Introduction
Colorectal cancer (CRC) is the third most common
cancer and the third-leading cause of cancer death in the United
States (1). Although most CRC
patients undergo surgical resection of tumors, regional and distant
metastases strongly impact 5-year survival rates (2). Unfortunately, treatments of patients
with metastatic CRC are not very efficient. Therefore, it is of
great importance to further explore the underlying molecular
mechanisms of CRC progression in order to develop new therapeutic
strategies, especially for patients with metastatic CRC.
Erythroblast transformation-specific (ETS)-related
gene (ERG) belongs to the ETS transcription factor family, and
plays an important physiological role in hematopoiesis (3), angiogenesis (4), and bone development (5). In addition, aberrant ERG expression
has been observed in solid tumors and leukemia (6,7).
Furthermore, ERG gene fusion proteins such as transmembrane
protease, serine 2 (TMPRSS2)-ERG have been associated with poor
prognosis in prostate cancer (8).
In prostate cancer, ectopic ERG expression may induce acquired
invasive traits and endothelial mesenchymal transition (EMT)
(9). Although an increasing number
of studies indicate that ectopic ERG expression is involved in the
pathogenesis of various solid tumors and hemopathies, further
studies are still required to fully elucidate the underlying
biological mechanisms of the ERG action in tumorigenesis.
microRNAs (miRNA or miR) are single strand
non-coding RNAs composed of 21–24 nucleotides, which exert their
function by interacting with the complementary sites in the 3′
untranslated region (UTR) of target mRNAs. miRNAs regulate multiple
cellular functions such as cell proliferation, apoptosis and
differentiation (10). In addition,
it has been shown that aberrant miRNA expression results in ectopic
expression of different gene products, which may consequently lead
to carcinogenesis and tumor progression (11).
miR-145 was first reported to be consistently
decreased in precancerous colorectal lesions as well as in
different stages of colorectal cancer tissue samples compared to
normal colorectal mucosa (12).
Since then, increasing evidence has shown that ERG expression is
reduced in different types of cancer, supporting the idea that
miR-145 acts as a tumor suppressor in breast cancer (13), prostate cancer (14) and neuroblastoma (15). In addition, it has been shown that
ectopic expression of miR-145 influences cell cycle distribution,
invasion and differentiation level of tumor cells in CRC (16). Contrary to these reports, Arndt
et al showed that miR-145 may have an oncogenic role in CRC
by suppressing E-cadherin expression and promoting
anchorage-independent growth in vitro (17). Therefore, depending on the cell
lines and cancer types, miR-145 may have either a tumor suppressive
or an oncogenic effect. Hence, the exact miR-145 function and
regulation in CRC is still unknown and demands further
investigation.
Therefore, we undertook to further investigate the
role of miR-145 in tumorigenesis as well as its relationship to ERG
in clinical samples of colorectal tumors and CRC cell lines in
vitro.
Materials and methods
Clinical specimens
In this study, 48 pairs of colorectal tumors and
corresponding normal mucous tissue (5 cm away from the cancer
lesions) were collected from colorectal cancer patients who
underwent colorectal resection at the Third Affiliated Hospital of
Guangzhou Medical University. Tissue samples were snap-frozen in
liquid nitrogen and then stored at −80°C until further use. The
pathological diagnosis of CRC specimens and confirmation of the
adjacent normal intestinal mucosa were performed by at least two
pathologists. The TNM classification was performed according to the
National Comprehensive Cancer Network (NCCN) guideline (18). None of the patients received
pre-operative chemotherapy or radiotherapy. Patients with other
malignancies were excluded. The Clinical Research Ethics Committee
of the Third Affiliated Hospital of Guangzhou Medical University
approved the research protocols. Written informed consents were
obtained from all patients.
Cell lines and treatment
Human embryonic kidney cell line HEK-293T and five
human CRC cell lines (RKO, DLD-1, HCT-116, SW620 and SW480) were
obtained from the American Type Culture Collection (ATCC;
Rockville, MD, USA). All cell lines were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco-BRL, Grand Island, NY, USA)
supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT,
USA) and 1% penicillin/streptomycin, and cultured in humidified
incubator at 37°C and supplemented with 5% CO2.
Total RNA isolation and quantitative
real-time PCR
Total RNA from clinical specimens and CRC cell lines
was isolated with TRIzol (Invitrogen Life Technologies, Carlsbad,
CA, USA) according to the manufacturer's protocol. A NanoDrop 2000
spectrophotometer (Thermo Fisher Scientific, Madrid, Spain) was
used to measure RNA concentration. Next, reverse transcription
reactions were performed with PrimeScript RT Reagent kit (Takara,
Dalian, China), and quantitative real-time PCR (qRT-PCR) was
performed using SYBR Premix DimerEraser (Takara) and the Step One
Plus Real-time PCR system (Applied Biosystems, Foster City, CA,
USA). For miR-145 detection, miRNA-specific primers were purchased
from Shanghai Generay Biotechnology Co., Ltd. (Shanghai, China),
and the relative miR-145 expression level was normalized to U6
expression. ERG expression was normalized to
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. Primer
sequences were as follows: miR-145 reverse transcription,
5′-TGGTGTCGTGGAGTCG-3′; miR-145 sense, 5′-ACACTCCAGCTGGGGTCCAGT
TTTCCCAGGAA-3′ and antisense,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGGGAT-3′; ERG forward,
5′-ATCGTGCCAGCAGATCCTAC-3′ and reverse,
5′-CCTTGGTCATCTTGCACAGTTC-3′; GAPDH forward,
5′-CGCTGAGTACGTCGTGGAGTC-3′ and reverse,
5′-GCTGATGATCTTGAGGCTGTTGTC-3′. The 2 −ΔΔCt method was
used for relative quantification. All reactions were performed in
triplicate.
Western blot analysis
Proteins from tissue samples and CRC cell lines were
extracted with lysis buffer and protein concentration was
determined by Enhanced BCA Protein Assay kit (both from Beyotime
Institute of Biotechnology, Haimen, China). Protein lysate was
separated on 10% SDS-PAGE gels, transferred onto a PVDF membrane
(Millipore, Billerica, MA, USA), and blocked with 5% non-fat dried
milk. Next, membranes were incubated with antibodies against ERG
(1:2,000) (ab133264; Abcam, Cambridge, MA, USA) or GAPDH (1:5,000)
(ProteinTech, Chicago, IL, USA) overnight at 4°C, washed with
Tris-buffered saline with Tween (TBST) four times, and incubated
with horse raddish peroxidase (HRP) labeled anti-rabbit secondary
antibody (1:5,000) for 1 h at room temperature. Enhanced
chemiluminescence (ECL) reagent (Pierce, Rockford, IL, USA) was
used to visualize protein bands according to the manufacturer's
protocol. The relative density of the western blot bands was
measured by ImageJ (19) and
normalized to sample T1.
Oligonucleotide transfection
For the oligonucleotide transfection experiments
miR-145 mimics, miR-145 inhibitors and corresponding normal
controls were purchased from RiboBio (Guangzhou, China). ERG siRNAs
and non-targeting siRNA were obtained from GenePharma (Shanghai,
China). siRNA sense and antisense sequences were as follows:
5′-GACGUCAACAUCUUGUUAUTT-3′ and 5′-GACGUCAACAUCUUGUUAUTT-3′.
