Introduction
Ovarian cancer is the most life-threatening
malignancy in reproductive tract and fourth leading cause of
cancer-related deaths in women. Majority of ovarian cancers stem
from epithelium, though there are also stromal and germ cell tumors
(1,2). Generally, ovarian tumors are silent at
early stage and remain unrecognized in most cases (>80%) until
ovarian carcinoma has metastasized to other areas out of the
ovaries. Most recently, 21,550 American new cases of ovarian cancer
and 14,600 deaths from the cancer in 2009 were reported by the
American Cancer Society (www.cancer.org). There is little improvement in 5-year
survival rates over the past 10 years, and long-term survival is
significantly unsatisfactory and among the worst of all the
anatomic sites of cancer.
Ral paralogs bind to an array of effectors such as
ZONAB, phospholipase D1 (PLD1), filamin, exocyst components (Exo84
and Sec5) and RalBP1 (RLIP) upon activation, leading to various
functions associated with Ral (3).
Earlier findings showed that RalBP1, the best characterized
effector of Ral, was correlated with Ral-mediated tumorigenesis
(4). Additionally, the human
bladder cancer tissues had higher expression of RalBP1 than normal
tissues, which was associated with Ral expression (5). The subcutaneous xenograft tumor growth
in cancer cell lines could be reduced by deletion of RalBP1 by
suppression of its function by antibody or by antisense reduction
(6). In RalBP1, there is
association between the exocyst complex component Sec5 and
transformation mediated by Ral (4).
There was also correlation between PLD1 and cell transformation by
oncogenes such as Ras (7).
MicroRNAs are non-protein-coding RNAs that play
post-transcriptional regulator of genes. microRNAs have been shown
to serve a role in almost all cellular processes in animals and
plants first discovered as critical regulators of developmental
timing in Caenorhabditis elegans (8,9).
MicroRNAs have ~22 nucleotides, derived from large major
transcripts which generate imperfect structures of stem loop.
MicroRNAs bind to target mRNAs and suppress transition or enhance
transcript degradation, thereby regulating the expression of genes
(10).
Notably, a great deal of microRNAs serve as crucial
regulator of gene expression and are generally present in the
non-protein-encoding regions (11).
More than 60% of protein-coding transcripts are regulated by small
non-coding RNAs known as microRNAs (12). Each microRNA regulates a range of
target genes at the post-transcriptional level. They are associated
with a variety of disorders and serve as tumor inhibitors and
oncogenes to affect tumorigenesis (13). For instance, microRNAs have been
associated with ovarian tumor development and progression (14). It is reported that germline
variations in processing genes and microRNAs known as messenger RNA
transcripts of their target genes play a role in tumor progression
as well as the risk of development of cancers, such as ovarian
cancer (15).
It has been previously shown that miR-143-3p is
differentially expressed in the A2780 cells collected from ovarian
carcinoma, and dysregulation of RalA-binding protein 1 (RALBP1) has
also been reported to be involved in the molecular mechanism of
apoptosis of A2780 cells (16,17).
By searching the online miRNA database, we found that RALBP1 is a
virtual target of miR-143-3p. In the present study, we validated
RALBP1 as a target of miR-143-3p and verified the involvement of
miR-143-3p and RALBP1 in the development of ovarian carcinoma.
Materials and methods
Subjects
The present study consisted of 35 patients treated
at the Department of Gynecology, Affiliated Hospital of Jining
Medical Unversity (Jining, China) between November 2013 and January
2015. All data processing and sample collection were approved by
the Ethics Committee of Affiliated Hospital of Jining Medical
Unversity. Participants or their first-degree relatives had already
signed the informed consents before start of the experiments after
carefully explained all potential risk factors. Cancerous tissue
samples and adjacent non-cancerous control were collected from each
participant. All of the specimens were obtained from surgical
resection. None of the patients was treated with any neoadjuvant
treatment, such as radiotherapy and chemotherapy, before surgical
resection and the followed tumor resection to collect clinical
data.
