Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignancies with a high patient mortality rate (1,2).
According to incomplete statistics, there are 360,000 diagnosed
cases of primary liver cancer resulting in 350,000 deaths each year
in China (3). Various new treatment
options have been proposed and surgical resection is the most
common strategy. Yet, the prognosis of HCC remains poor due to the
difficulty in early detection and drug resistance (4–6). Thus,
identification of biomarkers for the diagnosis and treatment of HCC
is crucial.
Cancer stem cells (CSCs) are a type of cells
associated with tumor initiation, metastasis and drug resistance
(7–9). They are characterized by self-renewal
capacity and the ability fo differentiate into multiple cell types.
Moreover, they have been identified as therapeutic targets of many
types of cancers, such as HCC (10). In a study by Sun et al, liver
cancer stem-like cells (LCSCs) from MHCC97H cells were enriched and
their aggressive phenotype compared with MHCC97H cells was
confirmed. They also demonstrated their role in increased
metastatic potential (11). By
immunohistochemical methods, Matthai et al found that the
LCSC marker, EpCAM, was down-regulated in HCC, which was not
associated with hepatitis B virus (HBV) infection (12). Moreover, various genetic factors,
such as gene mutation and exogenous stimulation, were also found to
affect the properties of CSCs and thus cause the progression of
cancers. Thus, investigation of the regulatory mechanisms of CSCs
is necessary in order to improve the early detection, as well as
the efficacious treatment of cancers.
With the development of high-throughput
technologies, such as microarray and RNA sequencing (RNA-Seq), more
and more regulators of CSCs have been identified. Through small
RNA-Seq, Jones et al found that CDX1-miR-215 regulated the
differentiation of colorectal CSCs (13). Wang et al demonstrated the
capability of N-cadherin in the translation of CSCs in prostate
cancer through ErbB signaling by micro-array and western blot
analysis (14). Long non-coding
RNAs (lncRNAs) are non-protein coding transcripts which regulate
gene expression in normal or cancer cells. In the past few years,
many lncRNAs have been studied in LCSCs. They were shown to play
important roles in the initiation, progression and metastatic of
HCC. For example, lncRNA CUDR was found to regulate the malignant
differentiation of LCSCs under the regulation of HULC and β-catenin
(15). LncRNA HOTAIR was found to
promote the growth of LCSCs via the downregulation of SETD2
(16). However, the exact
mechanisms of CSCs in liver cancer cells remain unknown.
In the present study, using the RNA-Seq dataset in
the Gene Expression Omnibus (GEO), we identified the differentially
expressed genes (DEGs) in LCSCs compared with HCC cells. Function
and pathway enrichment analysis indicated the Gene Ontology (GO)
terms and pathways which may be involved in the process of
translation from HCC cells into LCSCs. Moreover, an lncRNA-gene
regulatory network screened out several regulatory relationships in
LCSCs, which may contribute to the further understanding of the
regulatory mechanisms of HCC.
Materials and methods
RNA-Seq dataset
The RNA-Seq dataset, GSE70537, which was deposited
by Ding et al (8) was
downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo/). It contains two HCC
samples (Hep3B, Huh7) and two hepatic cancer stem cell (HCSC)
samples (Hep3B-C, Huh7-C). In this dataset, the total RNAs from the
four samples were extracted and sequenced via Illumina Genome
Analyzer II.
Differential expression analysis
TopHat (17) was
used to map the RNA-Seq reads to the GRCh37/hg19 genome with the
max mismatches of two. The sorted bam files from TopHat were used
to calculate the numbers of the reads which were mapped to the
exons of a gene based on Cufflinks (17) and normalized to reads per kilo-base
of the exon model per million mapped reads (RPKM), which is the
representation of the expression values of the genes. Finally,
through Cuffdiff in Cufflinks, DEGs in HCSCs compared with HCC
cells were obtained with the cutoff of
|log2(RPKMHCSC/RPKMHCC)| >1 and
p<0.05.
Functional enrichment analysis
For the DEGs in HCSCs, the Database for Annotation,
Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) (18) was used for the functional and
pathway enrichment analysis. In this study, GO terms and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathways meeting p<0.05
were considered to be enriched.
Screening of lncRNA-gene pairs
In the past few years, lncRNAs have been identified
as important regulators in many phenotypes by knocking down or
overexpressing them followed by microarray or RNA-Seq experiments.
LncRNA2Target (http://www.lncrna2target.org) (19) is a web-based tool that stores
lncRNA-gene pairs which were obtained from the independent low
throughput, such as qRT-PCR and western blot analysis, or high
throughput, such as gene microarray and RNA-Seq. In the present
study, various lncRNAs which may regulate the DEGs were obtained
from the LncRNA2Target database and the lncRNA-gene regulatory
network was constructed and visualized by Cytoscape software
(20).