Oligonucleotide transfection was performed according to the
manufacturer's protocol using Lipofectamine 2000 (Invitrogen Life
Technologies). In brief, 50 nmol/l of miR-145 mimics or 100 nmol/l
of miR-145 inhibitors were used in transfections. For knockdown of
endogenous ERG 100 nmol/l of ERG siRNA was used.
Luciferase reporter assay
We used Targetscan (http://www.targetscan.org) for the prediction of
putative miR-145 targets. Using this analysis the ERG (NG_029732)
3′-UTR region was predicted to have putative binding site
complementary to the seed region of miR-145. In order to construct
the plasmid containing this site, 1.5 kb sequences of the ERG
3′-UTR region were amplified from human genomic DNA, and inserted
into restriction sites (Sgf I and NotI) of
Dual-Luciferase reporter psiCHECK2 plasmid (Promega, Madison, WI,
USA) and this plasmid was denoted as psiCHECK2-WT. The
Dual-Luciferase reporter plasmid containing a mutated miR-145
binding site in the ERG 3′-UTR region was also constructed and
denoted as psiCHECK2-MUT. The primers used for these constructs
were as follows: psiCHECK2-WT forward,
5′-CGCGCGATCGCAGACCTGGCGGAGGCTTTTC-3′ and reverse,
5′-ATAAGAATGCGGCCGCGGCTCTCCCTTGCACAAGTTC-3′; psiCHECK2-MUTforward,
5′-TCTTTGTTTGTCAAATGAAAATTTTTTCCAGTTTTGTCTGATATTTAAGAGAAACATT-3′
and reverse,
5′-AATGTTTCTCTTAAATATCAGACAAAACTGGAAAAAATTTTCATTTGACAAACAAAGA-3′.
For Dual-Luciferase assays, 50 nM of miR-145 mimics or 100 nM of
miR-145 inhibitor were co-transfected with 0.5 µg of
psiCHECK2-WT or psiCHECK 2-MUT using Lipofectamine 2000 (Invitrogen
Life Technologies). The renilla luciferase signal and the firefly
luciferase signal were consecutively detected according to the
manufacturer's protocols for Dual-Luciferase Reporter assay system
(E1910) (Promega). The firefly luciferase signal was used to
normalize the renilla luciferase signal.
Scratch wound healing assay
For the scratch wound healing assay, cells
(1×106) were seeded homogeneously on 6-well plates and
cultured for 24 h to form a monolayer. Next, monolayers were
scratched carefully with a sterile plastic 200 µl pipette
tip. The floating cell debris was washed with DMEM. At 0 and 24 h
after scratch would formation, images were obtained using an
inverted microscope (Nikon, Tokyo, Japan) at a magnification of
200x and measured by Image-Pro Plus software (Media Cybernetics,
Inc., Rockville, MD, USA).
Invasion assay
For invasion assays, Falcon inserts were used with
Falcon Companion Tissue Culture Plates (Corning, Inc., Corning, N
Y, USA) (24 wells). The Falcon Cell Culture inserts with
8-µm pore membranes were coated with Matrigel (BD
Biosciences, San Jose, CA, USA) and placed in a 37°C incubator for
3 h to solidify. Homogeneous single cell suspensions with
serum-free medium were added to the upper chamber at a total of
5×105 cells per well. Medium containing 10% FBS was
added to the lower chambers and served as a chemoattractant. After
24 h, cells that remained on the upper surface of the membrane were
carefully removed by a cotton swab, and cells which invaded the
pores and adhered to the lower surface of the membranes were fixed
with methanol and stained with hematoxylin. Stained cells were
observed and counted (five random ×100 magnification fields per
well) under an inverted microscope. Each experiment was performed
in triplicate.
Statistical analysis
Statistical analysis was performed using SPSS 18.0
(IBM SPSS, Chicago, IL, USA). Results of all experiments are
presented as mean ± SD. Student's t-test was used to compare miRNA
expression in clinical tissue samples and CRC cell lines. Pearson's
correlation coefficient was used to measure the correlation between
miR-145 and ERG expression. P<0.05 was considered statistically
significant.
Results
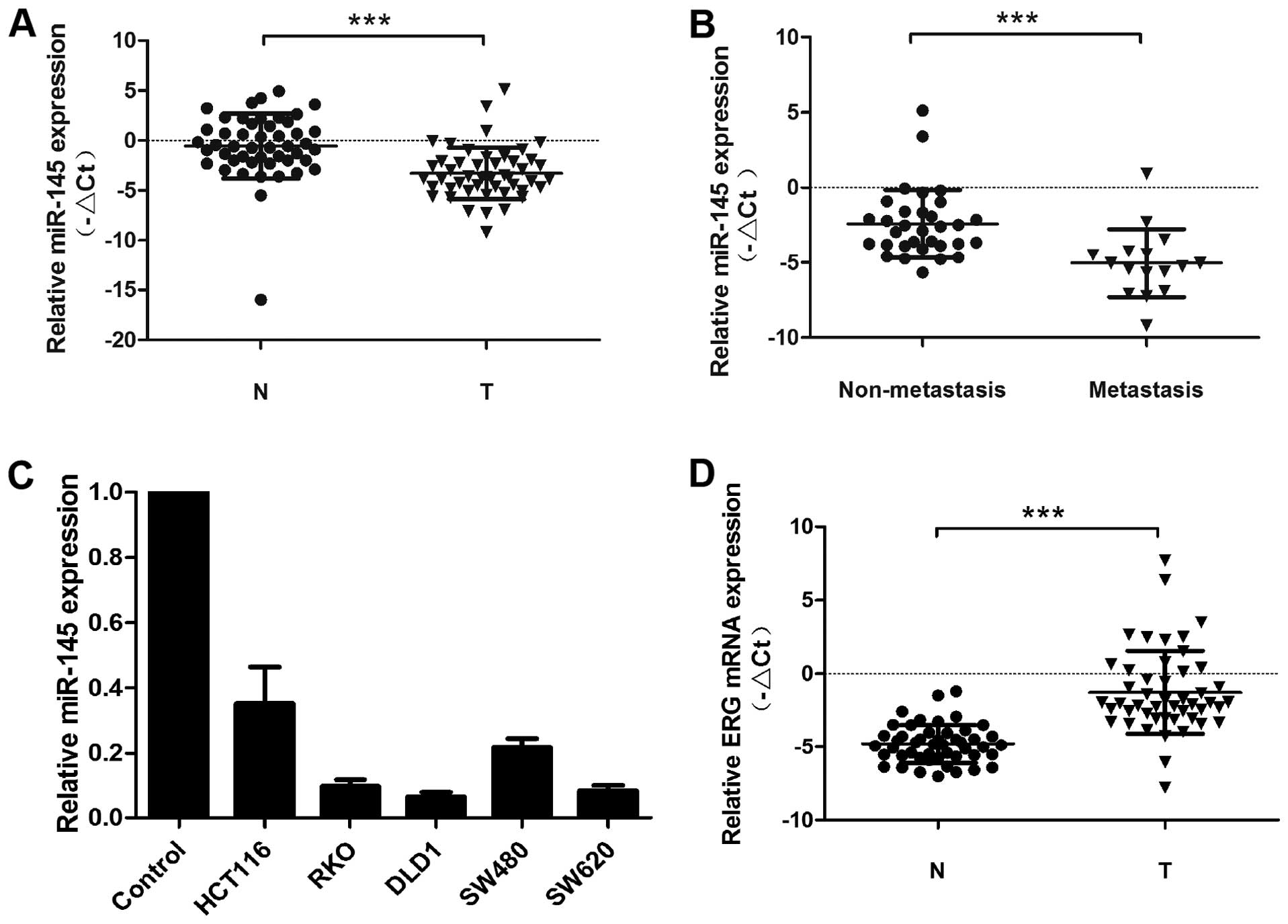
miR-145 is decreased in colorectal cancer
cell lines and clinical samples
In this study we examined miR-145 expression in 48
pairs of CRC tissues and adjacent non-cancerous mucosa samples by
qRT-PCR (Fig. 1A). In this sample
set, miR-145 expression was decreased in most of the CRC samples
compared to non-cancerous adjacent mucous tissue. Moreover, even
greater miR-145 downregulation was detected in CRC tumors obtained
from patients with lymph node metastasis compared to those without
lymph node metastasis (Fig. 1B).