RNA isolation and real-time PCR
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was
used to extract the total RNA from A2780 cells and tissue samples
based on the manufacturer's protocol. NanoDrop spectrophotometer
(ND-1000; NanoDrop Technologies, Wilmington, DE, USA) was used to
measure the quantity and quality of RNA, and the gel
electrophoresis was used to detect the integrity of RNA. For
detecting the miR-143-3p expression, miRcute miRNA Isolation kit
(Qiangen Biotech Co., Ltd., Beijing, China), miRcute miRNA
First-Strand cDNA synthesis kit and miRcute miRNA qPCR detection
kit (SYBR-Green) were used to detect the expression of RALBP1 mRNA
or miR-143-3p. MiRcute miRNA qPCR detection kit (SYBR-Green)
(Qiangen Biotech Co., Ltd.) was used to perform the reverse
transcription-polymerase chain reaction (RT-PCR) in accordance with
the protocol of the kit supplier with a final 20 µl PCR reaction
containing 10 µl miRcute miRNA premix, 0.4 µl reverse primers, 2 µl
of cDNA samples synthesized and 0.4 µl forward. CFX96™ Real-Time
System (Bio-Rad, Hercules, CA, USA) was used to carry out the
reactions. The internal control was U6. Bio-Rad CFX Manager
quantitative PCR software (Bio-Rad) was used to examine the change
in expression of miR-143-3p and RALBP1 mRNA, and the
2−ΔΔCt method was used to calculate the change in
expression of RALBP1 gene and miR-143-3p. Three independent tests
were performed.
Cell culture and transfection
Dulbeccos modified Eagles medium (DMEM) (Cellgro
Mediatech, Manassas, VA, USA) containing 1% non-essential amino
acids (Sigma-Aldrich, St. Louis, MO, USA), 2 mM L-glutamine, 50
µg/ml streptomycin sulfate, 50 U/ml penicillin and 10% fetal bovine
serum (FBS) (Invitrogen) was used to maintain the A2780 cells in an
atmosphere of 5% CO2/95% air at 37°C. When cells were
cultured to 80% confluency, transfection was performed using
Lipofectamine 2000 (Invitrogen) according to the manufacturer's
protocol. A2780 cells were analyzed 48 h after transfection.
Cell proliferation assay
A2780 cell proliferation was measured using the
Alamar Blue assay according to the quantitative metabolic
conversion of non-fluorescent resazurin and blue to fluorescent
resorufin and pink by living A2780 cells. An Alamar Blue
(Invitrogen) stock solution was added to the wells equal to 10% of
the total incubation volume for 2–6 h. Synergy HT Multi-Mode
Microplate Reader (Bio-Tek Instruments, Winooski, VT, USA) was used
to determine the reduction of resazurin in the cultures according
to the absorbance at 590 and 530 nm wavelengths. Each test was
performed at least three times.
Luciferase assay
The PCR fragment including the binding site was used
to make the luciferase constructs. The 3 untranslated region (3UTR)
of RALBP1 with the target site of miR-143-3p was inserted into the
XhoI and NotI sites of the psiCHECK-2 (Promega,
Madison, WI, USA) following the manufacturers recommendations.
Mutations were introduced using site-directed mutagenesis
(Stratagene, La Jolla, CA, USA). For the reporter assays,
Lipofectamine 2000 was used to co-transfect A2780 cells with
negative control (NC mimics), mutant miR-143-3p or wild-type
miR-143-3p mimics (SunBio Corporation, Shanghai, China) and
reporter plasmids (500 ng). Promega Luciferase Assay System was
used to determine luciferase activity according to the activity of
Renilla luciferase (control). All tests were repeated in
triplicate.
Western blot analysis
For analysis of the expression of RALBP1 protein,
the tissue sample or culture cells were lyzed using lysis buffer
including 1 mmol/l sodium orthovanadate, 2 mmol/l EDTA, 150 mmol/l
NaCl, 25 µg/ml leupeptin, 1% deoxycholate plus, 10 µg/ml aprotinin,
1% Triton X-100 and 50 mmol/l Tris-HCl pH 7.4. Then, the cellular
lysis were centrifuged for 15 min at 15,000 rpm, and the Bradford
assay (Bio-Rad) was used to analyze the quantity of the cellular
protein. SDS-PAGE (12%) was used to separate the lysates, and then
electro-transferred onto polyvinylidene difluoride (PVDF) membranes
(PerkinElmer, Waltham, MA, USA) for 2 h (90 V). Tris-buffered
saline containing 0.1% Tween-20 (TBST) with 5% non-fat dry milk
(avoid non-specificity binding) was used to block the membranes for
2 h. Then, monoclonal antibodies anti-human RALBP1 (1:1,000; Santa
Cruz Biotechnology, Shanghai, China) were used to incubate the
membranes for 12 h at 4°C in accordance with the manufacturer's
description, then the membranes were washed three times using PBST
(BioSharp, Hefei, China); next, secondary antibody at 1:5,000
dilution (Cell Signaling Technologies, Cambridge, MA, USA) in PBST
was used to treat the membranes for another 1 h, subsequently the
membranes were washed three times using PBST for 5 min. The
enhanced chemiluminescence (ECL) detection system (Pierce
Biotechnology, Inc., Rockford, IL, USA) was used to develop the
bands following the manufacturer's protocol. Each test was repeated
in triplicate.