Results
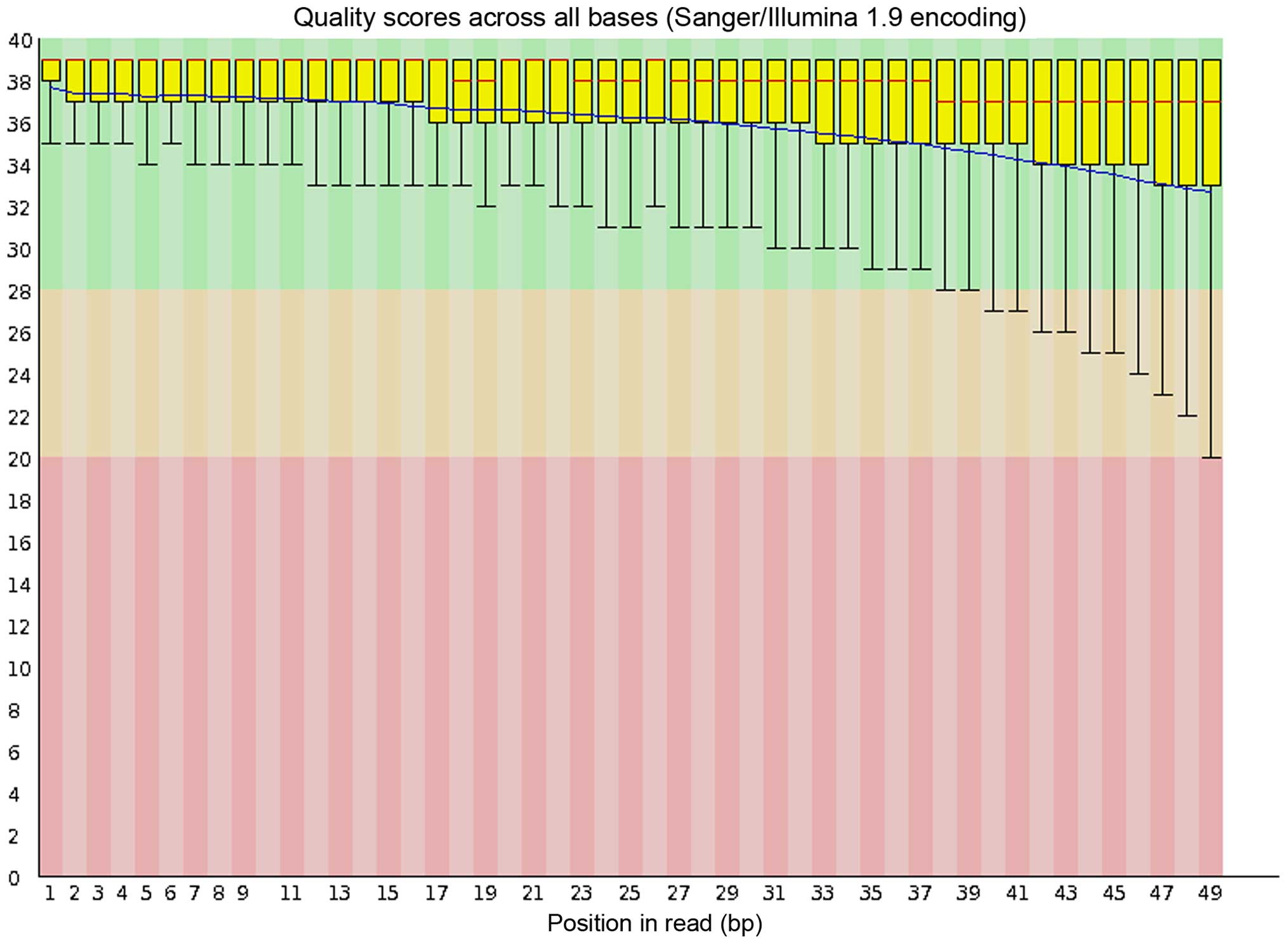
Quality control and mapping
As a representative, the quality control (QC)
outcome of the RNA-Seq reads of the Hep3B cells, which was obtained
from FastQC (http://www.bioinfor-matics.babraham.ac.uk/projects/fastqc/)
is shown in Fig. 1. An average of
95.5% (93.0–96.6%) mapping rate of each RNA-Seq sample was obtained
from TopHat and the mapping rate for each sample is shown in
Table I.
 | Table ITotal/mapped reads and mapping rates
in every sample. |
Table I
Total/mapped reads and mapping rates
in every sample.