miR-145 expression in colorectal cancer cell lines (RKO, DLD-1,
HCT-116, SW620 and SW480) was also decreased compared to adjacent
non-cancerous mucous tissue (Fig.
1C). In contrast to miR-145 expression, ERG expression levels
were upregulated in most CRC tissues compared to adjacent normal
mucosa (Fig. 1D). In addition, we
examined the relationship between miR-145 expression and
clinicopathological features of CRC patients, including gender,
age, lymph node metastasis, distant metastasis and TNM stage
(Table I). The mean value of
relative miR-145 expression (−2.7566) was used as a cut-off value
for the division of CRC patients into miR-145 low or high
expression groups, accordingly. Statistical analysis revealed that
low miR-145 expression correlated with poor outcome indicators
including lymph node metastasis (P<0.0001), distant metastasis
(P<0.0001) and TNM stage (P<0.0001).
 | Table ImiR-145 expression and
clinicopathological parameters of CRC patients and their
tumors. |
Table I
miR-145 expression and
clinicopathological parameters of CRC patients and their
tumors.
| Clinicopathological
parameters | No. of
patients
(N=48) | miR-145 expression
| P-value |
|---|
| Low | High |
|---|
| Age (years) |
|
≤65 | 13 | 8 | 5 | 0.0569 |
|
>65 | 35 | 17 | 18 | |
| Gender |
|
Female | 19 | 9 | 10 | 0.6675 |
|
Male | 29 | 16 | 13 | |
| LN involvement |
| No | 32 | 9 | 23 | <0.0001a |