Apoptosis analysis
Annexin V/propidium iodide staining with the
apoptosis detection kit (KenGEN, China) was used to detect A2780
cell apoptosis based on the manufacturer's protocol. Annexin V was
used to stain the A2780 cells subsequently, and then FACSCalibur
flow cytometer (BD Biosciences, New Jersey, NY, USA) was used to
analyze cell apoptosis. Each test was repeated in triplicate.
Statistical analysis
All data are presented as mean ± SD (standard
deviation). Every experiment was analyzed three times, with three
samples for each. Student's t-test was used to evaluate the
comparisons of treatment group. p-value <0.05 was considered to
indicate a statistically significant result. Each test was repeated
in triplicate.
Results
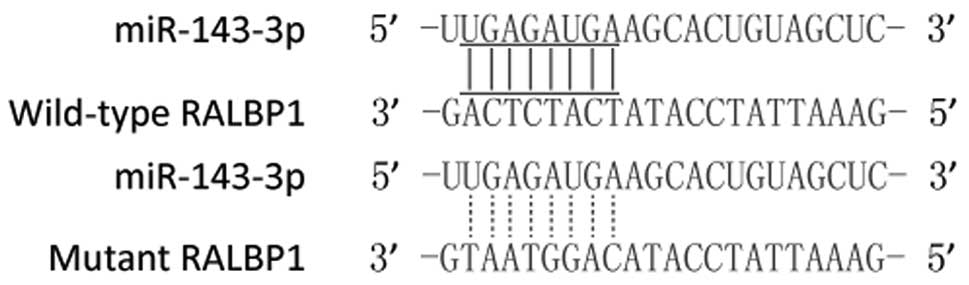
RALBP1 is a target of miR-143-3p
The relationship between the RALBP1-3'UTR and its
targeted miRNAs was predicted through bioinformatics analysis by
the bioinformatics algorithms TargetScan http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do) which showed
that the 3′-UTR of RALBP1 may be targeted by miR-143-3p with
potential ‘hits’ in the 3'UTR of RALBP1 (Fig. 1).
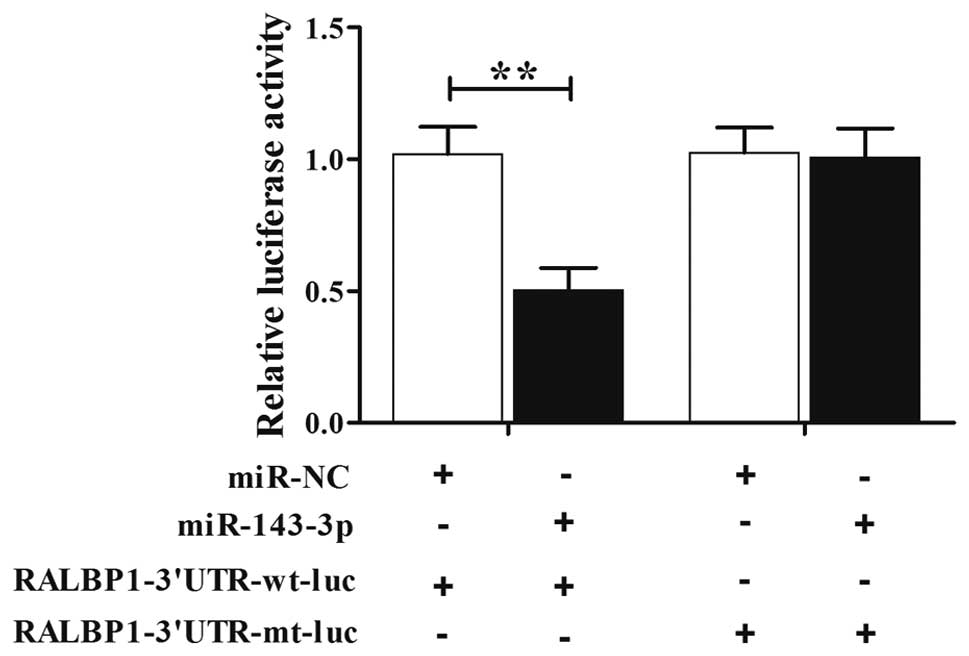
Luciferase assay was used to further confirm the
association between miR-143-3p and RALBP1. Then, the cells were
transfected with construct including wild-type or mutant RALBP1
3'UTR. As shown in Fig. 2, we found
that the luciferase activity of the cells co-transfected with
miR-143-3p mimics and wild-type RALBP1 3'UTR was lower than those
transfected with the scramble controls, while the introduction of
mutant RALBP1 with the potential ‘seed sequence’ in the 3'TUR of
RALBP1 almost completely abolished such repressive effect,
indicating that miR-143-3p negatively regulated RALBP1.