| Sample ID | Total reads | Mapped reads | Mapping rates
(%) |
|---|
| Hep3B | 9,347,804 | 8,693,154 | 93.0 |
| Huh7 | 11,367,875 | 10,981,181 | 96.6 |
| Hep3B-C | 7,842,223 | 7,524,237 | 95.9 |
| Huh7-C | 13,785,623 | 13,276,070 | 96.3 |
Differential expression analysis
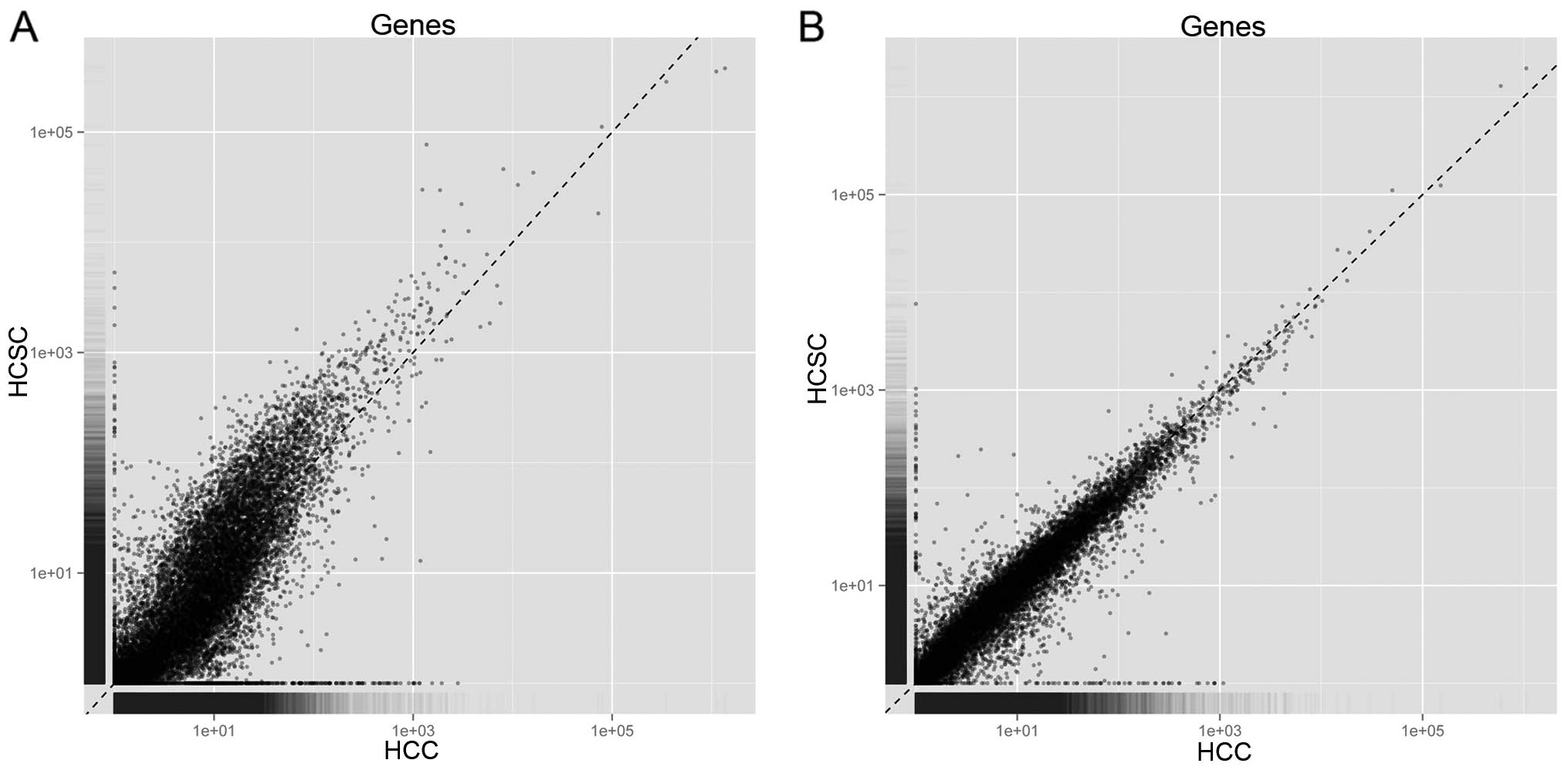
Based on Cufflinks, the mapped reads in each sample
were assembled into transcripts, and the number of reads mapped to
the genes was calculated. Fig. 2
shows the number of reads mapped to every gene between Hep3B and
Hep3B-C, as well as Huh7 and Huh7-C. Thus, it should be unbiased to
conduct differential expression analysis based on RPKM in the four
samples. By Cuffdiff, a number of 218 DEGs containing 82
downregulated and 136 upregulated DEGs were identified in the
Hep3B-C vs. Hep3B samples. In addition, a total of 591 DEGs
containing 298 down-regulated and 293 upregulated DEGs were
identified in the Huh7-C vs. Huh7 samples. A total of 22 overlaps
were found between these two lists of DEGs (Table II). Among these, 16 overlaps were
consistently upregulated or downregulated in the Hep3B-C and Huh7-C
samples.
 | Table IIOverlapped genes identified between
the DEGs in the Hep3B-C and Huh7-C samples. |
Table II
Overlapped genes identified between
the DEGs in the Hep3B-C and Huh7-C samples.