|
Yes | 16 | 16 | 0 | |
| Distant
metastasis |
| No | 36 | 13 | 23 | <0.0001a |
|
Yes | 12 | 12 | 0 | |
| TNM stage |
| I,
II | 29 | 6 | 23 | <0.0001a |
| III,
IV | 19 | 19 | 0 | |
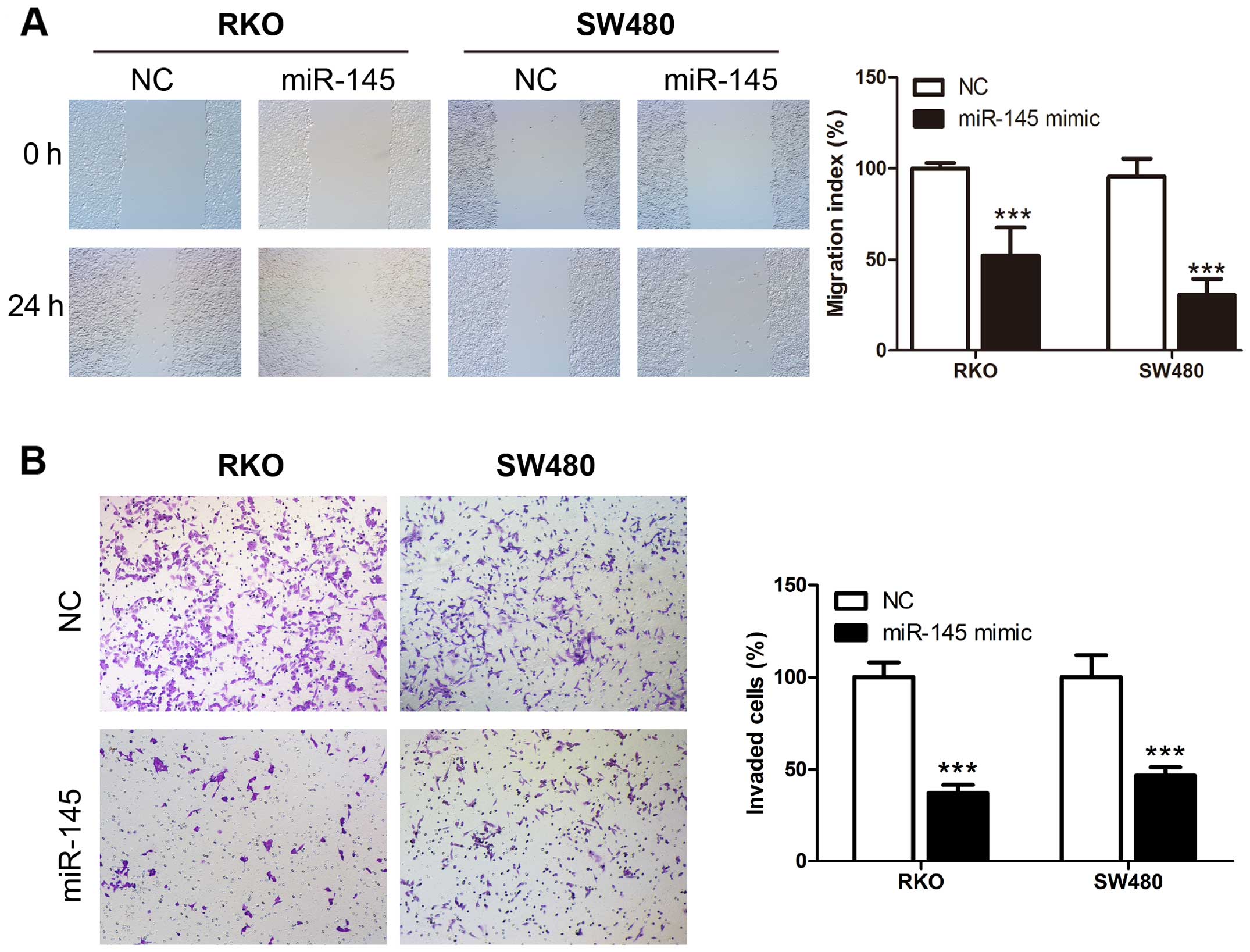
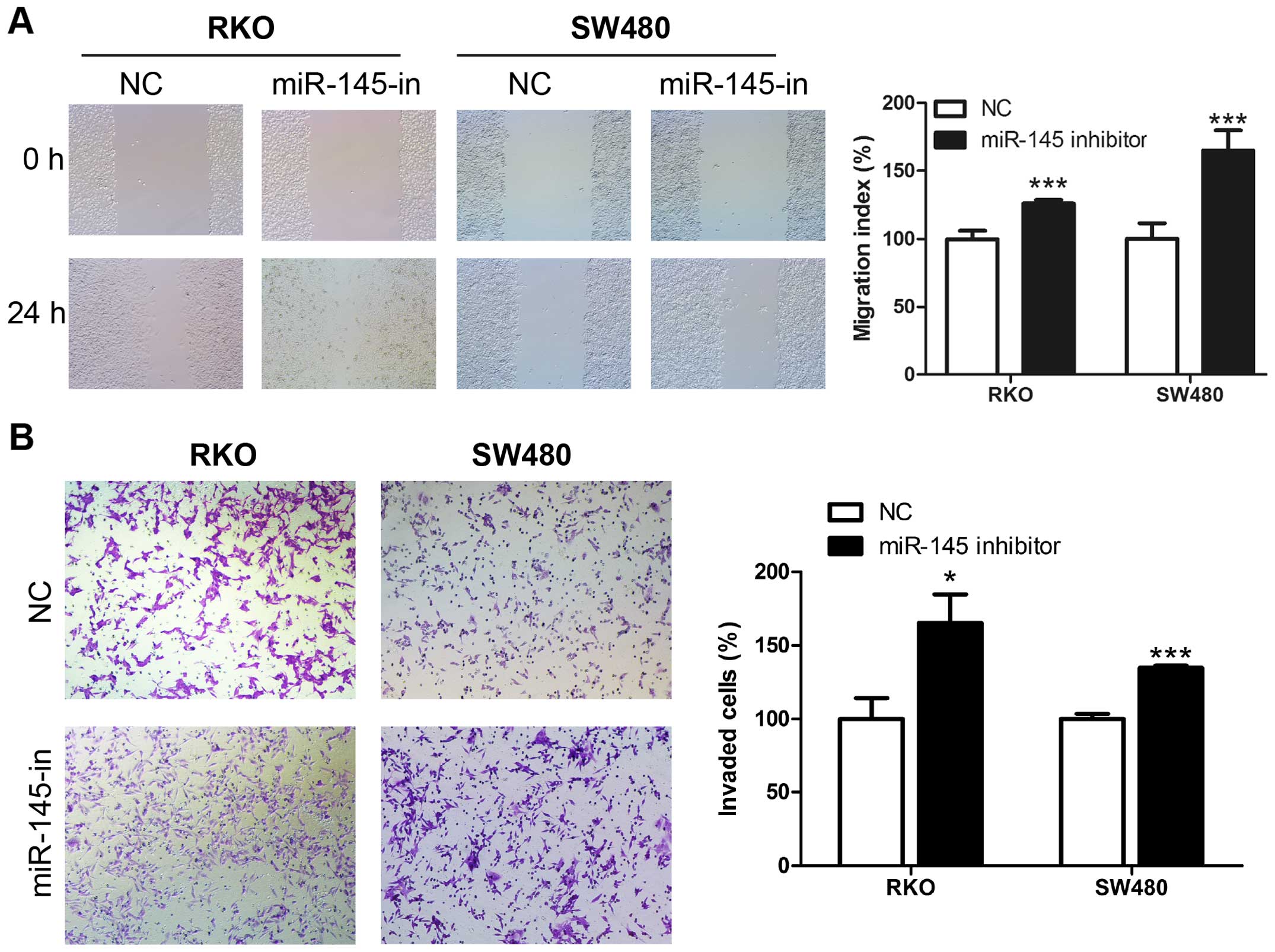
miR-145 suppresses migration and invasion
ability of human CRC cells
We examined miR-145 in human CRC cell lines using
the wound healing and Transwell assays. Transient transfection of
miR-145 mimic, miR-145 inhibitor and their corresponding control
RNAs was performed in CRC cell lines RKO and SW480. The wound
healing assay revealed that upregulation of miR-145 using the
miR-145 mimic transfection suppressed the migration rate of CRC
cells compared to control cells (Fig.
2A), while miR-145 knockdown in miR-145 inhibitor transfection
accelerated the migration rate compared to the control cells
(Fig. 3A). Matrigel Transwell
assays showed similar results. miR-145 overexpression in RKO and
SW480 significantly suppressed the invasion ability of the cells
(Fig. 2B), while the miR-145
knockdown increased the number of invaded cells (Fig. 3B). Collectively these results
indicate a possible tumor suppressive role of miR-145 in CRC
migration and invasion.
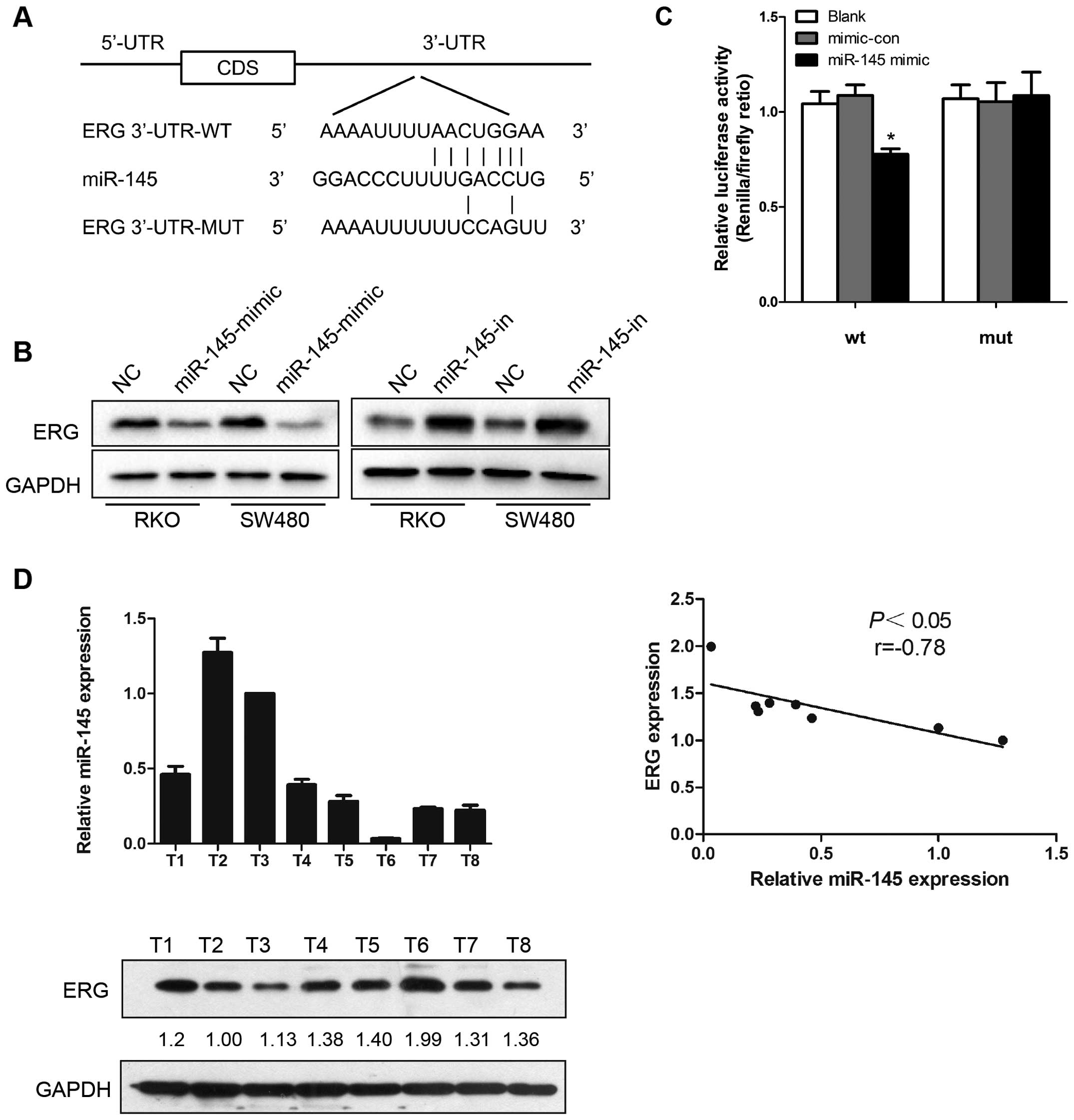
ERG is targeted by miR-145 in CRC
cells
As miRNA exerts its function by interacting with the