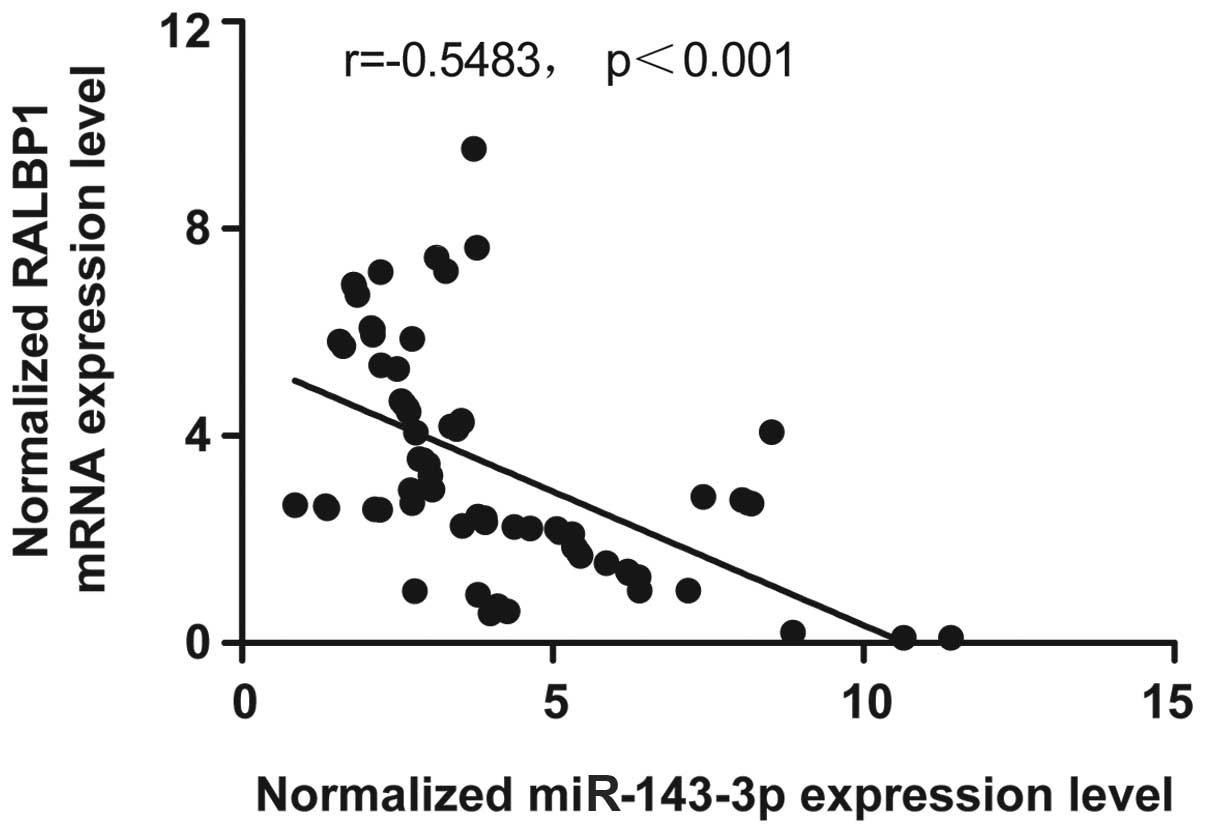
The negative regulatory relationship
between RALBP1 and miR-143-3p
The miRNA-mRNA regulatory relationship was confirmed
using RT-PCR. The results confirmed the negative regulatory
relationship between miR-143-3p and RALBP1, and the negative
correlation coefficient was −0.5483 (r=−0.5483), as shown in
Fig. 3.
