Introduction
Methylation of tumor suppressor gene promoters, one
of the most common events in various types of cancers, is generally
tumor-specific (1,2). Methylation of circulating tumor DNA
(ctDNA) can be detected in the plasma or serum of breast cancer
patients (3–9); thus, methylated ctDNA is a promising
cancer biomarker (2). Several genes
including GSTP1, RASSF1A and RARβ2 are
methylated in breast tumor tissue (3,10,11),
and the methylation of these genes can be detected in ctDNA of
breast cancer patients (3,4,6).
Although several studies have investigated the clinical application
of these methylated ctDNAs as prognostic indicators (4,5) or
monitoring markers of systemic therapy (7, 8), a
more sensitive and specific methylated ctDNA marker is needed.
SEPT9 belongs to a family of GTP-binding
proteins recognized as components of the cytoskeleton. These
proteins are involved in several cellular processes including
membrane trafficking, cytokinesis, angiogenesis, and cell
proliferation (12,13). SEPT9 consists of at least
seven transcripts with diverse functions (13,14).
Some of these transcripts possess tumor suppressor functions, while
others have oncogenic properties (15–18).
SEPT9_v2 has been shown to be epigenetically modified in
colorectal cancer, and an assay for detecting methylated
SEPT9_v2 ctDNA in plasma has been developed and validated
for clinical use as a marker for the early detection of colorectal
cancer (19–21). Recently, a more sensitive and
specific (48 and 92%, respectively) assay for methylated
SEPT9_v2 has been developed for colorectal cancer screening
(22). SEPT9 methylation can
be also detected in other types of cancers, including breast cancer
(23), suggesting that the assay
for methylated SEPT9_v2 in plasma may be useful in breast
cancer patients.
The aim of the present study was to determine
whether methylation of the SEPT9_v2 promoter was associated
with the expression of this gene in breast cancer cells. In
addition, we sought to clarify the clinicopathological
characteristics of breast tumors containing methylated
SEPT9_v2. We analyzed the methylation of the SEPT9_v2
promoter using next generation sequencing (NGS), which provides a
quantitative methylation index (MI) within a broad CpG area.
Lastly, we examined whether methylated SEPT9_v2 ctDNA can be
detected in the plasma of breast cancer patients, and explored its
utility as a novel blood biomarker for breast cancer diagnosis.
Materials and methods
Patients and breast tumor samples
Study I
Nineteen pairs of tumor and normal tissues were
obtained at surgery between 2001 and 2004 from primary breast
cancer (PBC) patients who received no preoperative chemotherapy or
hormonal therapy. The clinicopathological characteristics of these
patients are summarized in Table I.
Normal tissue samples were obtained from the quadrant not harboring
cancer. Tissue samples were snap-frozen in liquid nitrogen and kept
at −80°C until use.
 | Table IClinicopathological characteristics
of breast tumors (study I). |
Table I
Clinicopathological characteristics
of breast tumors (study I).
|
Characteristics | No. of
patients | % |
|---|
| All cases | 19 | |
| Age (years) |
| <50 | 9 | 52.6 |
| ≥50 | 10 | 47.4 |
| Menopausal
status |
| Pre | 10 | 52.6 |
| Post | 9 | 47.4 |
| Tumor size
(cm) |
| <2 | 5 | 26.3 |
| ≥2 | 14 | 73.7 |
| Lymph node
metastasis |
| Negative | 13 | 68.4 |
| Positive | 6 | 31.6 |
| Histological
type |
| IDC | 18 | 94.7 |
| Special type | 1 | 5.26 |
| Histological
grade |
| 1,2 | 13 | 68.4 |
| 3 | 6 | 31.6 |
| ER |
| Negative | 7 | 36.8 |
| Positive | 12 | 63.2 |
| PR |
| Negative | 10 | 52.6 |
| Positive | 9 | 47.4 |
| HER2 |
| Negative | 12 | 63.2 |
| Positive | 7 | 36.8 |
| Ki67 |
| Low | 5 | 26.3 |
| High | 1 | 5.26 |
| Unknown | 13 | 68.4 |
| Subtype
(IHC)a |
| Luminal A | 9 | 47.4 |
| Luminal B | 3 | 15.8 |
| HER2 | 4 | 21.1 |
| Triple
negative | 3 | 15.8 |
Study II
Tumor samples from stage II or III PBC patients
(n=107) were retrospectively included in the present study. These
patients had been treated at Osaka University Hospital between 2004
and 2009 with neoadjuvant chemotherapy (NAC) consisting of
paclitaxel (80 mg/m2) weekly for 12 cycles followed by
5-fluorouracil (500 mg/m2), epirubicin (75
mg/m2) and cyclophosphamide (500 mg/m2) every
three weeks for four cycles. Each patient underwent vacuum-assisted
biopsy of the tumors, and tumor samples were snap-frozen in liquid
nitrogen and kept at −80°C until use. Histological grade, estrogen
receptor (ER), progesterone receptor (PR) and HER2 status were
determined as previously described (11). Ki67 was defined as 'high' when ≥20%
of tumor cells was immunohistochemically positive (clone; MIB-1). A
pathological complete response (pCR) was defined as no evidence of
invasive cancer components in the breast irrespective of axilla
lymph node metastases. Intrinsic subtypes were determined by DNA
microarray using the PAM50 method (24,25).
The clinicopathological characteristics of these patients are
summarized in Table II.
 | Table IIComparison of the SEPT9_v2 MI
with clinicopathological parameters of breast tumors (study
II). |
Table II
Comparison of the SEPT9_v2 MI
with clinicopathological parameters of breast tumors (study
II).
|
Characteristics | No. of pts. | SEPT9_v2
|
|---|
| MI (mean ± SD) | P-value |
|---|
| All cases | 107 | | |
| Age (years) | | | 0.573 |
| <50 | 49 | 10.1±16.1 | |
| ≥50 | 58 | 11.7±12.8 | |
| Menopausal
status | | | 0.657 |
| Pre | 51 | 11.5±15.9 | |
| Post | 56 | 10.3±12.9 | |
| T stage | | | 0.539 |
| T1,2 | 84 | 10.3±12.6 | |
| T3, 4 | 23 | 13.0±19.8 | |
| Lymph node
metastasis | | | 0.486 |
| Negative | 30 | 12.5±15.5 | |
| Positive | 77 | 10.2±13.9 | |
| Stage | | | 0.467 |
| II | 88 | 10.2±12.5 | |
| III | 19 | 13.9±21.2 | |
| Histological
type | | | 0.123 |
| IDC | 96 | 10.3±14.7 | |
| Special type | 11 | 15.9±10.2 | |
| ER | | | <0.001 |
| Negative | 42 | 3.2±5.6 | |
| Positive | 65 | 15.8±16.0 | |
| PR | | | 0.002 |
| Negative | 65 | 7.4±13.3 | |
| Positive | 42 | 16.2±14.5 | |
| HER2 | | | 0.592 |
| Negative | 76 | 11.4±13.8 | |
| Positive | 31 | 9.6±15.7 | |
| Subtype
(IHC)a | | | <0.001b |
| Luminal A | 51 | 15.8±14.6 | |
| Luminal B | 14 | 16.1±21.0 | |
| HER2 | 17 | 4.3±6.3 | |
| Triple
negative | 25 | 2.4±5.1 | |
| Subtype
(PAM50) | | | <0.001b |
| Luminal A | 29 | 15.6±13.5 | |
| Luminal B | 21 | 14.3±16.8 | |
| HER2 | 16 | 15.2±20.7 | |
| Basal-like | 23 | 3.0±6.7 | |
| Normal-like | 18 | 5.4±5.7 | |
| Histological
grade | | | <0.001 |
| 1,2 | 86 | 12.7±15.3 | |
| 3 | 21 | 3.5±5.5 | |
| Ki67 | | | 0.116 |
| Low | 44 | 13.6±15.0 | |
| High | 62 | 9.1±13.8 | |
| Unknown | 1 | 0.7 | |
| Pathological
response | | | <0.001 |
| Non-pCR | 74 | 13.4±15.7 | |
| pCR | 33 | 5.2±8.47 | |
Study III
For the measurement of methylated SEPT9_v2
ctDNA in plasma, 2 ml plasma samples were obtained from healthy
controls (n=51), stage II or III PBC patients (n=82) and metastatic
breast cancer (MBC) patients (n=50) at Osaka University Hospital
and Osaka Police Hospital between 2012 and 2014. Among these
patients, frozen tumor tissues or formalin-fixed paraffin-embedded
(FFPE) tumor tissues were available from 49 PBC and 25 MBC
patients. These tumor tissues were subjected to the SEPT9_v2
methylation assay. These studies were approved by the Institutional
Review Board for Clinical Research, Osaka University Graduate
School of Medicine and the Osaka Police Hospital Ethical Committee.
Informed consent was obtained from each patient before
sampling.