3′-UTR of its target genes, bioinformatic algorithms were applied
for target prediction. ERG was predicted to be a potential target
of miR-145 (Fig. 4A). Based on this
prediction, western blot analysis was performed to investigate the
effects of ectopic expression of miR-145 in CRC cells. In our study
miR-145 upregulation suppressed ERG protein expression in human CRC
cell lines, RKO and SW480, while miR-145 knockdown resulted in
increased ERG protein expression (Fig.
4B). Results of Dual-Luciferase reporter assays in HEK-293T
cells revealed that miR-145 mimic transfection reduced the
luciferase activity, while mutation in the predicted targeting
region abrogated this suppressive effect (Fig. 4C). These results indicate that the
interaction between miR-145 and ERG depended on the miRNA
recognition element in the 3′-UTR region. Furthermore, miR-145 and
ERG protein expression in eight colorectal cancer tissue samples
was examined both by qRT-PCR and western blot analysis,
respectively, and an inverse correlation was observed between
miR-145 and ERG expression (Fig.
4D).
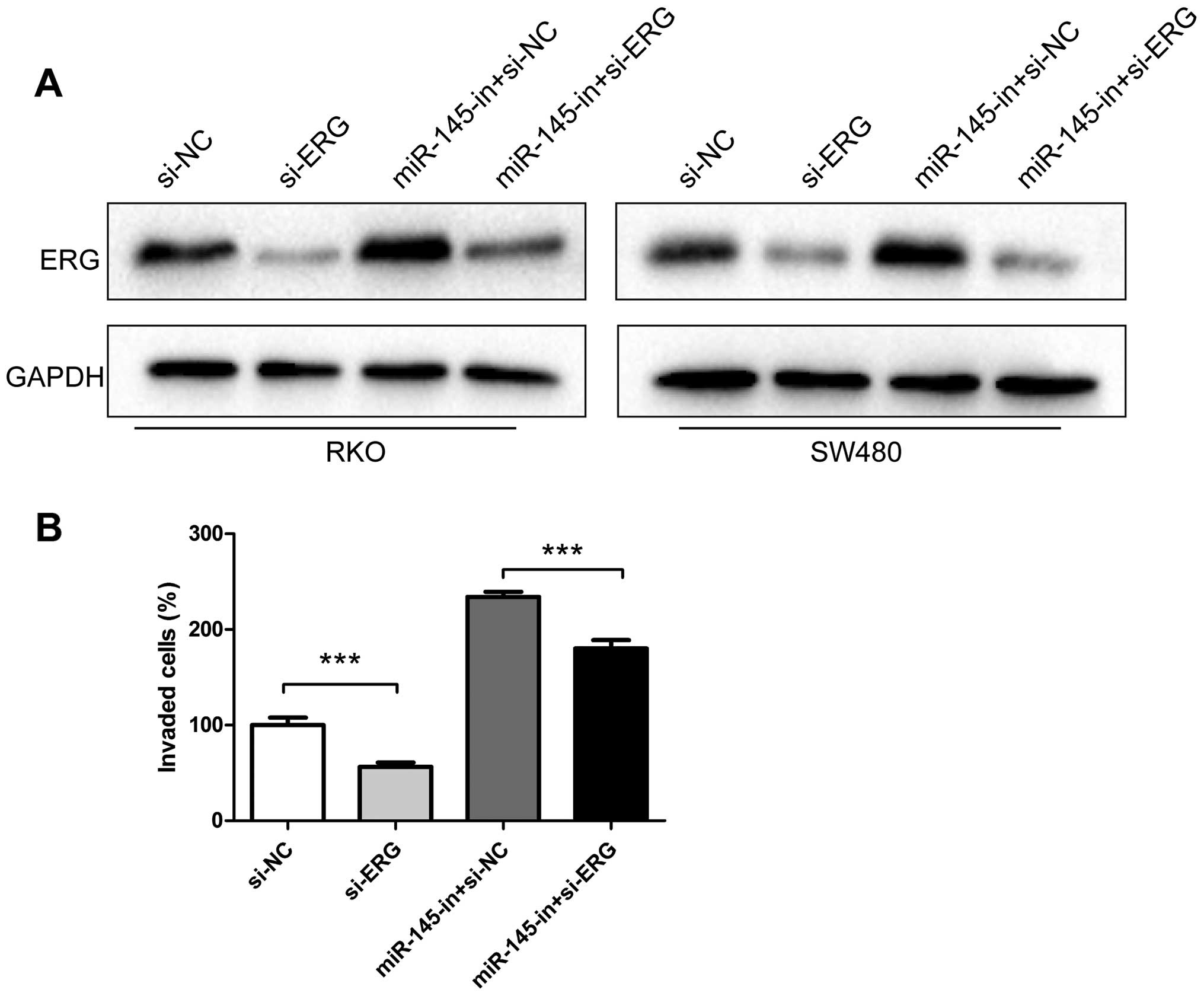
Downregulation of ERG partially reverses
the effect of miR-145 knockdown in CRC cells
To further investigate whether miR-145 exerts its
function in CRC cells through ERG regulation, siRNA targeting ERG
was synthetized and transfected into CRC cell lines RKO and SW480.