| Symbol | Log2FC_Hep3B-C | Log2FC_Huh7-C | Symbol | Log2FC_Hep3B-C | Log2FC_Huh7-C |
|---|
| FXYD3a | −5.98 | −3.09 | SPRY4b | 3.53 |
1.28 |
| CACNB2 | −4.71 |
2.53 | BMP4b | 3.63 |
1.35 |
| HDAC9 | −4.15 |
7.61 | BOP1b | 3.67 |
1.27 |
| IFI6a | −3.93 | −3.03 | HSD3B7 | 3.70 | −1.36 |
| TNS1a | −3.83 | −2.31 | NQO1b | 3.87 |
1.96 |
| PRTG | −3.61 |
1.89 | MVDb | 4.06 |
1.32 |
| NTS | −3.28 |
1.45 | UGT1A1b | 4.37 |
7.02 |
| KNG1a | −3.15 | −2.05 | SLC43A2b | 4.50 |
2.02 |
| MAT1A |
3.27 | −2.71 | HPDb | 4.56 |
2.20 |
| LY6Eb |
3.32 |
1.24 | FASNb | 4.61 |
1.51 |
| FDFT1b |
3.39 |
1.12 | VSNL1b | 4.88 |
3.95 |
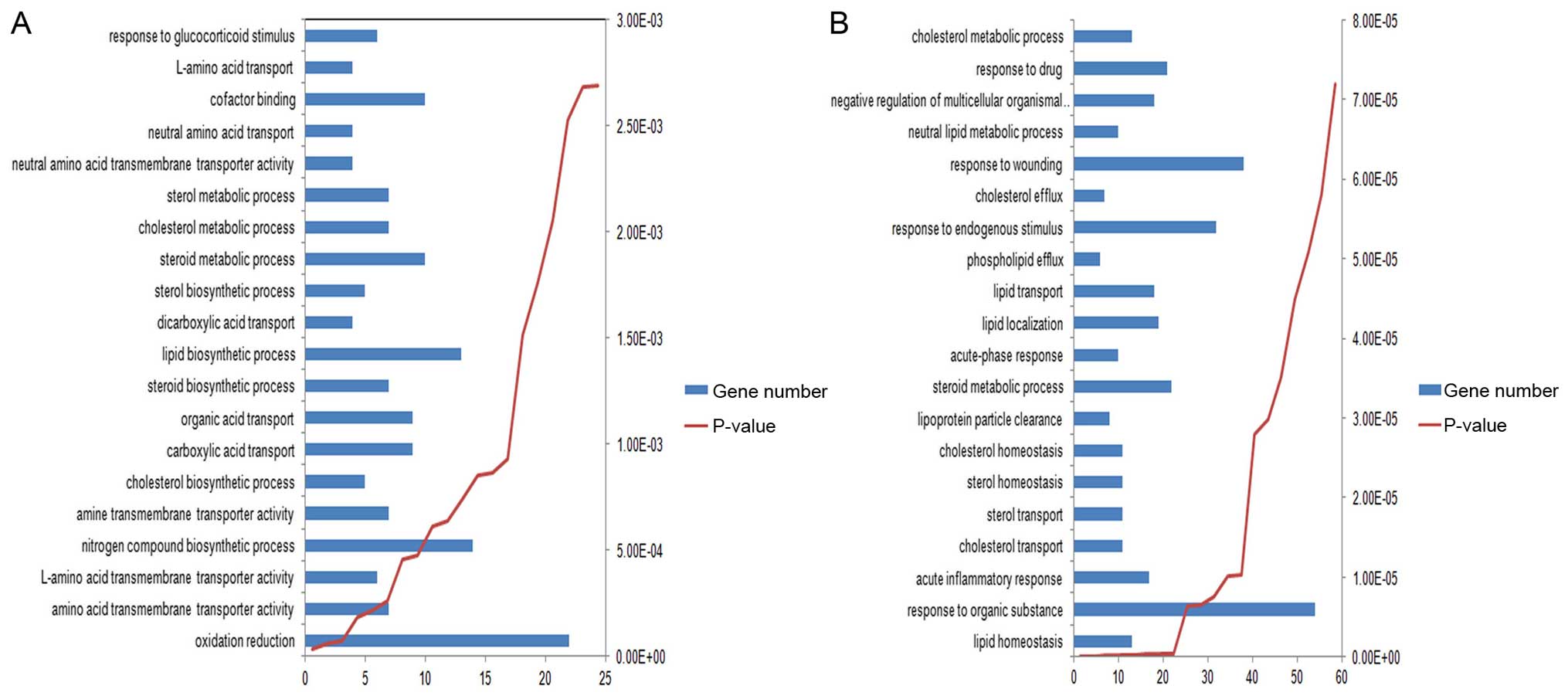
Enriched functions and pathways
Through functional and pathway enrichment analysis,
several GO terms were found to be co-enriched in the two lists of
DEGs, such as ʻsteroid metabolic process' and ʻcholesterol
metabolic process'. The top 20 most significantly enriched GO terms
in the two lists of DEGs are shown in Fig. 3. In addition, two pathways, which
include ʻarginine and proline metabolism' and ʻglycine, serine and
threonine metabolism', were found to be enriched in the DEGs of
Hep3B-C. The 7 enriched pathways of DEGs of Huh7-C are shown in
Table III.