DNA extraction and sodium bisulfite
treatment
Total DNA from cell lines was isolated using
TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) and
total DNA from snap-frozen breast tissue was extracted using the
DNeasy® Blood and Tissue kit (Qiagen, Valencia, CA,
USA). For DNA extraction from FFPE tumor tissues, three to five
10-μm sections/tumor were cut from the FFPE tumor tissues,
and the tumor area was dissected with a scalpel under a
stereoscopic assistance. Total DNA from the paraffin sections was
extracted using the QIAamp DNA FFPE kit (Qiagen). For the laser
captured microdissection (LCM), a 10 μm section of the FFPE
tumor tissues was mounted onto a polyethylene napthalate membrane
slide (Leica Microsystems GmbH, Wetzlar, Germany), and the
epithelium or stroma was separately collected with the laser
microdissection system LMD7000 (Leica) (26) and DNA was extracted using the QIAamp
DNA Micro kit (Qiagen). One microgram of genomic DNA was subjected
to sodium bisulfite treatment with the EpiTect®
Bisulfite kit (Qiagen). Plasma DNA was extracted using the
QIAamp® Circulating Nucleic Acid kit (Qiagen) from a 2
ml plasma sample and subjected to sodium bisulfite treatment as
previously described (4).
Quantitative SEPT9_v2 promoter
methylation analysis using NGS and methylation-specific polymerase
chain reaction
The NGS methylation assay was performed with the GS
Junior system (Roche Diagnostics, Basel, Switzerland) according to
the manufacturer's instructions. Data were analyzed using GS
Amplicon Variant Analyzer software ver. 2.7 (Roche Diagnostics).
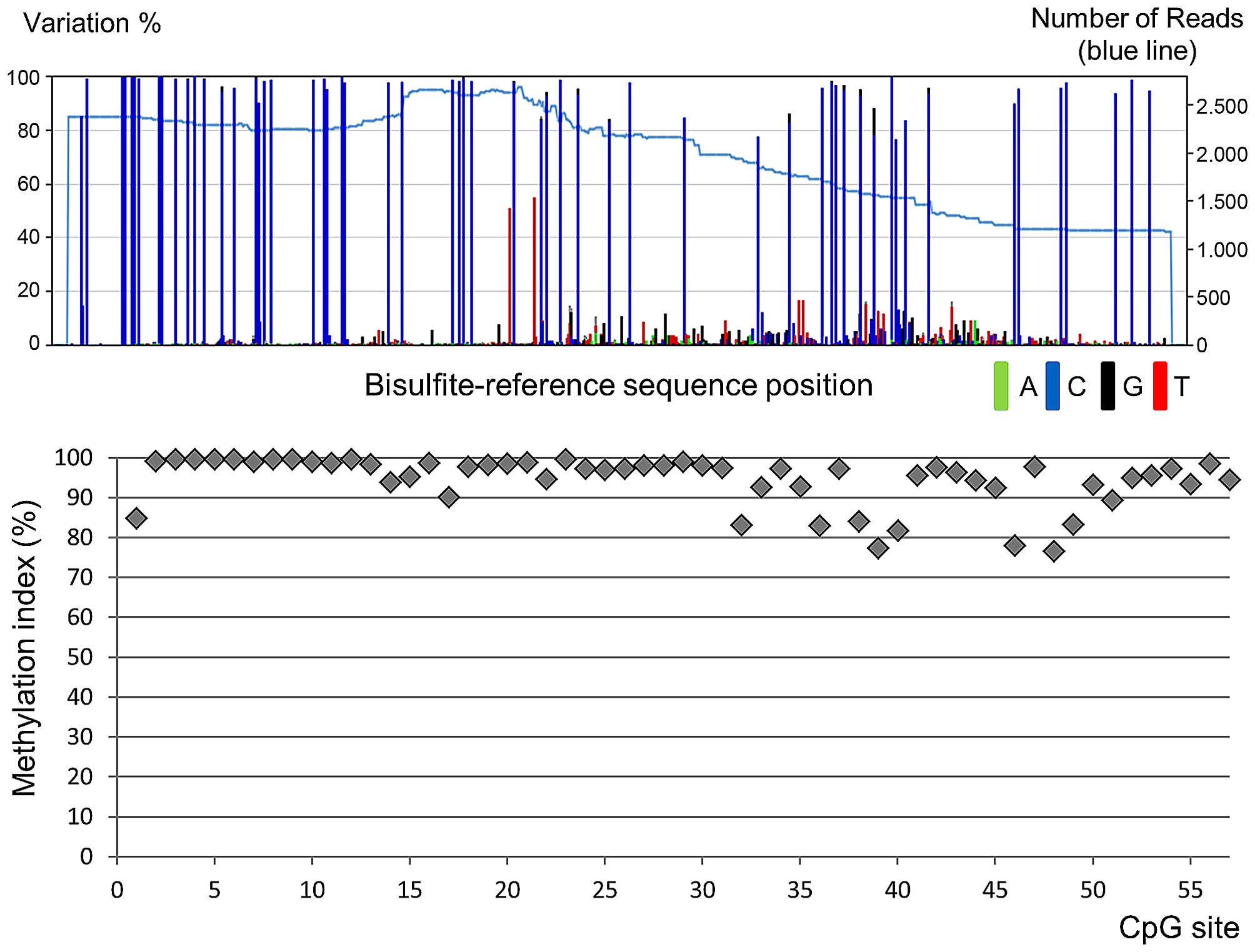
The MI was calculated by dividing the number of cytosines by the
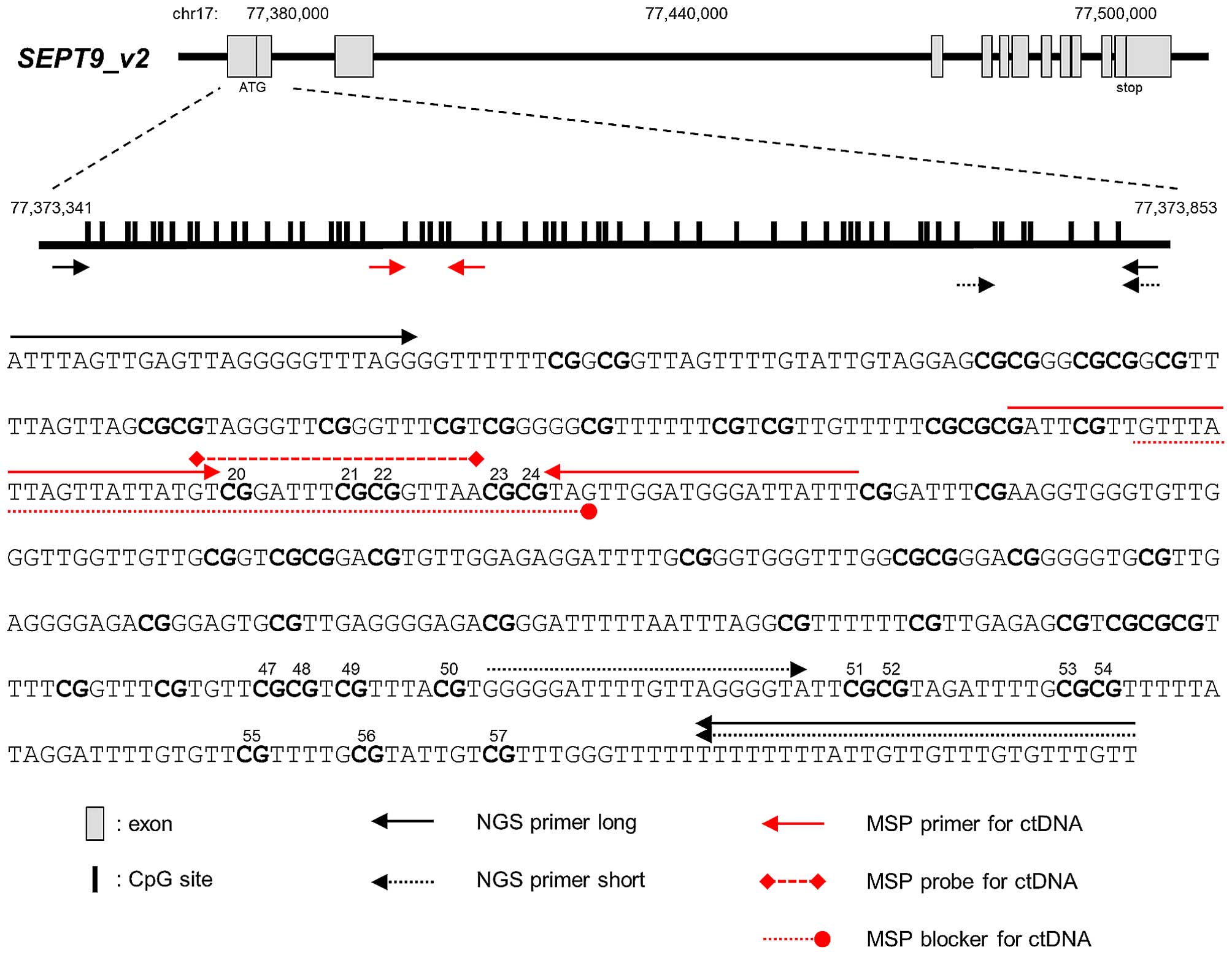
total reads at each CpG site. NGS primers used for SEPT9_v2
methylation of frozen tissue or cell lines were designed as
follows: forward, 5′-ATTTAGTTGAGTTAGGGGGTTTAGG-3′ and reverse,
5′-AACAAACACAAACAACAATAAAAAAAA-3′ (NGS primer long, Fig. 1). Among all 57 CpG sites, the
average MI of the 11 sites (47–57th CpGl; Fig. 1) showing the most significant
difference between cancer and normal tissue was used for the
statistical analysis of methylation. NGS primers used for DNA from
FFPE specimens were designed as follows: forward,
5′-GGGGGATTTTGTTAGGGGTA-3′ and reverse,
5′-AACAAACACAAACAACAATAAAAAAAA-3′ (NGS primer short, Fig. 1). The NGS short primer included
seven CpG sites, equivalent to 51st–57th CpG (Fig. 1). The cut-off of MI ≥10% was used to
define the Hypermethylation of SEPT9_v2 in the breast cancer
cell lines and breast tumors according to the previous studies
(27–30). For detecting the methylated
SEPT9_v2 in plasma and in the epithelial or stromal cells
obtained by LCM, a quantitative methylation-specific polymerase
chain reaction (MSP) assay was performed with the protocol modified
as previously described (20). The
SEPT9_v2 oligonucleotide sequence including the 20–24th CpG
sites (Fig. 1) for detection of
methylated SEPT9_v2 ctDNA in plasma were designed as
follows: forward, 5′-AAATAATCCCATCCAACTA-3′ and reverse,
5′-GATT-d-spacer-GTTGTTTATTAGTTATTATGT-3′ (Fig. 1) (20). The rCpG sequence was used as a
reference control as we previously reported (3). The SEPT9_v2 and rCpG PCR
reactions were performed using a 9 μl aliquot of the
bisulfite DNA eluate, in a 25 μl total volume using 96-well
plates. Methylated SEPT9_v2 ctDNA in plasma was defined as
positive when quantification cycles were <50 for SEPT9_v2
and <33.7 for rCpG as the loading reference (22).