In our experiments, siRNA transfection downregulated the ERG
protein level (Fig. 5A). Moreover,
miR-145 inhibitor transfection increased ERG levels, while miR-145
inhibitor and ERG siRNA co-transfection partially recovered the ERG
expression compared to cells transfected only with ERG siRNA
(Fig. 5A). In the invasion
experiments Matrigel Transwell assay revealed that ERG siRNA
suppressed the invasiveness of RKO and SW480. In our study ERG
siRNA transfection decreased the number of invaded cells compared
to the negative control, while transfection with miR-145 inhibitors
increased the number of invading cells. Co-transfection of miR-145
inhibitor and ERG siRNA recovered, to some degree, the invasive
properties of RKO cells promoted by the miR-145 inhibitor (Fig. 5B).
Discusion
Over the past few decades, research efforts and
improved understanding of the carcinogenesis and progression of
colorectal cancer have contributed to its better diagnosis opening
new avenues for targeted therapy. However, the efficacy of targeted
therapy remains unsatisfactory due to our incomplete understanding
of the tumor pathology and cell signaling pathways (20).
Previous studies have shown that miR-145 is commonly
downregulated in various types of cancer, including CRC, which
indicated the possible role of miR-145 in carcinogenesis and tumor
progression (21). With regards to
its expression, miR-145 was found to be persistently decreased in
adenoma and colorectal neoplasms (12). In addition, miR-145 downregulation
was reported in ovarian cancer (22), cervical cancer (23), lung cancer (24), and gastric cancer (25). Previous functional studies have
shown that miR-145 transfection represses CRC cell proliferation,
invasion and metastasis by directly suppressing fascin-1 (26) and paxillin (27). In addition, miR-145 transfection
negatively regulated IGF-1R expression and suppressed proliferation
of Caco-2 cells (28). Furthermore,
other recent studies reported that miR-145 could suppress RAD18
expression and consequently enhance DNA damage in CRC cells after
5-FU treatment, therefore implying that there is a drug-resistance
reversal effect of miR-145 (29).
Moreover, upregulation of miR-145 increased the sensitivity of
drug-resistant Colo205 cells to vemurafenib both in vitro
and in vivo (30). In a
study by Pagliuca et al, resaturation of miR-145 and miR-143
in colon cancer cells showed that by targeting CD44, KLF5, KRAS and
BRAF, miR-145 and miR-143 there was a resultant coordinated
decrease in proliferation, migration and chemoresistance of colon
cancer cells (31). Therefore,
downregulation of miR-145 might play an important role in the
tumorigenesis and progression of colorectal cancer and further
studies on miR-145 in cancer might bring new avenues for miR-145
targeted therapy.
Our previous study (32) examined the miRNA expression profiles
of 31 pairs of CRC tissues and adjacent non-cancerous tissues and
miR-145 was downregulated in most of the clinical samples. Thus, we
further explored the role of miR-145 in colorectal tumorigenesis.
Consistent with previous studies and our earlier findings (32), miR-145 expression was downregulated
in both clinical CRC samples, as well as in cell lines, compared to
normal colorectal tissue samples. Moreover, miR-145 expression in
primary tumors taken from patients with lymph node metastasis was
significantly lower than in tumors of patients without lymph node
metastasis. This finding indicates a potential tumor-suppressive
role of miR-145 in tumor metastasis. Moreover, miR-145 transfection
in RKO and SW480 CRC cell lines significantly reduced cell
migration and invasiveness. Contrary to these findings, miR-145
downregulation via the miR-145 inhibitor promoted migration and
invasion of CRC cells compared to untreated cells.
In contrast to a majority of studies, a few studies
have proposed that miR-145 is CRC oncogenic. For instance, Yuan
et al reported that miR-145 promoted HCT-8 colon cancer cell
metastasis by stabilizing Hsp-27, a protein associated with
metastasis (33). In another study
by Arndt et al miR-145 promoted cell proliferation and
anchorage-independent growth by downregulating E-cadherin in the
colorectal cancer cell line SW620 (17). The fact that a specific miRNA can
regulate various target genes may explain its dual and even
contradictory influence in tumor initiation and progression.
Indeed, diverse cellular contexts, tumor types and target gene
sequence variations may explain why a specific miRNA might have
diverse functions in tumor initiation and progression (34,35).
Based on the present results and our previous findings (36) we propose that miR-145 acts as a
tumor suppressor during the progression of CRC.
ERG is one of the transcription factors belonging to
the ETS family, which is one of the largest families of
transcriptional regulators with diverse functions and activities
(37). All members of the ETS
family functionally regulate carcinogenesis-related processes such
as cell proliferation, angiogenesis, apoptosis and metastasis
(38,39). Previous studies have demonstrated
that ETS genes function as proto-oncogenes in colon cancer, and
ETS-1 and -2 expression was associated with lymph node metastasis
and advanced tumor grade in colon cancer (40). Ectopic expression of ERG was
demonstrated in other tumors, such as acute myeloid leukemia and
Ewing's sarcoma (41). Furthermore,
invasive breast cancer mRNA expression datasets reflected a general
ERG-driven pattern of malignancy (39). Emerging evidence suggest that ERG
overexpression is involved in oncogenesis and progression of
various cancer types; however, little is known about the regulation
of ERG in CRC carcinogenesis. Interestingly, other members of the
ETS family have been reported to be associated with tumor grade and
metastasis status in colon cancer (42). Furthermore, aberrant expression of
ERG and ETS-2 were associated with the development of cervical
carcinoma (43). In addition,
Scheble et al suggested that the TMPRSS2-ERG gene fusion,
which can lead to overexpression of ERG, is specific for prostate
cancer, and no such gene fusion was observed in CRC (44). Recent studies identified various
miRNAs that can regulate ETS. For example, miR-196a and b act as
ERG regulators in acute leukemia (45). miR-145 directly targets ERG in
prostate cancer and suppresses proliferation of prostate cancer
cells (46). Furthermore, miR-145
inhibits expression of ETS-1 in gastric cancer cells and exerts its
suppressive effect towards invasion, metastasis and angiogenesis by
regulating the genes downstream of ETS-1, such as matrix
metalloproteinase-1 and -9 (47).
miR-139 suppresses ETS-1 expression in CRC cells and inhibits cell
proliferation and G1/S phase cell cycle transition (48). Thus, based on these findings we
hypothesized that some regulatory mechanisms other than
transcriptional regulation might contribute to the dysregulation of
ERG observed in various cancer types including CRC. Our current
results showed that ERG was upregulated in CRC cancer specimens
compared to corresponding adjacent normal tissue. Based on the
computational algorithm analysis, we speculated that miR-145
directly regulates ERG through interaction with the 3′-UTR of ERG
mRNA. Our results confirmed our hypothesis since the downregulation
of miR-145 increased ERG expression, while the restoration of
miR-145 resulted in decreased ERG protein expression in CRC cells.
Dual-Luciferase reporter assay suggested that miR-145-mediated
suppression of ERG is dependent on the 3′-UTR of ERG mRNA. miR-145
expression inversely correlated with ERG protein levels, further
confirming the negative regulation of ERG by miR-145. Moreover, ERG
knockdown suppressed the invasion of CRC cells in vitro,
demonstrating that ERG may serve as a proto-oncogene in CRC.