 | Table IIIPathways enriched in the DEGs of the
Huh7-C samples. |
Table III
Pathways enriched in the DEGs of the
Huh7-C samples.
| Pathway name | No. of genes | P-value | Genes |
|---|
| Complement and
coagulation cascades | 10 | 0.00111 | KNG1, C8B, F12,
A2M, C3, SERPINF2, CD46, F2, SERPING1, PROC |
| Axon guidance | 13 | 0.00352 | ROCK2, GNAI1,
EFNA1, ABLIM3, EPHB3, CXCL12, EPHB2, EPHB6, NFAT5, SEMA3D, SEMA3C,
ROBO3, SEMA3A |
| PPAR signaling
pathway | 8 | 0.0154 | APOA2, CD36, APOA1,
HMGCS2, APOC3, ACSL4, CPT1A, PCK1 |
| TGF-β signaling
pathway | 9 | 0.0172 | INHBB, BMP4, EP300,
ID2, ROCK2, INHBE, ID1, ZFYVE9, FST |
| Terpenoid backbone
biosynthesis | 4 | 0.0177 | HMGCS2, MVD, HMGCR,
HMGCS1 |
| Leukocyte
transendothelial migration | 10 | 0.0354 | ITGAL, VAV3, CLDN4,
ROCK2, GNAI1, PECAM1, CLDN2, ESAM, CXCL12, CLDN14 |
| Synthesis and
degradation of ketone bodies | 3 | 0.0437 | HMGCS2, OXCT1,
HMGCS1 |
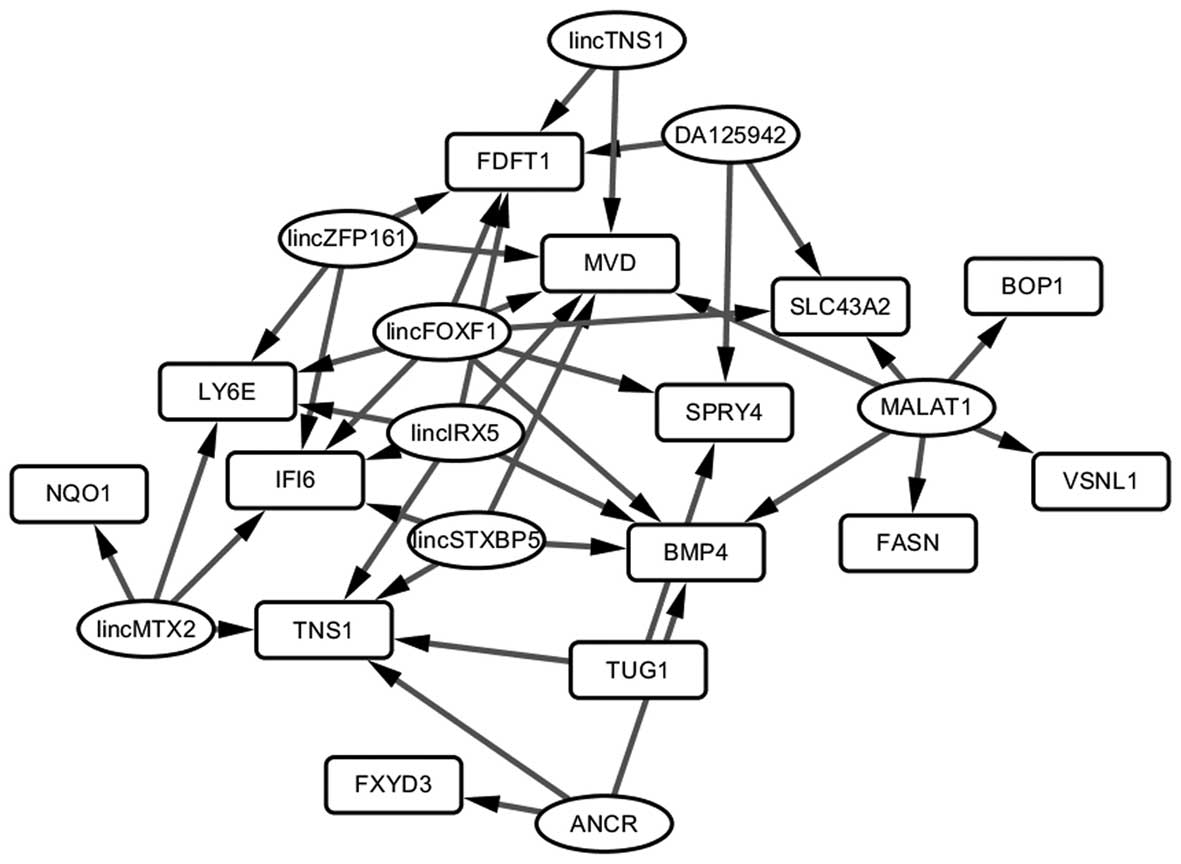
lncRNA-gene pairs
In the LncRNA2Target database, 10 lncRNAs were found
to regulate 13 of the 16 DEGs with consistent differential
expression in the Hep3B-C and Huh7-C samples. Among them, 42
lncRNA-gene pairs were identified. By Cytoscape, the lncRNA-gene
regulatory network (Fig. 4) was
constructed. lincFOXF1 was the hub node in the network and
regulated 7 genes. It may act as a target which influences the
transformation of HCC cells to HCSCs.