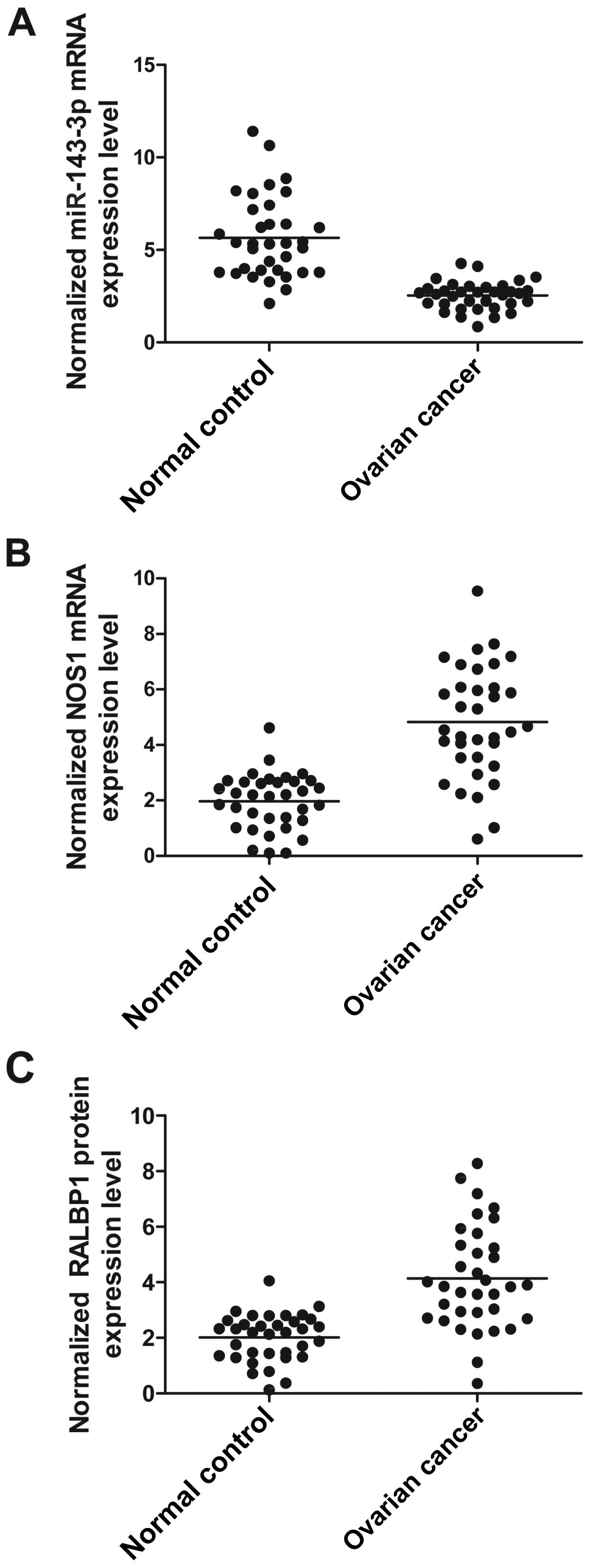
Upregulation of miR-143-3p reduces the
expression of RALBP1
Using real-time PCR and western blot analyses, we
found the expression of miR-143-3p (Fig. 4A) was much lower in participants
with compared with normal control, while the expression of RALBP1
mRNA (Fig. 4B) and protein
(Fig. 4C) were evidently
overexpressed in participants with ovarian cancer compared with
normal control, indicated that downregulated expression of
miR-143-3p and overexpression of RALBP1 can induce the development
of ovarian cancer.