In situ hybridization for SEPT9_v2
mRNA
The QuantiGene®ViewRNA In Situ
hybridization Tissue Assay kit (Affymetrix, Santa Clara, CA, USA)
was used according to the manufacturer's protocol. FFPE sections (4
μm) of tumor tissue were incubated for 20 min at 98°C with a
pretreatment solution followed by protease digestion for 10 min.
The SEPT9_v2- or glyceraldehyde 3-phosphate dehydrogenase
(GAPDH)-specific view RNA™ Probe set (Affymetrix) was
hybridized for 3 h. This probe set was designed to hybridize the
SEPT9_v2-specific sequence with a length of 342 bp, which
comprises 25% of the length regularly required for in situ
hybridization (ISH) (chromosome 17; 77,373,207 to 77,373,548). ISH
images were captured by a fluorescent microscope (Bz-9000; Keyence,
Osaka, Japan). Signal intensity was semi-quantitatively determined
based on the number of cytoplasmic fluorescent dots in five
non-overlapping fields at high-power magnification (×400).
Isolation of breast tumor cells by
magnetic-activated cell sorting
Breast tumor cells were isolated from the FFPE tumor
tissue by magnetic-activated cell sorting (MACS) using the EasySep
Human EpCAM Positive Selection Cocktail, the EasySep Human MUC1
Positive Selection Cocktail, and EasySep Magnetic Particles (Stem
Cell Technologies, Vancouver, BC, Canada) as previously described
(31). Total DNA was extracted from
the isolated tumor cells using the QIAamp® DNA FFPE
Tissue kit (Qiagen).
Demethylation study with
5-aza-2′-deoxycytidine in cell lines
Twelve breast cancer cell lines (MCF7, ZR75-1, T47D,
ZR75-30, MDA-MB-361, BT474, SKBR3, AU565, MDA-MB-453, MDA-MB-231,
MDA-MB-468 and BT-20) and one normal breast epithelial cell line
(HMEC) were cultured according to ATCC culture guides. For
demethylation studies, cultured cells were treated with 10
μmol/l 5-aza-2′-deoxycytidine (Sigma-Aldrich, St. Louis, MO,
USA) or with dimethyl sulfoxide as control for 72 h. The medium was
changed every 24 h.
RNA extraction and quantitative real-time
PCR
Total RNA was isolated from cell lines using
TRIzol® reagent (Invitrogen). One microgram of total RNA
was reverse-transcribed for single stranded cDNA using random
primers and the ReverTra Ace® qPCR RT kit (Toyobo,
Osaka, Japan). The reverse-transcription reaction was performed at
65°C for 5 min and subsequently at 37°C for 15 min and 98°C for 5
min. mRNA was quantitated using the LightCycler 480 Real-Time PCR
system (Roche Applied Science, Mannheim, Germany) at 95°C (10 min),
followed by 50 cycles of 95°C (15 sec), 60°C (60 sec) and 1 cycle
of 50°C (10 sec). SEPT9_v2 and GAPDH
TaqMan® Gene Expression Assays (assay identification
nos. are Hs01107941_m1 and Hs02758991_g1, respectively; Applied
Biosystems, Foster City, CA, USA) were used for quantitative
real-time PCR. The expression of SEPT9_v2 was normalized to
that of GAPDH, and each assay was performed in duplicate.
Each 5-aza-2′-deoxycytidine-treated breast cancer cell line was
normalized to its control, which was set to a value of 1.
Statistical analyses
JMP statistical software (version 10; SAS Institute,
Cary, NC, USA) was used for statistical analyses. Associations
between the various parameters and the SEPT9_v2 MI
were evaluated using Student's t-test for two groups or the
Kruskal-Wallis test for more than two groups. The paired t-test was
used for comparison of frozen cancer and normal tissue MIs in
matched-pair samples. Dunnett's test was used for comparison of
SEPT9_v2 MIs in each of the subtypes. Univariate and
multivariate analyses of various parameters for their association
with pCR were conducted using the logistic regression model. All
statistical analyses were two-sided and P-values <0.05 were
considered statistically significant.
Results
Methylation of the SEPT9_v2 promoter and
its impact on gene expression in breast cancer cell lines
The NGS methylation assay was performed on the
SEPT9_v2 promoter in 12 breast cancer cell lines and a
normal human mammary epithelial cell line. A representative NGS
result is shown in Fig. 2. The
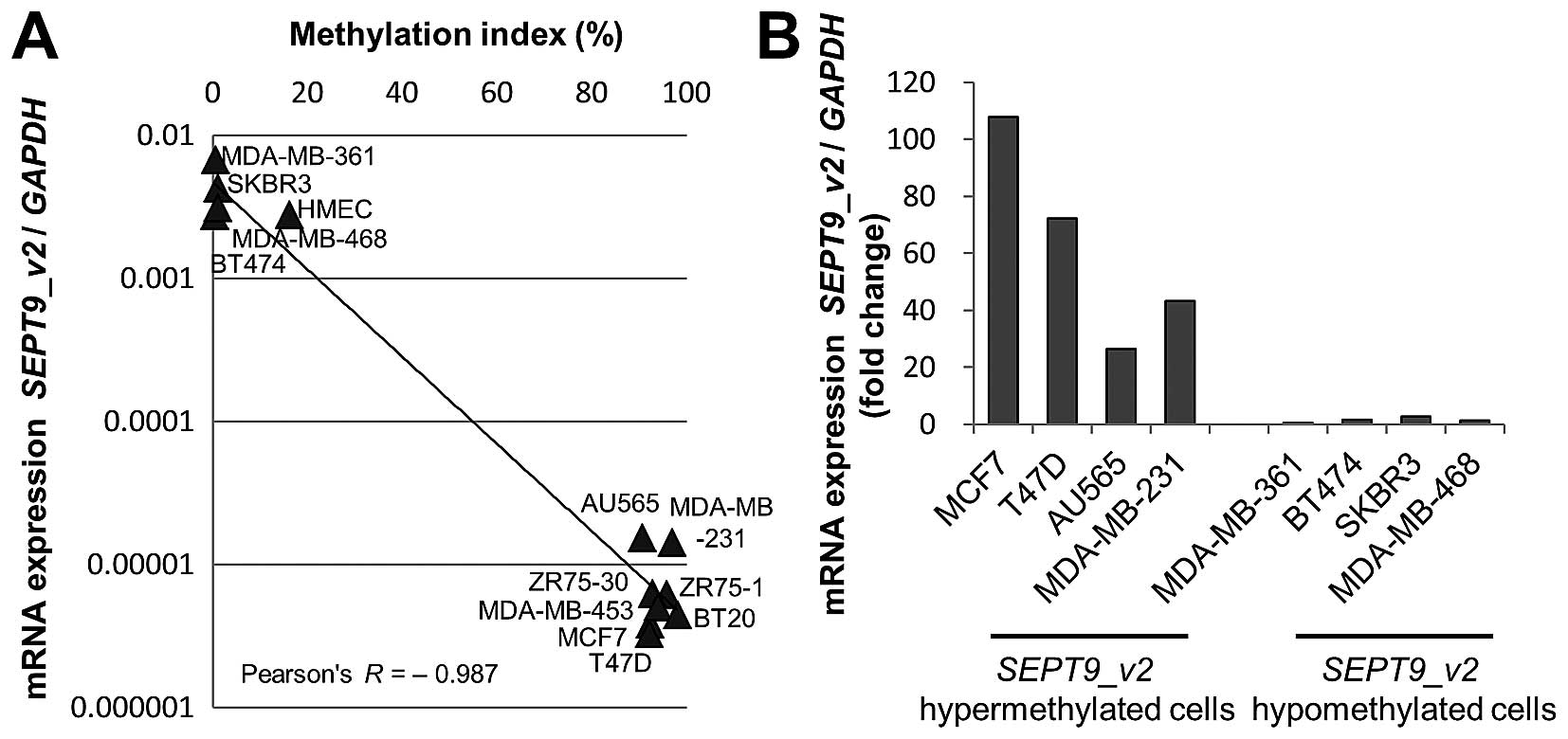
SEPT9_v2 gene promoter was hypermethylated in eight and
hypomethylated in four breast cancer cell lines and normal human
mammary epithelial cells (Fig.