Furthermore, co-transfection of ERG siRNA and miR-145 inhibitor
showed that ERG downregulation partially, but not completely,
reversed the tumor promoting effect caused by the miR-145
inhibitor. Taken together, these results indicate that ERG acts as
a proto-oncogene in CRC cells.
Although it is not yet clear whether ERG is
subjected to transcriptional regulation in CRC, this current study
indicates that there is post-transcriptional regulation of ERG by
miR-145 in CRC. Previous studies have shown that miR-145 inhibits
colon cancer cell growth by targeting Friend leukemia virus
integration-1 (FLI-1), another member of the ETS family of
transcription factors (49).
Furthermore, Ban et al reported feedback regulation between
EWS-FLI-1 and Hsa-miR-145 in Ewing's sarcoma (50). Therefore, along with our results, it
is reasonable to speculate that miR-145 may regulate different
members of the ETS transcription factor family and consequently
modulate diverse pathological processes in CRC. Further studies are
needed to understand the roles of miR-145 and ERG in CRC
metastasis, which might provide us with a novel therapy for the
treatment of colorectal cancer.
In conclusion, our results show that miR-145 is
persistently downregulated during CRC progression. In addition,
miR-145 might suppress CRC cell proliferation and invasion through
direct repression of the proto-oncogene ERG. Therefore, restoration
of miR-145 and suppression of ERG might be utilized as a potential
new therapeutic strategy in colorectal cancer in the future.
Acknowledgments
We appreciate the help generously offered by S.W.,
Z.L., R.C., X.W. and N.H. for the clinical sample collection. We
also thank the facility and academic advice given by the Key
Laboratory for Major Obstetric Diseases of Guangdong province and
the Key Laboratory of Reproduction and Genetics of Guangdong Higher
Education Institutes. This research was Supported by Science and
Technology Planning Project of Guangdong Province, China (grant no.
2016B090918130); Medical Science and Technology Research Foundation
of Guangdong Province (grant no. A2016500) and Youth Program of
Guangzhou Medical University (grant no. 2014A10). We also greatly
appreciate the funding support provided by Dr Anwei Chen.
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Loughran SJ, Kruse EA, Hacking DF, de
Graaf CA, Hyland CD, Willson TA, Henley KJ, Ellis S, Voss AK,
Metcalf D, et al: The transcription factor Erg is essential for
definitive hematopoiesis and the function of adult hematopoietic
stem cells. Nat Immunol. 9:810–819. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Birdsey GM, Dryden NH, Amsellem V,
Gebhardt F, Sahnan K, Haskard DO, Dejana E, Mason JC and Randi AM:
Transcription factor Erg regulates angiogenesis and endothelial
apoptosis through VE-cadherin. Blood. 111:3498–3506. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iwamoto M, Tamamura Y, Koyama E, Komori T,
Takeshita N, Williams JA, Nakamura T, Enomoto-Iwamoto M and
Pacifici M: Transcription factor ERG and joint and articular
cartilage formation during mouse limb and spine skeletogenesis. Dev
Biol. 305:40–51. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao X, Li LY, Zhou FJ, Xie KJ, Shao CK, Su
ZL, Sun QP, Chen MK, Pang J, Zhou XF, et al: ERG rearrangement for
predicting subsequent cancer diagnosis in high-grade prostatic
intraepithelial neoplasia and lymph node metastasis. Clin Cancer
Res. 18:4163–4172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marcucci G, Maharry K, Whitman SP,
Vukosavljevic T, Paschka P, Langer C, Mrózek K, Baldus CD, Carroll
AJ, Powell BL, et al Cancer and Leukemia Group B Study: High
expression levels of the ETS-related gene, ERG, predict adverse
outcome and improve molecular risk-based classification of
cytogenetically normal acute myeloid leukemia: A Cancer and
Leukemia Group B Study. J Clin Oncol. 25:3337–3343. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Markert EK, Mizuno H, Vazquez A and Levine
AJ: Molecular classification of prostate cancer using curated
expression signatures. Proc Natl Acad Sci USA. 108:21276–21281.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Becker-Santos DD, Guo Y, Ghaffari M,
Vickers ED, Lehman M, Altamirano-Dimas M, Oloumi A, Furukawa J,
Sharma M, Wang Y, et al: Integrin-linked kinase as a target for
ERG-mediated invasive properties in prostate cancer models.
Carcinogenesis. 33:2558–2567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
O'Donnell KA, Wentzel EA, Zeller KI, Dang
CV and Mendell JT: c-Myc-regulated microRNAs modulate E2F1
expression. Nature. 435:839–843. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Michael MZ, O' Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
13
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Watahiki A and Wang Y, Morris J, Dennis K,
O'Dwyer HM, Gleave M, Gout PW and Wang Y: MicroRNAs associated with
metastatic prostate cancer. PLoS One. 6:e249502011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang H, Pu J, Qi T, Qi M, Yang C, Li S,
Huang K, Zheng L and Tong Q: MicroRNA-145 inhibits the growth,
invasion, metastasis and angiogenesis of neuroblastoma cells
through targeting hypoxia-inducible factor 2 alpha. Oncogene.
33:387–397. 2014. View Article : Google Scholar
|
|
16
|
Panza A, Votino C, Gentile A, Valvano MR,
Colangelo T, Pancione M, Micale L, Merla G, Andriulli A, Sabatino
L, et al: Peroxisome proliferator-activated receptor γ-mediated
induction of microRNA-145 opposes tumor phenotype in colorectal
cancer. Biochim Biophys Acta. 1843:1225–1236. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Arndt GM, Dossey L, Cullen LM, Lai A,
Druker R, Eisbacher M, Zhang C, Tran N, Fan H, Retzlaff K, et al:
Characterization of global microRNA expression reveals oncogenic
potential of miR-145 in metastatic colorectal cancer. BMC Cancer.
9:3742009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Benson AB III, Arnoletti JP, Bekaii-Saab
T, Chan E, Chen YJ, Choti MA, Cooper HS, Dilawari RA, Engstrom PF,
Enzinger PC, et al National Comprehensive Cancer Network: Anal
Carcinoma, version 2.2012: Featured updates to the NCCN guidelines.
J Natl Compr Canc Netw. 10:449–454. 2012.PubMed/NCBI
|
|
19
|
Schindelin J, Rueden CT, Hiner MC and
Eliceiri KW: The ImageJ ecosystem: An open platform for biomedical
image analysis. Mol Reprod Dev. 82:518–529. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sameer AS: Colorectal cancer: Molecular
mutations and polymorphisms. Front Oncol. 3:1142013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo X, Burwinkel B, Tao S and Brenner H:
MicroRNA signatures: Novel biomarker for colorectal cancer? Cancer
Epidemiol Biomarkers Prev. 20:1272–1286. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive microRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu X, Sempere LF, Galimberti F,
Freemantle SJ, Black C, Dragnev KH, Ma Y, Fiering S, Memoli V, Li
H, et al: Uncovering growth-suppressive MicroRNAs in lung cancer.