Discussion
The process of transformation of HCC cells to
hepatocellular carcinoma stem cells (HCSCs) is associated with the
initiation, progression, metastasis and poor prognosis of HCC
(21,22). Yet, the specific mechanism remains
unclear. In the present study, through reanalysis of RNA-Seq
dataset in GEO, we identified various key factors that may
contribute to the development of HCSCs and serve as new therapeutic
targets in the treatment of HCC.
Hep3B and Huh7 are two types of HCC cells which have
been used in many studies (23,24).
In the dataset of this study, HCSCs (Hep3B-C, Huh7-C) were cultured
from Hep3B and Huh7 cells. Then, the total RNA of these four
samples was extracted and sequenced by Illumina Genome Analyzer II.
Here, we identified various DEGs in the Hep3B-C vs. Hep3B and
Huh7-C vs. Huh7 samples, respectively. A total of 22 overlaps were
obtained between these two lists of DEGs. In addition, 16 genes
were found to display consistent differential expression in the
Hep3B-C and Huh7-C samples. These results indicate that various
common mechanisms exist in the development of HCSC samples from
different HCC cells which is consistent with previous studies
(25,26). FXYD3 was one of the most
downregulated genes in both HCSC samples among the 16 identified
genes. FXYD3, also known as MTA8 and PLML,
encodes a cell membrane protein which regulates the function of ion
pumps and ion channels. It also plays an important role in tumor
progression (27–29). The upregulation of FXYD3 is
associated with many types of cancers, such as rectal cancer,
breast cancer, endometrial cancer and intrahepatic
cholangiocarcinoma (30–33). In the present study, FXYD3
was found to be downregulated in the Hep3B-C and Huh7-C samples,
which has not been demonstrated by any other previous study. This
indicates its role in the transformation of HCSCs from HCC cells
and the association with the development of HCC.
The pathways of ʻsubstance metabolism and
transport', including ʻsteroid metabolic process' and ʻcholesterol
metabolic process' were found to be enriched in the two lists of
DEGs. As steroid is an important metabolite, the dysregulation of
its functions is associated with the generation of CSCs in many
types of cancers and could affect progression (34–36).
In this study, MVD, which is related to cholesterol
biosynthesis, was found to be present in both of these two
pathways: ʻsteroid biosynthetic/metabolic process' and ʻterpenoid
backbone biosynthesis' in the Huh7-C sample. To date, there is no
study that shows the relationship between MVD and the
development of HCC. Thus, this gene may be a potential novel
biomarker.
lncRNAs are a type of non-coding RNAs which populate
in the cancer genome. They play an important role in gene
translation and have been identified as potential biomarkers in
many types of cancers (37).
Moreover, the rapid growth of high-throughput technologies, such as
microarray and RNA-Seq, has accelerated the mining of novel lncRNAs
followed by the vast emergence of new resources. For example,
NONCODE 2016 (http://www.bioinfo.org/noncode/), which was developed
by Zhao et al, provides the largest amount of lncRNA
collections and annotation information, including conservation
annotation and references (38). As
a manually curated lncRNA database with experimental support,
Lnc2Cancer (http://www.bio-bigdata.net/lnc2cancer) could provide
the relationship between lncRNAs and various human cancers
(39). In the present study,
LncRNA2Target curated lncRNA-gene pairs through manipulating low-
and high-throughput experiments to screen the potential lncRNAs for
the 16 DEGs which displayed consistent differential expression in
the two types of HCSCs. In the lncRNA-gene regulatory network, we
found that lincFOXF1 regulated 7 DEGs and was identified as a hub
node. lincFOXF1 is also known as FENDRR and its
downregulation was found to be associated with gastric cancer cell
metastasis and poor prognosis (40). It also plays an important role in
mammalian embryogenesis (41), and
has been closely associated with stem cell development. Thus, it
may also affect the transformation of CSCs and provide valuable
targets for cancer treatment. Moreover, the targets of lincFOXF1,
including BMP4, FDFT1, IFI6, LY6E,
MVD, SLC43A2 and SPRY4, have been validated as
playing roles in cancer progression and metastasis, and several
were novel targets identified in the present study. Yet, further
studies are still needed.
In conclusion, through the analysis of the RNA-Seq
dataset in GEO, we identified various potential targets that may
contribute to the generation of HCSCs and are associated with the
initiation, progression and metastasis of HCC. These targets may
facilitate the diagnosis and treatment of HCC and improve the
prognosis of HCC patients.
Acknowledgments
We would like to thank all the members of our
research group for their enthusiastic participation in this
study.