3A).
The expression of SEPT9_v2 mRNA was examined
by quantitative real-time PCR using SEPT9_v2-specific
primers and probes. There was an inverse correlation between the
expression of SEPT9_v2 mRNA and the MI (Pearson's
correlation coefficient=−0.987) (Fig.
3A). We then treated eight of these cell lines with a
demethylating agent (10 μM 5-aza-2′-deoxycytidine) and
compared mRNA expression between treated and untreated cells.
Treatment with 5-aza-2′-deoxycytidine induced 20- to 110-fold
upregulation of mRNA expression in all four hypermethylated breast
cancer cell lines (MCF7, T47D, AU565 and MDA-MB-231) (Fig. 3B). There was no upregulation in the
four hypomethylated breast cancer cell lines (MDA-MB-361, BT474,
SKBR3 and MDA-MB-468), demonstrating that the SEPT9_v2 gene
was re-expressed by demethylation of its promoter region.
Methylation and expression of SEPT9_v2 in
human breast cancer tissues
To study the methylation status of SEPT9_v2
in human breast cancer and normal breast tissues, the NGS
methylation assay was performed using 19 paired tumor and normal
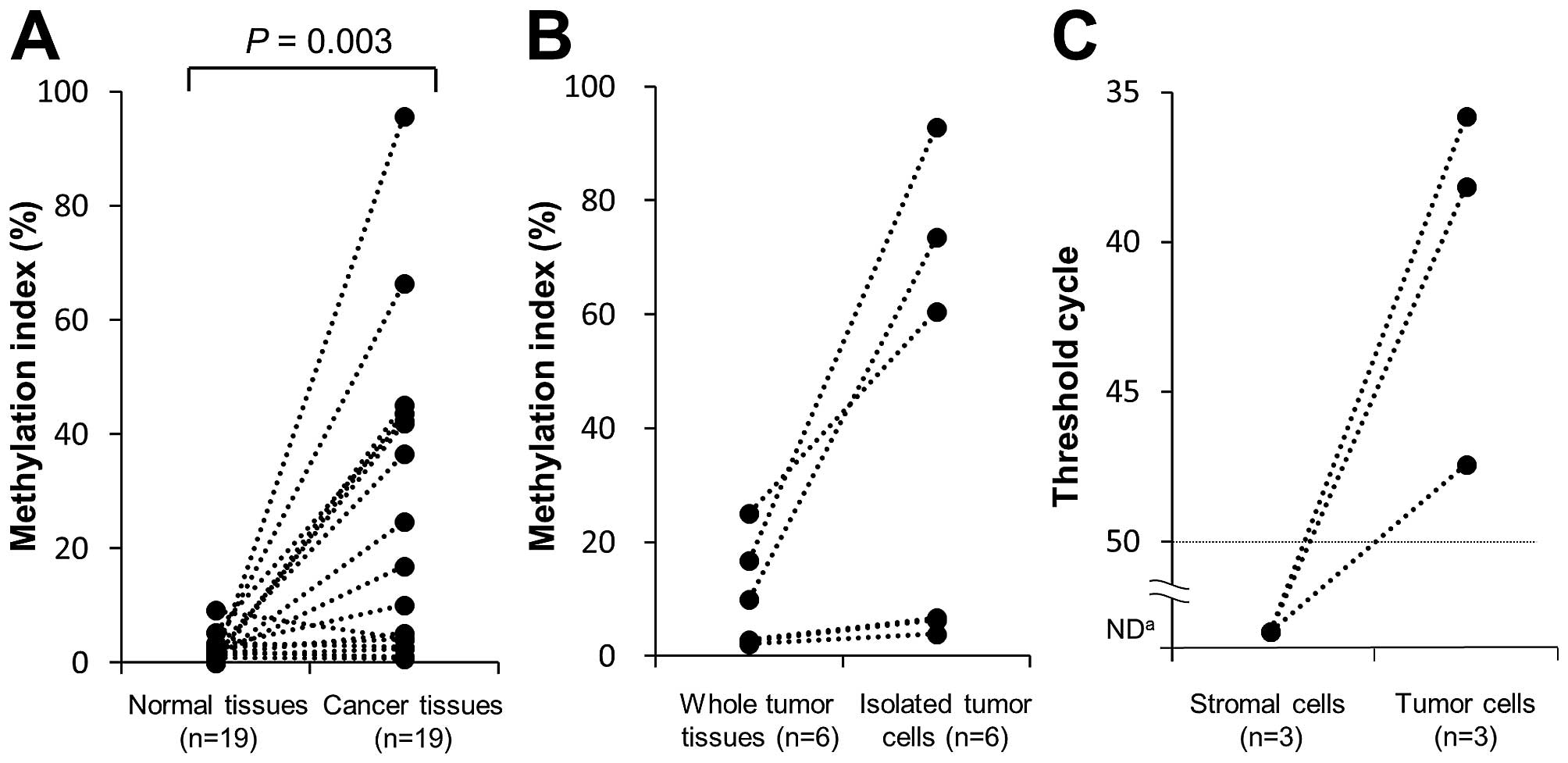
tissues (study I). The MI was significantly higher in tumor than
normal tissues (median values=10.0 and 1.7%, respectively; P=0.003)
(Fig. 4A). The proportion of
SEPT9_v2 tumors that were hypermethylated (MI ≥10%) was
53%.
For more accurate assessment of the cancer
cell-specific methylation status, we isolated cells from FFPE tumor
tissue by the MACS method. The isolated tumor cells were subjected
to the NGS methylation assay. Six tumor tissue samples with a
relatively low-MI (<30%) were analyzed since the low-MI was
speculated to be due to contamination by normal stromal and
inflammatory cells. The MI was clearly high in the tumor cells
isolated from the three tumors with a relatively high-MI. However,
the MI was low in tumor cells isolated from the remaining three
tumors with a very low-MI (Fig.
4B).