Effect of miR-143-3p on the expression
of RALBP1
To further confirm whether RALBP1 negatively
correlated with miR-143-3p, we investigated the mRNA and protein
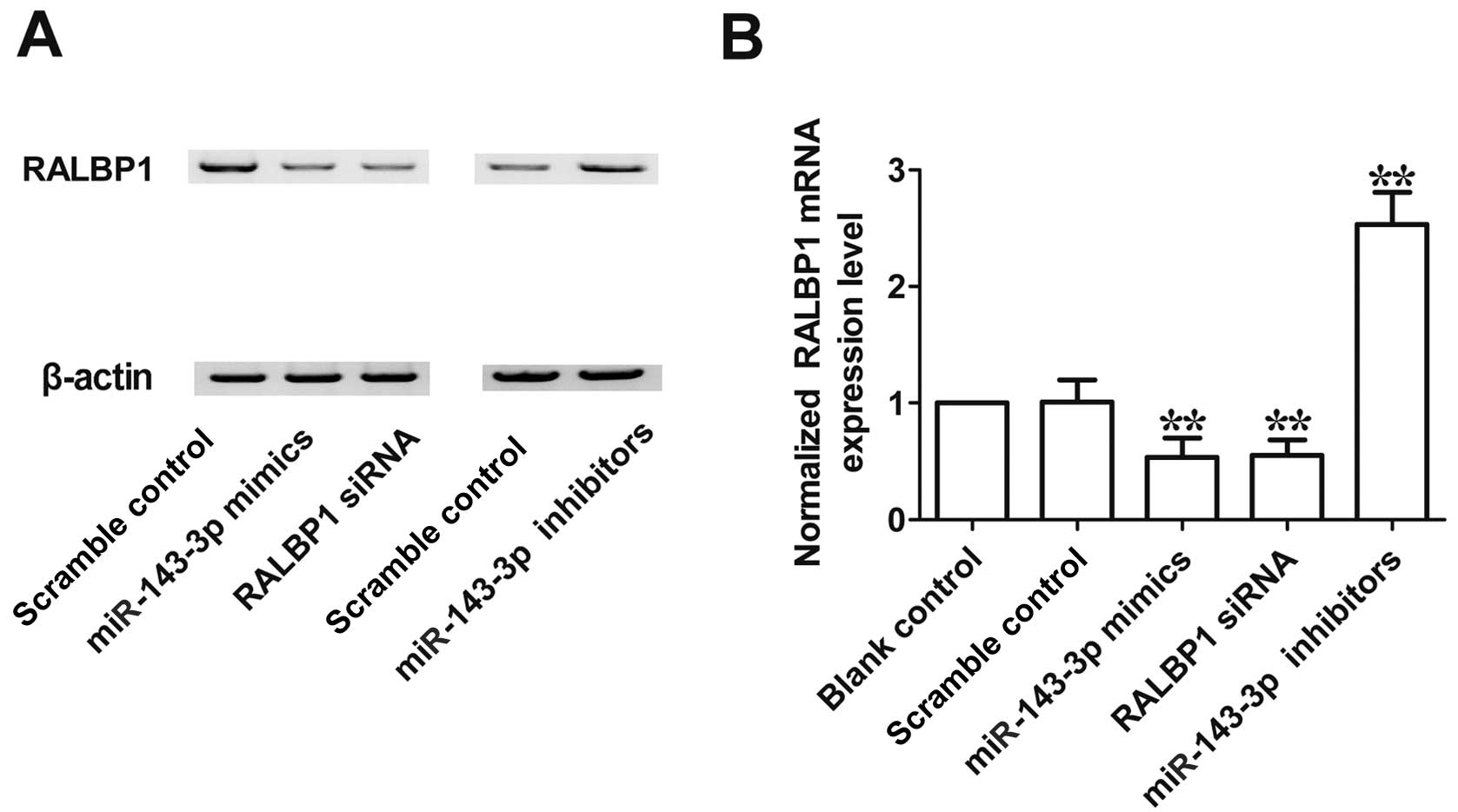
expression level of RALBP1 in A2780 cells. As shown in Fig. 5, the expression level of RALBP1 mRNA
(Fig. 5A) and protein (Fig. 5B) in cells transfected with
miR-143-3p mimics or RALBP1 siRNA was significantly lower than in
the scramble control, while the RALBP1 mRNA (Fig. 5A) and protein (Fig. 5B) level were evidently upregulated
following transfection with miR-143-3p inhibitor. These
observations indicated that there was negative regulatory
relationship between miR-143-3p and RALBP1.