References
|
1
|
Chen X, Liu HP, Li M and Qiao L: Advances
in non-surgical management of primary liver cancer. World J
Gastroenterol. 20:16630–16638. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Margini C and Dufour JF: The story of HCC
in NAFLD: From epidemiology, across pathogenesis, to prevention and
treatment. Liver Int. 36:317–324. 2016. View Article : Google Scholar
|
|
3
|
Chen JG and Zhang SW: Liver cancer
epidemic in China: Past, present and future. Semin Cancer Biol.
21:59–69. 2011. View Article : Google Scholar
|
|
4
|
D'Alessandro R, Messa C, Refolo MG and
Carr BI: Modulation of sensitivity and resistance to multikinase
inhibitors by microenvironmental platelet factors in HCC. Expert
Opin Pharmacother. 16:2773–2780. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zou H, Feng X and Cao JG: Twist in
hepatocellular carcinoma: Pathophysiology and therapeutics. Hepatol
Int. 9:399–405. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Frau M, Biasi F, Feo F and Pascale RM:
Prognostic markers and putative therapeutic targets for
hepatocellular carcinoma. Mol Aspects Med. 31:179–193. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Todoroki K, Ogasawara S, Akiba J, Nakayama
M, Naito Y, Seki N, Kusukawa J and Yano H:
CD44v3+/CD24− cells possess cancer stem
cell-like properties in human oral squamous cell carcinoma. Int J
Oncol. 48:99–109. 2016.
|
|
8
|
Ding M, Li J, Yu Y, Liu H, Yan Z, Wang J
and Qian Q: Integrated analysis of miRNA, gene, and pathway
regulatory networks in hepatic cancer stem cells. J Transl Med.
13:2592015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moitra K: Overcoming multidrug resistance
in cancer stem cells. BioMed Res Int. 2015:6357452015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Khorrami S, Zavaran Hosseini A, Mowla SJ
and Malekzadeh R: Verification of ALDH activity as a biomarker in
colon cancer stem cells-derived HT-29 cell line. Iran J Cancer
Prev. 8:e34462015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun J, Luo Q, Liu L, Zhang B, Shi Y, Ju Y
and Song G: Biomechanical profile of cancer stem-like cells derived
from MHCC97H cell lines. J Biomech. 49:45–52. 2016. View Article : Google Scholar
|
|
12
|
Matthai SM and Ramakrishna B: Cancer stem
cells in hepatocellular carcinoma - an immunohistochemical study
with histopathological association. Indian J Med Res. 142:391–398.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jones MF, Hara T, Francis P, Li XL, Bilke
S, Zhu Y, Pineda M, Subramanian M, Bodmer WF and Lal A: The CDX1-
microRNA-215 axis regulates colorectal cancer stem cell
differentiation. Proc Natl Acad Sci USA. 112:E1550–E1558. 2015.
View Article : Google Scholar
|
|
14
|
Wang M, Ren D, Guo W, Huang S, Wang Z, Li
Q, Du H, Song L and Peng X: N-cadherin promotes
epithelial-mesenchymal transition and cancer stem cell-like traits
via ErbB signaling in prostate cancer cells. Int J Oncol.
48:595–606. 2016.
|
|
15
|
Gui X, Li H, Li T, Pu H and Lu D: Long
noncoding RNA CUDR regulates HULC and beta-catenin to govern human
liver stem cell malignant differentiation. Mol Ther. 23:1843–1853.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H, An J, Wu M, Zheng Q, Gui X, Li T, Pu
H and Lu D: LncRNA HOTAIR promotes human liver cancer stem cell
malignant growth through downregulation of SETD2. Oncotarget.
6:27847–27864. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
19
|
Jiang Q, Wang J, Wu X, Ma R, Zhang T, Jin
S, Han Z, Tan R, Peng J, Liu G, et al: LncRNA2Target: A database
for differentially expressed genes after lncRNA knockdown or
overexpression. Nucleic Acids Res. 43:D193–D196. 2015. View Article : Google Scholar :
|
|
20
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li J, Yu Y, Wang J, Yan Z, Liu H, Wang Y,
Ding M, Cui L, Wu M, Jiang X, et al: Establishment of a novel
system for the culture and expansion of hepatic stem-like cancer
cells. Cancer Lett. 360:177–186. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
You N, Zheng L, Liu W, Zhong X, Wang W and
Li J: Proliferation inhibition and differentiation induction of
hepatic cancer stem cells by knockdown of BC047440: A potential
therapeutic target of stem cell treatment for hepatocellular
carcinoma. Oncol Rep. 31:1911–1920. 2014.PubMed/NCBI
|
|
23
|
Yao CJ, Yeh CT, Lee LM, Chuang SE, Yeh CF,
Chao WJ, Lai TY and Lai GM: Elimination of cancer stem-like ʻside
population' cells in hepatoma cell lines by chinese herbal mixture
ʻtien-hsien liquid'. Evid Based Complement Alternat Med.