Relationship between SEPT9_v2 methylation
and clinicopathological characteristics
The NGS methylation assay of SEPT9_v2 was
carried out using the vacuum-assisted biopsy specimens obtained
before NAC (study II). The relationship between the extent of
SEPT9_v2 methylation and various clinicopathological
parameters including response to NAC was examined (Table II). SEPT9_v2
hypermethylation (MI ≥10%) was observed in 37% (40/107) of the
specimens. hypermethylation was significantly associated with
hormone receptor positivity, low histological grade and non-pCR
(Table II). The MI was
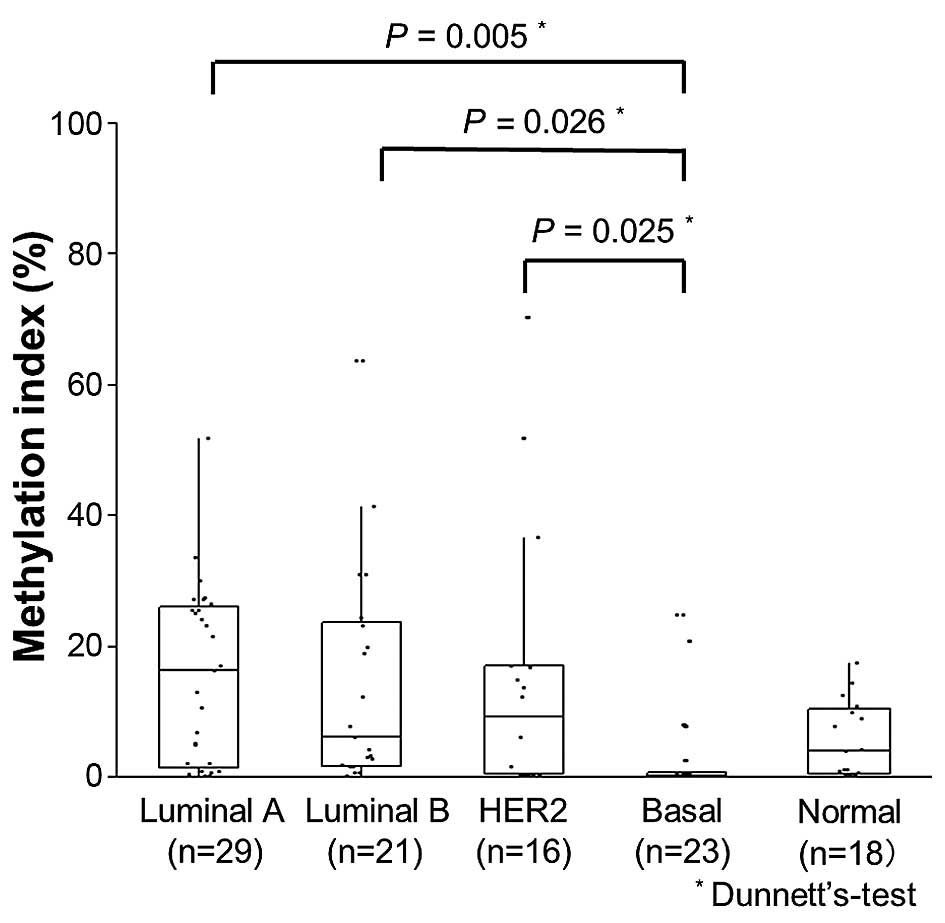
significantly lower in basal type tumors (3.0%) than in luminal A
(P=0.005), luminal B (P=0.026) or HER2 type (P=0.025) cancers
(Fig. 5).
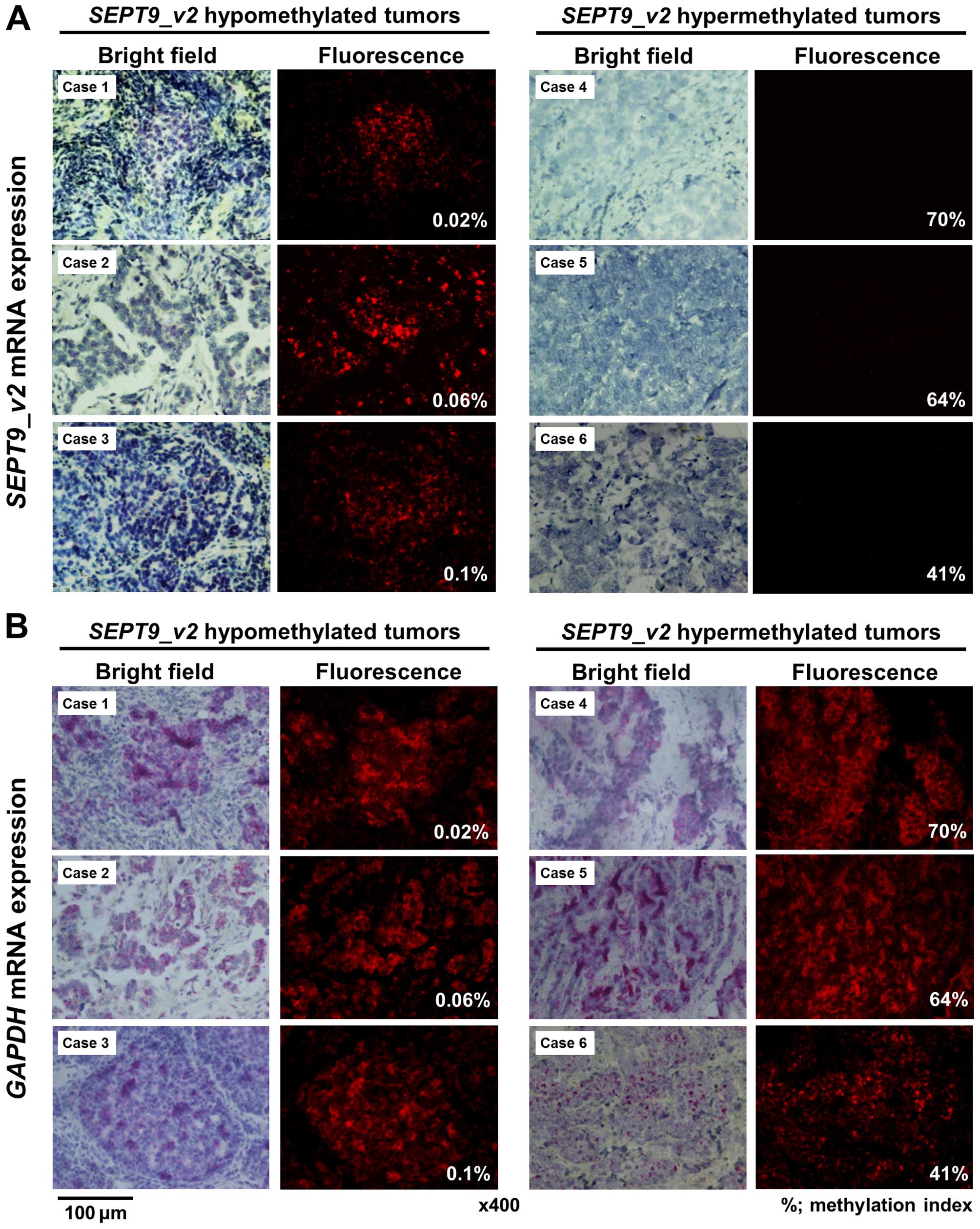
To determine whether methylation was related to gene
expression, we performed ISH of SEPT9_v2 and GAPDH
mRNA in the SEPT9_v2 hypermethylated (n=10) and
hypomethylated tumors (n=10). GAPDH mRNA was expressed in
both the SEPT9_v2 hypermethylated and hypomethylated tumors
(Fig. 6B). In contrast, ISH signals
of SEPT9_v2 mRNA in tumor cells were negative or barely
detectable in all 10 hypermethylated tumors, but were clearly
detectable in eight of 10 hypomethylated tumors (Fig. 6A).
Relationship between SEPT9_v2 methylation
and the response to NAC
Clinicopathological parameters were evaluated by
univariate analysis for their association with pCR (Table III). Age, Ki67, ER, PR, HER2 and
SEPT9_v2 methylation status were significantly associated
with pCR. Multivariate analysis showed that ER, but not
SEPT9_v2 methylation, was a significant and independent
predictor for pCR. The SEPT9_v2 methylation status of breast
tumors before and after NAC was also investigated in 20 patients
with residual tumors (non-pCR) after NAC including 10 patients with
SEPT9_v2 hypermethylated tumors and 10 patients with
hypomethylated tumors before NAC. The methylation status of the
residual tumors was completely the same as that of the tumors
before NAC, i.e., all of 10 SEPT9_v2 hypermethylated tumors
before NAC showed hypermethylation in the residual tumors after NAC
and all of 10 SEPT9_v2 hypomethylated tumors before NAC
showed hypomethylation in the residual tumors after NAC (data not
shown).
 | Table IIIUnivariate and multivariate analysis
of clinicopathological parameters for pCR. |
Table III
Univariate and multivariate analysis
of clinicopathological parameters for pCR.