Introduction of miR-143-3p
significantly affects the viability and apoptosis in A2780
cells
To study the molecular mechanism underlying the
observed viability-reduced and apoptosis-promoting effect of
miR-143-3p, we performed flow cytometry analysis in the cells
transfected with miR-143-3p mimics, miR-143-3p inhibitor and RALBP1
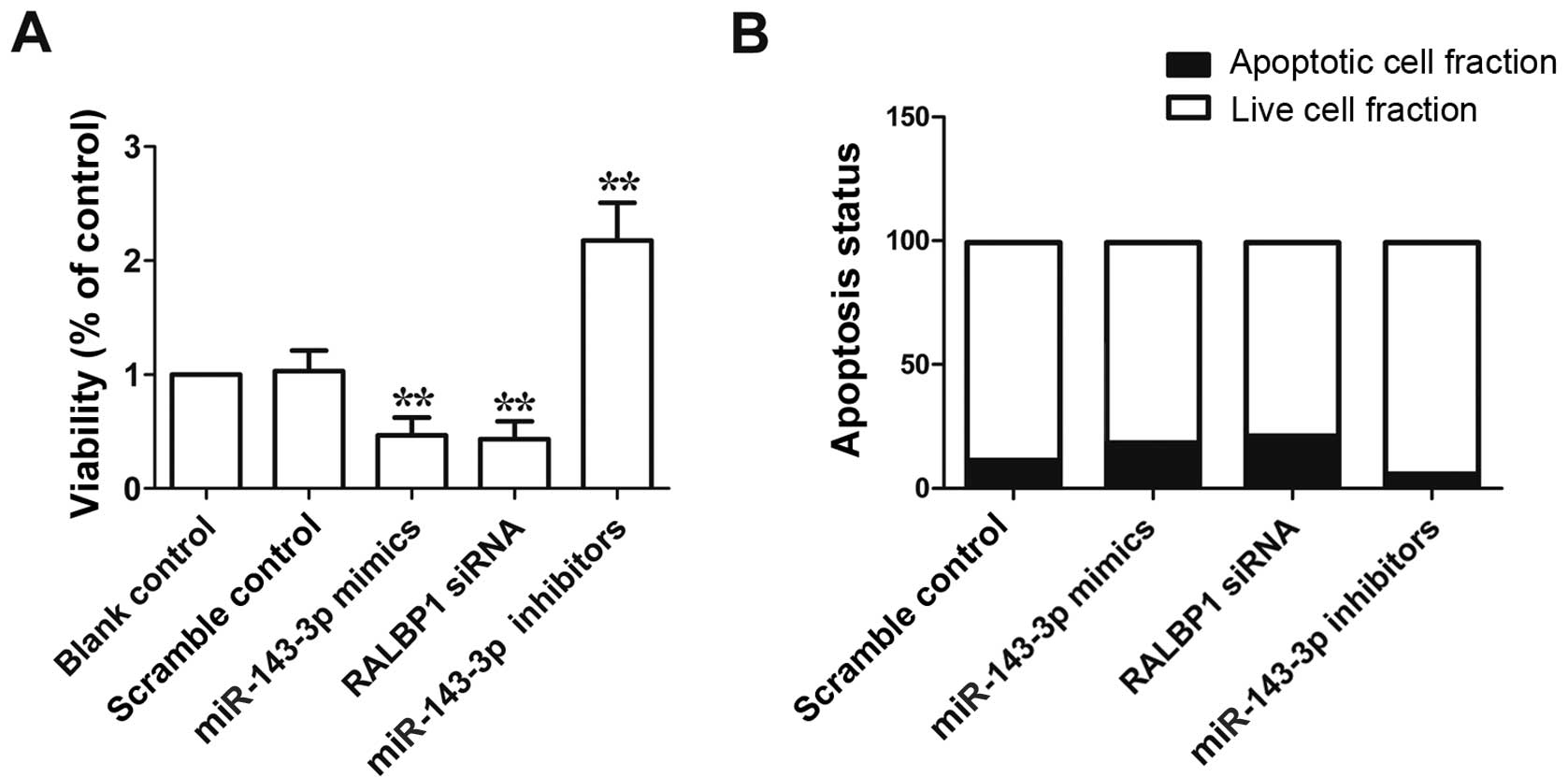
siRNA, as shown in Fig. 6A, the
viability of cells transfected with miR-143-3p mimics or RALBP1
siRNA was evidently reduced compared with scramble control, while
the viability of cells transfected with miR-143-3p inhibitor was
markedly higher than scramble control. As shown in Fig. 6B, apoptosis of cells transfected
with miR-143-3p mimics or RALBP1 siRNA was evidently promoted
compared with scramble control, while apoptosis of cells
transfected with miR-143-3p inhibitor was markedly reduced compare
to scramble control. These data indicated the viability-reduced and
apoptosis-promoting effect of miR-143-3p.