2012:6170852012. View Article : Google Scholar
|
|
24
|
Chen YL, Yuan RH, Yang WC, Hsu HC and Jeng
YM: The stem cell E3-ligase Lin-41 promotes liver cancer
progression through inhibition of microRNA-mediated gene silencing.
J Pathol. 229:486–496. 2013. View Article : Google Scholar
|
|
25
|
Yang Z, Zhang L, Ma A, Liu L, Li J, Gu J
and Liu Y: Transient mTOR inhibition facilitates continuous growth
of liver tumors by modulating the maintenance of CD133+
cell populations. PLoS One. 6:e284052011. View Article : Google Scholar
|
|
26
|
Nishina S, Shiraha H, Nakanishi Y, Tanaka
S, Matsubara M, Takaoka N, Uemura M, Horiguchi S, Kataoka J,
Iwamuro M, et al: Restored expression of the tumor suppressor gene
RUNX3 reduces cancer stem cells in hepatocellular carcinoma by
suppressing Jagged1-Notch signaling. Oncol Rep. 26:523–531.
2011.PubMed/NCBI
|
|
27
|
Zhu ZL, Zhao ZR, Zhang Y, Yang YH, Wang
ZM, Cui DS, Wang MW, Kleeff J, Kayed H, Yan BY, et al: Expression
and significance of FXYD-3 protein in gastric adenocarcinoma. Dis
Markers. 28:63–69. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang MW, Gu P, Zhang ZY, Zhu ZL, Geng Y,
Kayed H, Zentgraf H and Sun XF: FXYD3 expression in gliomas and its
clinicopathological significance. Oncol Res. 18:133–139. 2009.
View Article : Google Scholar
|
|
29
|
Widegren E, Onnesjö S, Arbman G, Kayed H,
Zentgraf H, Kleeff J, Zhang H and Sun XF: Expression of FXYD3
protein in relation to biological and clinicopathological variables
in colorectal cancers. Chemotherapy. 55:407–413. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Herrmann P and Aronica SM: Estrogen and
tamoxifen up-regulate FXYD3 on breast cancer cells: Assessing the
differential roles of ER α and ZEB1. Springerplus. 4:2452015.
View Article : Google Scholar
|
|
31
|
Li Y, Zhang X, Xu S, Ge J, Liu J, Li L,
Fang G, Meng Y, Zhang H and Sun X: Expression and clinical
significance of FXYD3 in endometrial cancer. Oncol Lett. 8:517–522.
2014.PubMed/NCBI
|
|
32
|
Loftås P, Onnesjö S, Widegren E, Adell G,
Kayed H, Kleeff J, Zentgraf H and Sun XF: Expression of FXYD-3 is
an independent prognostic factor in rectal cancer patients with
preoperative radiotherapy. Int J Radiat Oncol Biol Phys.
75:137–142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Subrungruanga I, Thawornkunob C,
Chawalitchewinkoon-Petmitrc P, Pairojkul C, Wongkham S and Petmitrb
S: Gene expression profiling of intrahepatic cholangiocarcinoma.
Asian Pac J Cancer Prev. 14:557–563. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hilton HN and Clarke CL: Impact of
progesterone on stem/progenitor cells in the human breast. J
Mammary Gland Biol Neoplasia. 20:27–37. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Finlay-Schultz J and Sartorius CA: Steroid
hormones, steroid receptors, and breast cancer stem cells. J
Mammary Gland Biol Neoplasia. 20:39–50. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Callari M, Guffanti A, Soldà G, Merlino G,
Fina E, Brini E, Moles A, Cappelletti V and Daidone MG: In-depth
characterization of breast cancer tumor-promoting cell
transcriptome by RNA sequencing and microarrays. Oncotarget.
7:976–994. 2016.
|
|
37
|
Sahu A, Singhal U and Chinnaiyan AM: Long
noncoding RNAs in cancer: From function to translation. Trends
Cancer. 1:93–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y,
Li Z, Bu D, Sun N, Zhang MQ, et al: NONCODE 2016: An informative
and valuable data source of long non-coding RNAs. Nucleic Acids
Res. 44:D203–D208. 2016. View Article : Google Scholar :
|
|
39
|
Ning S, Zhang J, Wang P, Zhi H, Wang J,
Liu Y, Gao Y, Guo M, Yue M, Wang L, et al: Lnc2Cancer: A manually
curated database of experimentally supported lncRNAs associated
with various human cancers. Nucleic Acids Res. 44:D980–D985. 2016.
View Article : Google Scholar :
|
|
40
|
Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin
L, Chen WM, Han L, Zhang EB, Kong R, et al: Decreased expression of
the long non-coding RNA FENDRR is associated with poor prognosis in
gastric cancer and FENDRR regulates gastric cancer cell metastasis
by affecting fibronectin1 expression. J Hematol Oncol. 7:632014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Grote P and Herrmann BG: The long
non-coding RNA Fendrr links epigenetic control mechanisms to gene
regulatory networks in mammalian embryogenesis. RNA Biol.
10:1579–1585. 2013. View Article : Google Scholar : PubMed/NCBI
|