|
Characteristics | Univariate analysis
| Multivariate
analysis
|
|---|
| Odds ratio | 95% CI | P-value | Odds ratio | 95% CI | P-value |
|---|
Age
(years)
(≥50 vs. <50) | 3.14 | 1.32–7.98 | 0.009 | 2.74 | 0.98–8.10 | 0.054 |
T stage
(T1,2 vs. T3,4) | 1.80 | 0.64–5.90 | 0.274 | | | |
Lymph node
status
(positive vs. negative) | 1.68 | 0.66–4.68 | 0.286 | | | |
Ki67
(positive vs. negative) | 3.04 | 1.25–8.02 | 0.013 | 1.49 | 0.48–4.68 | 0.489 |
ER
(negative vs. positive) | 10.5 | 4.16–29.0 | <0.001 | 7.97 | 2.15–39.40 | 0.001 |
PR
(negative vs. positive) | 5.60 | 2.09–17.9 | <0.001 | 0.73 | 0.12–3.58 | 0.697 |
HER2
(positive vs. negative) | 2.47 | 1.02–5.99 | 0.044 | 1.74 | 0.62–4.88 | 0.291 |
SEPT9_v2
methylation
(<10 vs. ≥10%) | 2.99 | 1.20–8.25 | 0.018 | 1.61 | 0.48–5.48 | 0.440 |
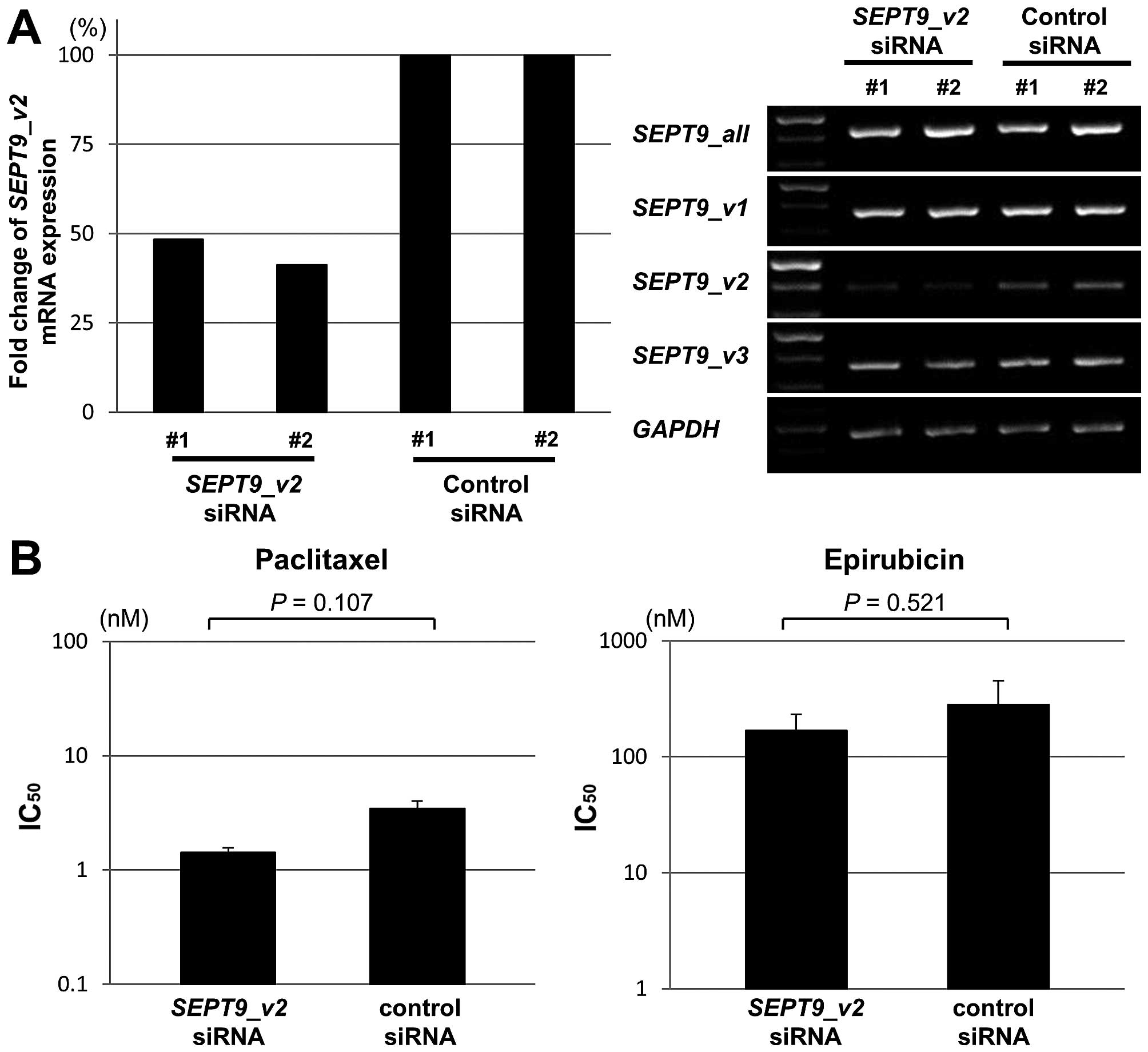
To further investigate the effect of SEPT9_v2
on chemosensitivity, we carried out a knockdown assay with
SEPT9_v2 siRNA for MDA-MB-468 cells, in which
SEPT9_v2 is highly expressed and sensitive to both
paclitaxel and epirubicin. SEPT9_v2 siRNA specifically
decreased the expression of SEPT9_v2 in MDA-MB-468 cells
(Fig. 7A) and the IC50
value for both paclitaxel and epirubicin was not significantly
different between SEPT9_v2 siRNA-treated and control cells
(Fig. 7B).
Detection of methylated SEPT9_v2 ctDNA in
breast cancer patients
The presence of methylated SEPT9_v2 ctDNA was
assessed in plasma from 82 PBC and 50 MBC patients, and 51 healthy
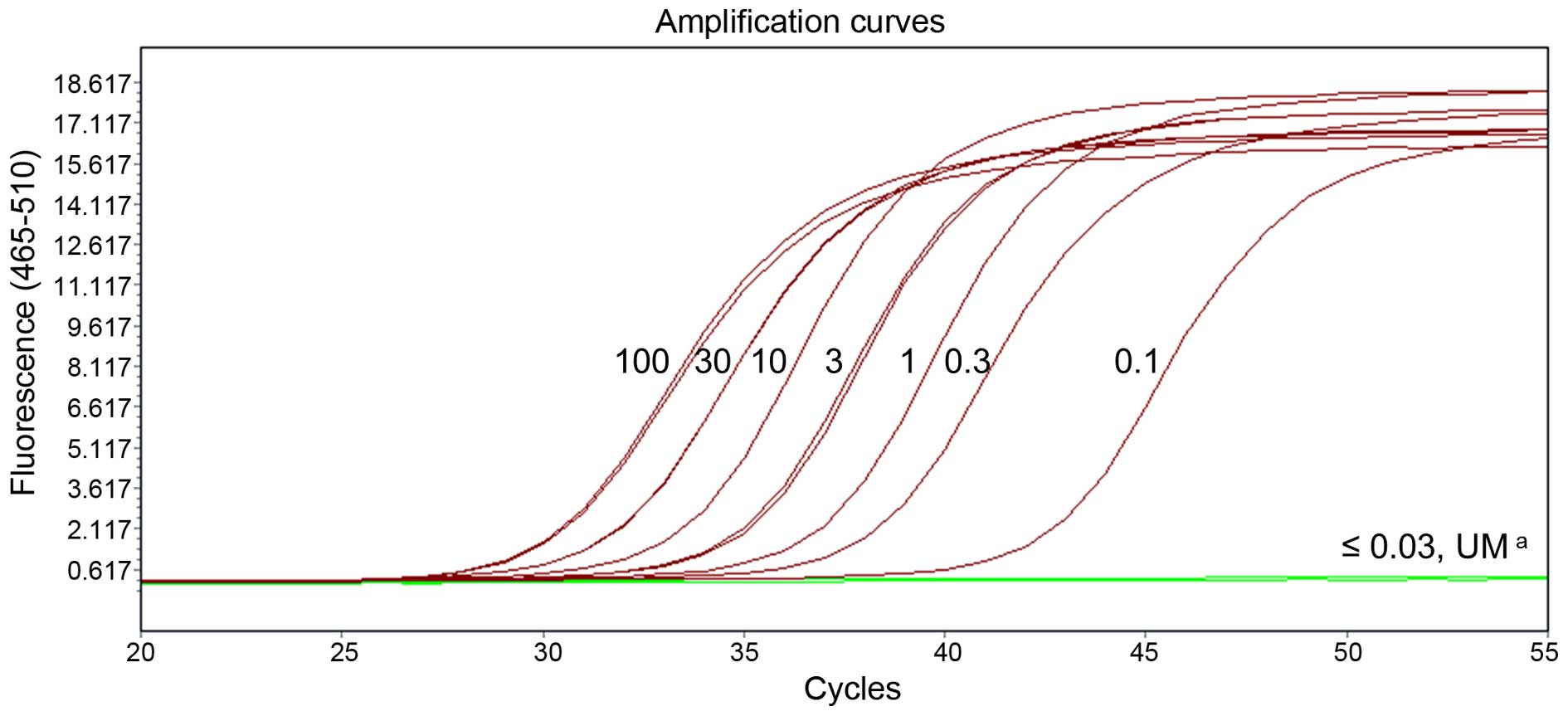
controls (study III). An amplification curve of the eight standards
was obtained by diluting the methylated human control DNA to 100,
30, 10, 3, 1, 0.3, 0.1 and 0.03 ng/well (Fig. 8). The limit of detection for
methylated SEPT9_v2 DNA was 0.1 ng/well. Methylated
SEPT9_v2 ctDNA was detected in 11% (9/82) of PBC patients,
and 52% (26/50) of MBC patients, but not in any of the healthy
controls (Table IV). In addition,
the methylation status of SEPT9_v2 was investigated in the
primary tumors available from PBC patients (n=49) and MBC patients
(n=25). SEPT9_v2 hypermethylation in the primary tumors was
found in 67% (33/49) of PBC patients and 68% (17/25) of MBC
patients. Methylated SEPT9_v2 ctDNA was significantly
(P<0.05) more frequently observed in the PBC and MBC patients
with SEPT9_v2 hypermethylated tumors [34% (17/50)] than
those with SEPT9_v2 hypomethylated tumors [8% (2/24)]
(Table IV), indicating a positive
correlation of SEPT9_v2 hypermethylation in primary tumors
with the presence of SEPT9_v2 ctDNA. However, in order to
confirm that SEPT9_v2 methylated ctDNA actually originated
from tumor cells, tumor cells and stromal cells were separately
collected by means of LCM and subjected to the methylation assay in
three patients positive for SEPT9_v2 ctDNA. In all three
patients, SEPT9_v2 methylation was detectable only in tumor
cells but not in the stromal cells (Fig. 4C).
