Langhans SA: Crosstalk of
oncogenic signaling pathways during epithelial-mesenchymal
transition. Front Oncol. 4:3582014. View Article : Google Scholar
|
|
31
|
Nagathihalli NS and Merchant NB:
Src-mediated regulation of E-cadherin and EMT in pancreatic cancer.
Front Biosci. 17:2059–2069. 2012. View
Article : Google Scholar
|
|
32
|
Li MX and Liu BC: Epithelial to
mesenchymal transition in the progression of tubulointerstitial
fibrosis. Chin Med J. 120:1925–1930. 2007.PubMed/NCBI
|