|
1
|
Mendell JT: MicroRNAs: Critical regulators
of development, cellular physiology and malignancy. Cell Cycle.
4:1179–1184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Szabo G and Bala S: MicroRNAs in liver
disease. Nat Rev Gastroenterol Hepatol. 10:542–552. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu C, Iqbal J, Teruya-Feldstein J, Shen
Y, Dabrowska MJ, Dybkaer K, Lim MS, Piva R, Barreca A, Pellegrino
E, et al: MicroRNA expression profiling identifies molecular
signatures associated with anaplastic large cell lymphoma. Blood.
122:2083–2092. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hummel R, Wang T, Watson DI, Michael MZ,
Van der Hoek M, Haier J and Hussey DJ: Chemotherapy-induced
modification of microRNA expression in esophageal cancer. Oncol
Rep. 26:1011–1017. 2011.PubMed/NCBI
|
|
9
|
Pathak S, Meng WJ, Nandy SK, Ping J,
Bisgin A, Helmfors L, Waldmann P and Sun XF: Radiation and SN38
treatments modulate the expression of microRNAs, cytokines and
chemokines in colon cancer cells in a p53-directed manner.
Oncotarget. 6:44758–44780. 2015.PubMed/NCBI
|
|
10
|
Shoshan E, Mobley AK, Braeuer RR, Kamiya
T, Huang L, Vasquez ME, Salameh A, Lee HJ, Kim SJ, Ivan C, et al:
Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma
growth and metastasis. Nat Cell Biol. 17:311–321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hudson J, Duncavage E, Tamburrino A,