 | Table IVDetection sensitivity of methylated
SEPT9_v2 in plasma of primary and metastatic breast cancer
patients (study III). |
Table IV
Detection sensitivity of methylated
SEPT9_v2 in plasma of primary and metastatic breast cancer
patients (study III).
| Total | Methylated
SEPT9_v2 in plasma
|
|---|
| Positive No.
(%) | Negative No.
(%) |
|---|
| Healthy
control | 51 | 0 (0) | 51 (100) |
| PBC patients | 82 | 9 (11) | 73 (89) |
| MBC patients | 50 | 26 (52) | 24 (48) |
| PBC + MBC
patientsa with | 74 | | |
| SEPT9_v2
hypermethylated tumors | 50 | 17 (34) | 33 (66)b |
| SEPT9_v2
hypomethylated tumors | 24 | 2 (8) | 22 (92) |
Discussion
In the present study, SEPT9_v2
hypermethylation was observed in 8 (67%) of the 12 breast cancer
cell lines and 53% of breast tumor tissues examined (study I), but
not in a normal human mammary epithelial cell line or normal breast
tissues. The lower MI in tumor tissues compared to breast cancer
cell lines may have resulted from the contamination of tumor tissue
by normal stromal or inflammatory cells, since the tumor cells
isolated by the MACS method showed higher MIs than the tumor tissue
from which they were derived and since SEPT9_v2 methylation
was observed in tumor cells but not in stromal cells separately
obtained by LCM. These results clearly indicate that tumor cells
actually harbor methylated SEPT9_v2.
We found that the expression of SEPT9_v2 mRNA
was inversely correlated with the SEPT9_v2 MI in breast
cancer cell lines, and that treatment of SEPT9_v2
hypermethylated breast cancer cell lines with a demethylating
reagent resulted in the reactivation of SEPT9_v2 mRNA
expression. These results clearly demonstrate that SEPT9_v2
expression is epigenetically regulated by promoter methylation in
breast cancer cell lines, consistent with the studies on colorectal
cancers (32). In addition, we
confirmed such epigenetic regulation in breast tumor tissue using
ISH to demonstrate that SEPT9_v2 mRNA expression was
silenced in SEPT9_v2 hypermethylated tumors.
The SEPT9_v2 MI was significantly lower in
basal type tumors than in other intrinsic tumor subtypes. This is
consistent with the study that basal type tumors are globally
hypomethylated as compared with other subtypes (33). Although SEPT9_v2
hypermethylation was significantly associated with non-pCR, this
does not necessarily mean that such hypermethylation plays a
significant role in chemotherapy resistance. Multivariate analysis
failed to demonstrate that SEPT9_v2 hypermethylation was an
independent predictor for non-pCR. In addition, we observed that
knockdown of SEPT9_v2 mRNA by siRNA had no effect on
chemosensitivity even though the potential involvement of
SEPT9_V1 and SEPT9_V4 in chemoresistance has been
reported (34–36). However, the methylation status of
the residual tumors after NAC was completely the same as that of
the tumors before NAC, indicating that chemotherapy did not affect
the SEPT9_v2 methylation status. Thus, it is possible that
SEPT9_v2 hypermethylation is indirectly associated with
non-pCR via its strong association with the ER, which is a
well-established predictor for non-pCR (37–39).
Taken together, SEPT9_v2 is unlikely to play a significant
role in chemoresistance, and is not a clinically useful predictor
for non-pCR.
Methylated SEPT9_v2 ctDNA appears to be one
of the most successful markers for the early detection of
colorectal cancer. Since a recent study showed that this ctDNA can
be detected in lung cancer patients (40), its clinical utility for other types
of cancers needs to be clarified. The present study revealed that
methylated SEPT9_v2 ctDNA was detectable in 11% of PBC
patients and 52% of MBC patients and that it was significantly more
frequently observed in the PBC/MBC patients with hypermethylated
tumors than those with hypomethylated tumors. However, the
sensitivity of this methylated ctDNA for PBC and MBC was lower than
that for primary and metastatic colorectal cancer (45 and 77%,
respectively) (22). This lower
sensitivity can be explained by the lower proportion of methylated
SEPT9_v2 breast cancer [50% (100/200), all tumors combined
in the present study] compared to colorectal cancer (82%) (41). Another potential explanation is the
different methods of assay that were used. The methylated
SEPT9_v2 ctDNA detection kit, known as Epi
proColon® 2.0 (Epigenomics AG, Berlin, Germany), can
detect 14 pg/ml of this ctDNA. This high sensitivity is achieved by
a triplicate assay using a greater volume of plasma (3.5 ml). The
present study was carried out by a single assay using less plasma
(2 ml) since our retrospective samples contained insufficient
volumes.
In conclusion, although methylation of many other
genes has been reported in breast cancer (3–9),
methylation status of SEPT9_v2 and its correlation with mRNA
expression has yet to be studied in human breast cancers. Then, we
analyzed this issue, and were able to show that hypermethylation of
SEPT9_v2 was seen in a high proportion of breast tumors and
that its methylation was clearly correlated with the silencing of
SEPT9_v2 mRNA expression. However, we could show for the
first time that SEPT9_v2 methylated ctDNA was detectable in
a significant proportion of PBC and MBC patients, suggesting a
possibility that SEPT9_v2 methylated ctDNA may serve as a
novel tumor marker. Our present observation needs to be further
investigated in a future study including a larger number of
patients.
Acknowledgments
The present study was supported in part by
Grants-in-Aid from the Knowledge Cluster Initiative of the Ministry
of education, Culture, Sports, Science and Technology of Japan.
References
|
1
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rodríguez-Paredes M and Esteller M: Cancer
epigenetics reaches mainstream Oncology. Nat Med. 17:330–339. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamamoto N, Nakayama T, Kajita M, Miyake
T, Iwamoto T, Kim SJ, Sakai A, Ishihara H, Tamaki Y and Noguchi S:
Detection of aberrant promoter methylation of GSTP1, RASSF1A, and
RARβ2 in serum DNA of patients with breast cancer by a newly
established one-step methylation-specific PCR assay. Breast Cancer
Res Treat. 132:165–173. 2012. View Article : Google Scholar
|
|
4
|
Fujita N, Nakayama T, Yamamoto N, Kim SJ,
Shimazu K, Shimomura A, Maruyama N, Morimoto K, Tamaki Y and
Noguchi S: Methylated DNA and total DNA in serum detected by
one-step methylation-specific PCR is predictive of poor prognosis
for breast cancer patients. Oncology. 83:273–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fujita N, Kagara N, Yamamoto N, Shimazu K,
Shimomura A, Shimoda M, Maruyama N, Naoi Y, Morimoto K, Oda N, et
al: Methylated DNA and high total DNA levels in the serum of
patients with breast cancer following neoadjuvant chemotherapy are
predictive of a poor prognosis. Oncol Lett. 8:397–403.
2014.PubMed/NCBI
|
|
6
|
Hoque MO, Feng Q, Toure P, Dem A,
Critchlow CW, Hawes SE, Wood T, Jeronimo C, Rosenbaum E, Stern J,
et al: Detection of aberrant methylation of four genes in plasma
DNA for the detection of breast cancer. J Clin Oncol. 24:4262–4269.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Avraham A, Uhlmann R, Shperber A, Birnbaum
M, Sandbank J, Sella A, Sukumar S and Evron E: Serum DNA
methylation for monitoring response to neoadjuvant chemotherapy in
breast cancer patients. Int J Cancer. 131:E1166–E1172. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sharma G, Mirza S, Parshad R, Gupta SD and
Ralhan R: DNA methylation of circulating DNA: A marker for
monitoring efficacy of neoadjuvant chemotherapy in breast cancer
patients. Tumour Biol. 33:1837–1843. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fackler MJ, Lopez Bujanda Z, Umbricht C,
Teo WW, Cho S, Zhang Z, Visvanathan K, Jeter S, Argani P, Wang C,
et al: Novel methylated biomarkers and a robust assay to detect
circulating tumor DNA in metastatic breast cancer. Cancer Res.