Clin Cancer Res. 15:1177–1183. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Takagi T, Iio A, Nakagawa Y, Naoe T,
Tanigawa N and Akao Y: Decreased expression of microRNA-143 and
-145 in human gastric cancers. Oncology. 77:12–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dong R, Liu X, Zhang Q, Jiang Z, Li Y, Wei
Y, Li Y, Yang Q, Liu J, Wei JJ, et al: miR-145 inhibits tumor
growth and metastasis by targeting metadherin in high-grade serous
ovarian carcinoma. Oncotarget. 5:10816–10829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qin J, Wang F, Jiang H, Xu J, Jiang Y and
Wang Z: MicroRNA-145 suppresses cell migration and invasion by
targeting paxillin in human colorectal cancer cells. Int J Clin Exp
Pathol. 8:1328–1340. 2015.PubMed/NCBI
|
|
28
|
Shi B, Sepp-Lorenzino L, Prisco M, Linsley
P, de Angelis T and Baserga R: Micro RNA 145 targets the insulin
receptor substrate-1 and inhibits the growth of colon cancer cells.
J Biol Chem. 282:32582–32590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu RL, Dong Y, Deng YZ, Wang WJ and Li
WD: Tumor suppressor miR-145 reverses drug resistance by directly
targeting DNA damage-related gene RAD18 in colorectal cancer.
Tumour Biol. 36:5011–5019. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peng W, Hu J, Zhu XD, Liu X, Wang CC, Li
WH and Chen ZY: Overexpression of miR-145 increases the sensitivity
of vemurafenib in drug-resistant colo205 cell line. Tumour Biol.
35:2983–2988. 2014. View Article : Google Scholar
|
|
31
|
Pagliuca A, Valvo C, Fabrizi E, di Martino
S, Biffoni M, Runci D, Forte S, De Maria R and Ricci-Vitiani L:
Analysis of the combined action of miR-143 and miR-145 on oncogenic
pathways in colorectal cancer cells reveals a coordinate program of
gene repression. Oncogene. 32:4806–4813. 2013. View Article : Google Scholar
|
|
32
|
Xu XH, Wu XB, Wu SB, Liu HB and Chen Rand
Li Y: Identification of miRNAs differentially expressed in clinical
stages of human colorectal carcinoma-an investigation in Guangzhou,
China. PLoS One. 9:e940602014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yuan W, Sui C, Liu Q, Tang W, An H and Ma
J: Up-regulation of microRNA-145 associates with lymph node
metastasis in colorectal cancer. PLoS One. 9:e1020172014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu H and Kohane IS: Tissue and process
specific microRNA-mRNA co-expression in mammalian development and
malignancy. PLoS One. 4:e54362009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chaudhuri K and Chatterjee R: MicroRNA
detection and target prediction: Integration of computational and
experimental approaches. DNA Cell Biol. 26:321–337. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu X, Xu X, Li S, Wu S, Chen R, Jiang Q,
Liu H, Sun Y, Li Y and Xu Y: Identification and validation of
potential biomarkers for the detection of dysregulated microRNA by
qPCR in patients with colorectal adenocarcinoma. PLoS One.
10:e01200242015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Seth A and Watson DK: ETS transcription
factors and their emerging roles in human cancer. Eur J Cancer.
41:2462–2478. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dejana E, Taddei A and Randi AM: Foxs and
Ets in the transcriptional regulation of endothelial cell
differentiation and angiogenesis. Biochim Biophys Acta.
1775:298–312. 2007.PubMed/NCBI
|
|
39
|
Mochmann LH, Neumann M, von der Heide EK,
Nowak V, Kühl AA, Ortiz-Tanchez J, Bock J, Hofmann WK and Baldus
CD: ERG induces a mesenchymal-like state associated with
chemoresistance in leukemia cells. Oncotarget. 5:351–362. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ito Y, Takeda T, Okada M and Matsuura N:
Expression of ets-1 and ets-2 in colonic neoplasms. Anticancer Res.
22:1581–1584. 2002.PubMed/NCBI
|
|
41
|
Salek-Ardakani S, Smooha G, de Boer J,
Sebire NJ, Morrow M, Rainis L, Lee S, Williams O, Izraeli S and
Brady HJ: ERG is a megakaryocytic oncogene. Cancer Res.
69:4665–4673. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Peng C, Gao H, Niu Z, Wang B, Tan Z, Niu
W, Liu E, Wang J, Sun J, Shahbaz M, et al: Integrin αvβ6 and
transcriptional factor Ets-1 act as prognostic indicators in
colorectal cancer. Cell Biosci. 4:532014. View Article : Google Scholar
|
|
43
|
Simpson S, Woodworth CD and DiPaolo JA:
Altered expression of Erg and Ets-2 transcription factors is
associated with genetic changes at 21q22.2–22.3 in immortal and
cervical carcinoma cell lines. Oncogene. 14:2149–2157. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Scheble VJ, Braun M, Beroukhim R, Mermel
CH, Ruiz C, Wilbertz T, Stiedl AC, Petersen K, Reischl M, Kuefer R,
et al: ERG rearrangement is specific to prostate cancer and does
not occur in any other common tumor. Mod Pathol. 23:1061–1067.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Coskun E, von der Heide EK, Schlee C,
Kühnl A, Gökbuget N, Hoelzer D, Hofmann WK, Thiel E and Baldus CD:
The role of microRNA-196a and microRNA-196b as ERG regulators in
acute myeloid leukemia and acute T-lymphoblastic leukemia. Leuk
Res. 35:208–213. 2011. View Article : Google Scholar
|
|
46
|
Hart M, Wach S, Nolte E, Szczyrba J, Menon
R, Taubert H, Hartmann A, Stoehr R, Wieland W, Grässer FA, et al:
The proto-oncogene ERG is a target of microRNA miR-145 in prostate
cancer. FEBS J. 280:2105–2116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zheng L, Pu J, Qi T, Qi M, Li D, Xiang X,
Huang K and Tong Q: miRNA-145 targets v-ets erythroblastosis virus
E26 oncogene homolog 1 to suppress the invasion, metastasis, and
angiogenesis of gastric cancer cells. Mol Cancer Res. 11:182–193.
2013. View Article : Google Scholar
|
|
48
|
Gu J, Chen Y, Huang H, Yin L, Xie Z and
Zhang MQ: Gene module based regulator inference identifying miR-139
as a tumor suppressor in colorectal cancer. Mol Biosyst.
10:3249–3254. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang J, Guo H, Zhang H, Wang H, Qian G,
Fan X, Hoffman AR, Hu JF and Ge S: Putative tumor suppressor
miR-145 inhibits colon cancer cell growth by targeting oncogene
Friend leukemia virus integration 1 gene. Cancer. 117:86–95. 2011.
View Article : Google Scholar
|
|
50
|
Ban J, Jug G, Mestdagh P, Schwentner R,
Kauer M, Aryee DN, Schaefer KL, Nakatani F, Scotlandi K, Reiter M,
et al: Hsa-mir-145 is the top EWS-FLI1-repressed microRNA involved
in a positive feedback loop in Ewing's sarcoma. Oncogene.
30:2173–2180. 2011. View Article : Google Scholar : PubMed/NCBI
|