Salerno P, Xi L, Raffeld M, Moley J and Chernock RD: Overexpression
of miR-10a and miR-375 and downregulation of YAP1 in medullary
thyroid carcinoma. Exp Mol Pathol. 95:62–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sand M, Skrygan M, Sand D, Georgas D, Hahn
SA, Gambichler T, Altmeyer P and Bechara FG: Expression of
microRNAs in basal cell carcinoma. Br J Dermatol. 167:847–855.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
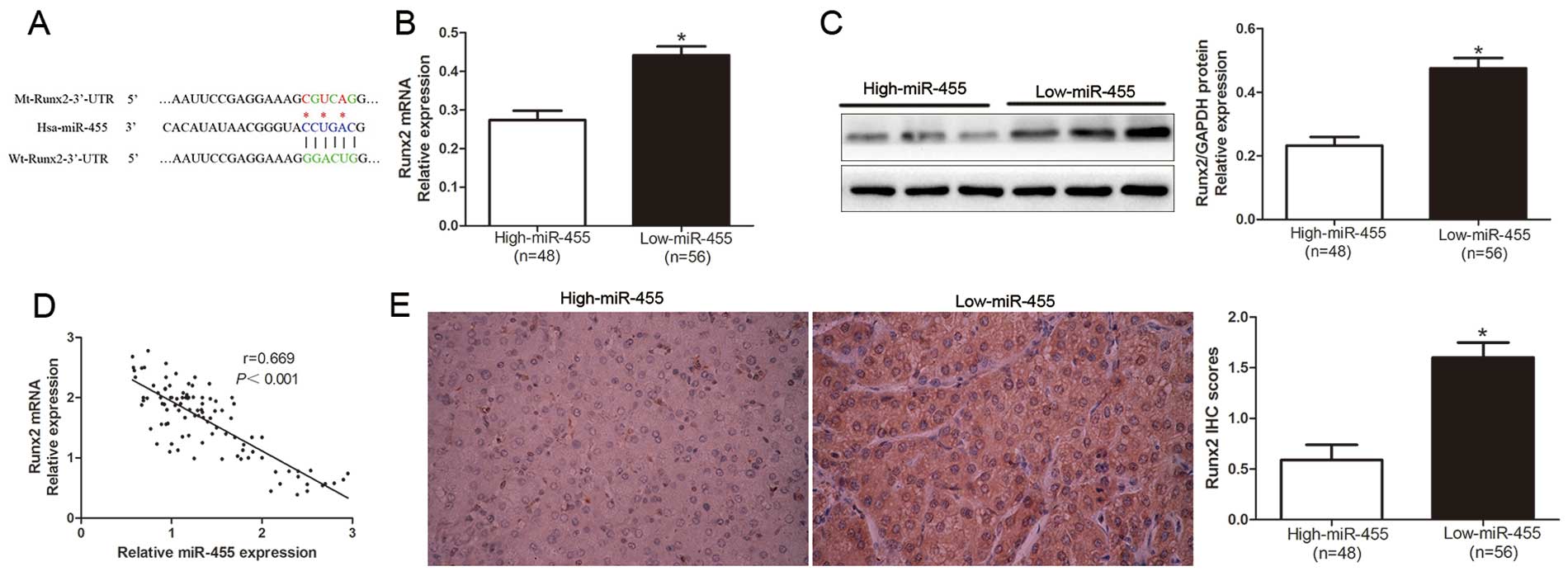
Zhang Z, Hou C, Meng F, Zhao X, Zhang Z,
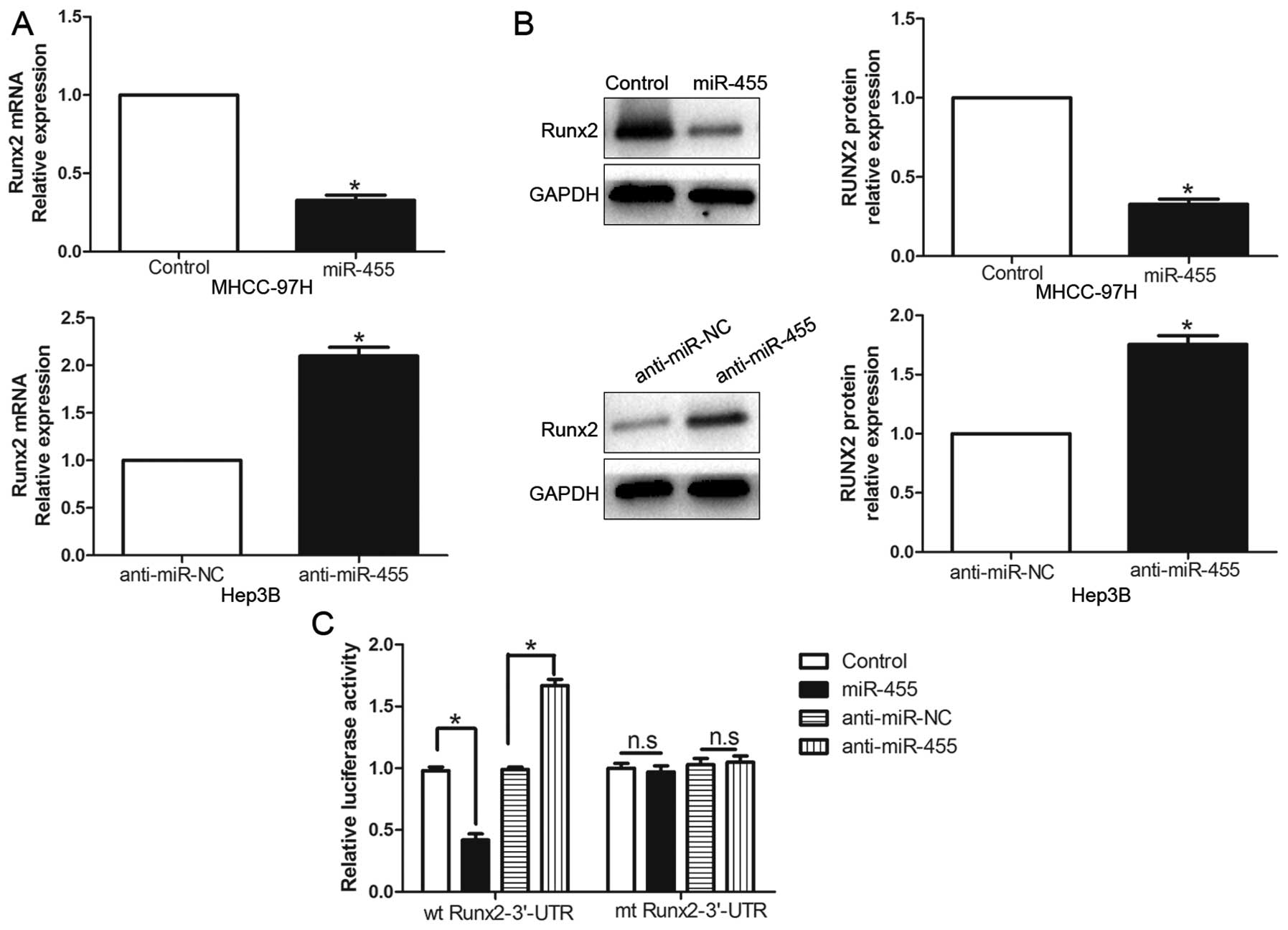
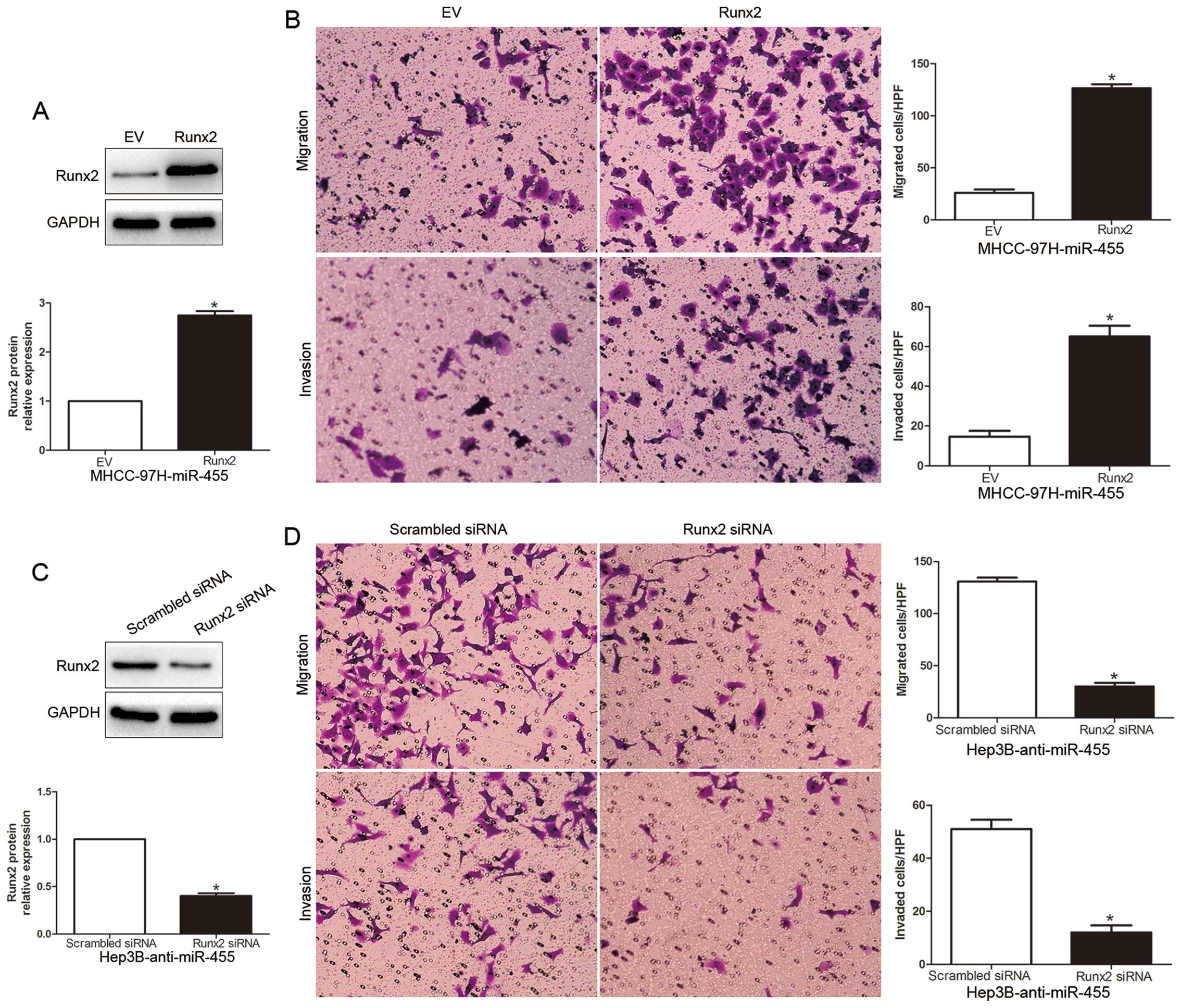
Huang G, Chen W, Fu M and Liao W: MiR-455-3p regulates early
chondrogenic differentiation via inhibiting Runx2. FEBS Lett.