74:2160–2170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Arai T, Miyoshi Y, Kim SJ, Taguchi T,
Tamaki Y and Noguchi S: Association of GSTP1 CpG islands
hypermethylation with poor prognosis in human breast cancers.
Breast Cancer Res Treat. 100:169–176. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miyake T, Nakayama T, Naoi Y, Yamamoto N,
Otani Y, Kim SJ, Shimazu K, Shimomura A, Maruyama N, Tamaki Y, et
al: GSTP1 expression predicts poor pathological complete response
to neoadjuvant chemotherapy in ER-negative breast cancer. Cancer
Sci. 103:913–920. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mostowy S and Cossart P: Septins: The
fourth component of the cytoskeleton. Nat Rev Mol Cell Biol.
13:183–194. 2012.PubMed/NCBI
|
|
13
|
Robertson C, Church SW, Nagar HA, Price J,
Hall PA and Russell SE: Properties of SEPT9 isoforms and the
requirement for GTP binding. J Pathol. 203:519–527. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peterson EA and Petty EM: Conquering the
complex world of human septins: Implications for health and
disease. Clin Genet. 77:511–524. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kalikin LM, Sims HL and Petty EM: Genomic
and expression analyses of alternatively spliced transcripts of the
MLL septin-like fusion gene (MSF) that map to a 17q25 region of
loss in breast and ovarian tumors. Genomics. 63:165–172. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Russell SE, McIlhatton MA, Burrows JF,
Donaghy PG, Chanduloy S, Petty EM, Kalikin LM, Church SW, McIlroy
S, Harkin DP, et al: Isolation and mapping of a human septin gene
to a region on chromosome 17q, commonly deleted in sporadic
epithelial ovarian tumors. Cancer Res. 60:4729–4734.
2000.PubMed/NCBI
|
|
17
|
Montagna C, Lyu MS, Hunter K, Lukes L,
Lowther W, Reppert T, Hissong B, Weaver Z and Ried T: The Septin 9
(MSF) gene is amplified and overexpressed in mouse mammary gland
adenocarcinomas and human breast cancer cell lines. Cancer Res.
63:2179–2187. 2003.PubMed/NCBI
|
|
18
|
Connolly D, Abdesselam I, Verdier-Pinard P
and Montagna C: Septin roles in tumorigenesis. Biol Chem.
392:725–738. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lofton-Day C, Model F, Devos T, Tetzner R,
Distler J, Schuster M, Song X, Lesche R, Liebenberg V, Ebert M, et
al: DNA methylation biomarkers for blood-based colorectal cancer
screening. Clin Chem. 54:414–423. 2008. View Article : Google Scholar
|
|
20
|
deVos T, Tetzner R, Model F, Weiss G,
Schuster M, Distler J, Steiger KV, Grützmann R, Pilarsky C,
Habermann JK, et al: Circulating methylated SEPT9 DNA in plasma is
a biomarker for colorectal cancer. Clin Chem. 55:1337–1346. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Warren JD, Xiong W, Bunker AM, Vaughn CP,
Furtado LV, Roberts WL, Fang JC, Samowitz WS and Heichman KA:
Septin 9 methylated DNA is a sensitive and specific blood test for
colorectal cancer. BMC Med. 9:1332011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Church TR, Wandell M, Lofton-Day C, Mongin
SJ, Burger M, Payne SR, Castaños-Vélez E, Blumenstein BA, Rösch T,
Osborn N, et al PRESEPT Clinical Study Steering Committee,
Investigators and Study Team: Prospective evaluation of methylated
SEPT9 in plasma for detection of asymptomatic colorectal cancer.
Gut. 63:317–325. 2014. View Article : Google Scholar :
|
|
23
|
Connolly D, Yang Z, Castaldi M, Simmons N,
Oktay MH, Coniglio S, Fazzari MJ, Verdier-Pinard P and Montagna C:
Septin 9 isoform expression, localization and epigenetic changes
during human and mouse breast cancer progression. Breast Cancer
Res. 13:R762011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Naoi Y, Kishi K, Tanei T, Tsunashima R,
Tominaga N, Baba Y, Kim SJ, Taguchi T, Tamaki Y and Noguchi S:
Development of 95-gene classifier as a powerful predictor of
recurrences in node-negative and ER-positive breast cancer
patients. Breast Cancer Res Treat. 128:633–641. 2011. View Article : Google Scholar
|
|
25
|
Parker JS, Mullins M, Cheang MC, Leung S,
Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al:
Supervised risk predictor of breast cancer based on intrinsic
subtypes. J Clin Oncol. 27:1160–1167. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mishima C, Kagara N, Tanei T, Naoi Y,
Shimoda M, Shimomura A, Shimazu K, Kim SJ and Noguchi S: Mutational
analysis of MED12 in fibroadenomas and phyllodes tumors of the
breast by means of targeted next-generation sequencing. Breast
Cancer Res Treat. 152:305–312. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pfütze K, Benner A, Hoffmeister M, Jansen
L, Yang R, Bläker H, Herpel E, Ulrich A, Ulrich CM, Chang-Claude J,
et al: Methylation status at HYAL2 predicts overall and
progression-free survival of colon cancer patients under 5-FU
chemotherapy. Genomics. 106:348–354. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee ES, Issa JP, Roberts DB, Williams MD,
Weber RS, Kies MS and El-Naggar AK: Quantitative promoter
hypermethylation analysis of cancer-related genes in salivary gland
carcinomas: Comparison with methylation-specific PCR technique and
clinical significance. Clin Cancer Res. 14:2664–2672. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nygren AO, Ameziane N, Duarte HM,
Vijzelaar RN, Waisfisz Q, Hess CJ, Schouten JP and Errami A:
Methylation-specific mlPA (MS-MLPA): Simultaneous detection of CpG
methylation and copy number changes of up to 40 sequences. Nucleic
Acids Res. 33:e1282005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Toyota M, Kopecky KJ, Toyota MO, Jair KW,
Willman CL and Issa JP: Methylation profiling in acute myeloid
leukemia. Blood. 97:2823–2829. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Otani Y, Miyake T, Kagara N, Shimoda M,
Naoi Y, Maruyama N, Shimomura A, Shimazu K, Kim SJ and Noguchi S:
BRCA1 promoter methylation of normal breast epithelial cells as a
possible precursor for BRCA1-methylated breast cancer. Cancer Sci.
105:1369–1376. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tóth K, Galamb O, Spisák S, Wichmann B,
Sipos F, Valcz G, Leiszter K, Molnár B and Tulassay Z: The
influence of methylated septin 9 gene on RNA and protein level in
colorectal cancer. Pathol Oncol Res. 17:503–509. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Network CGA; Cancer Genome Atlas Network:
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Amir S and Mabjeesh NJ: SEPT9_V1 protein
expression is associated with human cancer cell resistance to
microtubule-disrupting agents. Cancer Biol Ther. 6:1926–1931. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Froidevaux-Klipfel L, Poirier F, Boursier
C, Crépin R, Poüs C, Baudin B and Baillet A: Modulation of septin
and molecular motor recruitment in the microtubule environment of
the Taxol-resistant human breast cancer cell line MDA-MB-231.
Proteomics. 11:3877–3886. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chacko AD, McDade SS, Chanduloy S, Church
SW, Kennedy R, Price J, Hall PA and Russell SE: Expression of the
SEPT9_i4 isoform confers resistance to microtubule-interacting
drugs. Cell Oncol. 35:85–93. 2012. View Article : Google Scholar
|
|
37
|
Carey LA, Dees EC, Sawyer L, Gatti L,
Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ml and Perou
CM: The triple negative paradox: Primary tumor chemosensitivity of
breast cancer subtypes. Clin Cancer Res. 13:2329–2334. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rouzier R, Perou CM, Symmans WF, Ibrahim
N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P,
et al: Breast cancer molecular subtypes respond differently to
preoperative chemotherapy. Clin Cancer Res. 11:5678–5685. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ignatiadis M and Sotiriou C: Luminal
breast cancer: from biology to treatment. Nat Rev Clin Oncol.
10:494–506. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Powrózek T, Krawczyk P, Kucharczyk T and
Milanowski J: Septin 9 promoter region methylation in free
circulating DNA-potential role in noninvasive diagnosis of lung
cancer: Preliminary report. Med Oncol. 31:9172014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ahmed D, Danielsen SA, Aagesen TH,
Bretthauer M, Thiis-Evensen E, Hoff G, Rognum TO, Nesbakken A,
Lothe RA and Lind GE: A tissue-based comparative effectiveness
analysis of biomarkers for early detection of colorectal tumors.
Clin Transl Gastroenterol. 3:e272012. View Article : Google Scholar
|