589:3671–3678. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boisen MK, Dehlendorff C, Linnemann D,
Nielsen BS, Larsen JS, Osterlind K, Nielsen SE, Tarpgaard LS,
Qvortrup C, Pfeiffer P, et al: Tissue microRNAs as predictors of
outcome in patients with metastatic colorectal cancer treated with
first line capecitabine and oxaliplatin with or without
bevacizumab. PLoS One. 9:e1094302014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bera A, VenkataSubbaRao K, Manoharan MS,
Hill P and Freeman JW: A miRNA signature of chemoresistant
mesenchymal phenotype identifies novel molecular targets associated
with advanced pancreatic cancer. PLoS One. 9:e1063432014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hamilton MP, Rajapakshe K, Hartig SM, Reva
B, McLellan MD, Kandoth C, Ding L, Zack TI, Gunaratne PH, Wheeler
DA, et al: Identification of a pan-cancer oncogenic microRNA
superfamily anchored by a central core seed motif. Nat Commun.
4:27302013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hiroki E, Akahira J, Suzuki F, Nagase S,
Ito K, Suzuki T, Sasano H and Yaegashi N: Changes in microRNA
expression levels correlate with clinicopathological features and
prognoses in endometrial serous adenocarcinomas. Cancer Sci.
101:241–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li YJ, Ping C, Tang J and Zhang W:
MicroRNA-455 suppresses non-small cell lung cancer through
targeting ZEB1. Cell Biol Int. 40:621–628. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chai J, Wang S, Han D, Dong W, Xie C and
Guo H: MicroRNA-455 inhibits proliferation and invasion of
colorectal cancer by targeting RAF proto-oncogene
serine/threonine-protein kinase. Tumour Biol. 36:1313–1321. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song G, Gu L, Li J, Tang Z, Liu H, Chen B,
Sun X, He B, Pan Y, Wang S, et al: Serum microRNA expression
profiling predict response to R-CHOP treatment in diffuse large B
cell lymphoma patients. Ann Hematol. 93:1735–1743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang H and Wang Y: Five miRNAs considered
as molecular targets for predicting neuroglioma. Tumour Biol.
37:1051–1059. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baniwal SK, Khalid O, Gabet Y, Shah RR,
Purcell DJ, Mav D, Kohn-Gabet AE, Shi Y, Coetzee GA and Frenkel B:
Runx2 transcriptome of prostate cancer cells: Insights into
invasiveness and bone metastasis. Mol Cancer. 9:2582010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Akech J, Wixted JJ, Bedard K, van der Deen
M, Hussain S, Guise TA, van Wijnen AJ, Stein JL, Languino LR,
Altieri DC, et al: Runx2 association with progression of prostate
cancer in patients: Mechanisms mediating bone osteolysis and
osteoblastic metastatic lesions. Oncogene. 29:811–821. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brusgard JL, Choe M, Chumsri S, Renoud K,
MacKerell AD Jr, Sudol M and Passaniti A: RUNX2 and TAZ-dependent
signaling pathways regulate soluble E-cadherin levels and
tumorsphere formation in breast cancer cells. Oncotarget.
6:28132–28150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tandon M, Chen Z and Pratap J: Runx2
activates PI3K/Akt signaling via mTORC2 regulation in invasive
breast cancer cells. Breast Cancer Res. 16:R162014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van der Deen M, Akech J, Wang T, Fitz
Gerald TJ, Altieri DC, Languino LR, Lian JB, van Wijnen AJ, Stein
JL and Stein GS: The cancer-related Runx2 protein enhances cell
growth and responses to androgen and TGFβ in prostate cancer cells.
J Cell Biochem. 109:828–837. 2010.PubMed/NCBI
|
|
27
|
Cohen-Solal KA, Boregowda RK and Lasfar A:
RUNX2 and the PI3K/AKT axis reciprocal activation as a driving
force for tumor progression. Mol Cancer. 14:1372015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Niu DF, Kondo T, Nakazawa T, Oishi N,
Kawasaki T, Mochizuki K, Yamane T and Katoh R: Transcription factor
Runx2 is a regulator of epithelial-mesenchymal transition and
invasion in thyroid carcinomas. Lab Invest. 92:1181–1190. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tandon M, Chen Z, Othman AH and Pratap J:
Role of Runx2 in IGF-1Rβ/Akt- and AMPK/Erk-dependent growth,
survival and sensitivity towards metformin in breast cancer bone
metastasis. Oncogene. 35:4730–4740. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wysokinski D, Blasiak J and Pawlowska E:
Role of RUNX2 in breast carcinogenesis. Int J Mol Sci.
16:20969–20993. 2015. View Article : Google Scholar : PubMed/NCBI
|