Discussion
We discovered that a miRNA cluster known as
miR-145-5p/miR-143-3p may have positive regulatory role in the cell
proliferation of CIK cells using chromosome clustering and GO
analysis. Notably, this miRNA cluster has been predicted to target
proto-oncogene such as c-Myc, Bcl-2 and Ras family. Nevertheless,
we only observed significant upregulation of c-Myc and Bcl-2 during
CIK production. During CIK preparation, the two genes that are
anti-apoptotic may be implicated in the proliferation of cells
(18). It has been shown that the
downregulated expression levels of miR-143-3p and miR-143-5p were
observed in gastric cancer, which complied with results described
by other study teams (19,20). Recently, investigators analyzed 70
paired samples of benign tissues and gastric cancers using
real-time RT-PCR and chip assays (20). They discovered greatest
downregulation of miR-143 among miRNAs in gastric cancers, when
compared with benign tissues. There was correlation between the
expression level of miR-143 and the progression of gastric cancer
and stage IV cancers had significantly lower level of miR-143 than
stage I and II cancers. Takagi et al found downregulation of
miR-143 in gastric cancer cell lines and that the viability of
gastric cancer cells was suppressed by transfection with miR-143-3p
which targeted ERK5 and AKT consistent with our observations
(19). Nevertheless, the role of
miR-143-5p in gastric cancer remains unknown. In the present study,
computational analysis [bioinformatics algorithms TargetScan
(http://www.targetscan.org/) and miRanda
(http://www.microrna.org/microrna/home.do)] was used to
identify that RALBP1 was a target gene of miR-143-3p, and further
confirmed using luciferase assay, the results shown that luciferase
activity of the cells co-transfected with wild-type RALBP1 3'UTR
was lower than those transfected with the scramble controls, while
the introduction of mutant RALBP1 with the potential ‘seed
sequence’ in the 3'TUR of RALBP1 almost completely abolished such
repressive effect. The miR-143-3p level was determined using
western blotting and real-time PCR, we found that the expression of
miR-143-3p mRNA was markedly downregulated in participants with
ovarian cancer compared with normal control, while the expression
of RALBP1 mRNA and protein were evidently overexpressed in
participants with ovarian cancer compared with normal control.
RLIP76 has been reported to be a novel R-Ras
effector linking R-Ras to the activation of Arf6 and subsequently,
to Rac1 resulting in cell migration and spreading dependent on
adhesion. Additionally, the various roles of RLIP76 may contribute
to multiple functions of R-Ras. For instance, studies of Keely
et al in 1999, and Holly et al in 2005, showed that
the activation of Rac GTPase dependent on RLIP76 may account for
the function of R-Ras to enhance neurite outgrowth and cell
migration, which were Rac-dependent processes (21,22).
Moreover, in 1997, Radhakrishna and Donaldson demonstrated that the
adhesion-induced active Arf6 GTPase, known as a regulator of
vesicle trafficking, was triggered by RLIP76 (23); therefore, regulation of Arf6 may
account for the function of RLIP76 to regulate endocytosis
(24). In fact, there is physical
correlation between RLIP76 and μ2 chain of AP-2, an adaptor with
membrane recruitment regulated by Arf6, demonstrating that
endocytosis mediated by clathrin can be coordinately regulated by
the two proteins clathrin-mediated (25). Their function to facilitate cell
proliferation can be limited by endocytosis of growth factor
receptors; hence, the ability of R-Ras limiting the proliferation
of smooth muscle cells and endothelial cells can be explained by
the interaction of RLIP76 with R-Ras (26).
Molecules that regulate signaling networks of
angiogenesis are potential therapeutic targets. Ral-interacting
protein of 76 kDa (RLIP76, also RalBP1) has been identified as a
potential target, contributing to its physiological and cellular
properties that are still being studied, and to the substantial
reduction in tumors obtained by blockade of RLIP76 in numerous
tumor models (reviewed in ref. 27). As a pleiotropic protein, RLIP76
emerged as a Ral GTPase effector protein connecting Ral to Rho
pathways via its activity of RhoGAP (28). In addition, RLIP76 serves as an
ATP-dependent glutathione-conjugated transporter for small
molecules, such as endogenous metabolites and anticancer drugs, and
in cell spreading and migration, mitochondrial fission and
endocytosis (29–32). A number of signaling molecules can
bind to the sites of the protein (28); therefore, RLIP76 seems to back up a
regulatory scaffolding function for signaling. Majority of human
tissues such as kidney, muscle, lung, ovary, heart and liver, and
majority of human tumor cell lines can express RLIP76, and
overexpression of RLIP76 is observed in many cancers including
melanomas, ovarian carcinomas and lung cancer (27). There is a correlation between
blockade of RLIP76 with antisense or targeting antibodies and
enhanced sensitivity to chemotherapy and radiotherapy, which
results in substantial tumor regression in B16 melanomas, prostate
cancer, colon and non-small cell lung carcinomas in mice (6,33,34).
Nevertheless, tumor regression observed in the studies may have
been obtained from effects in either the animal host cells or the
tumor cells, or both. Therefore, RLIP76 is necessary for cancer
survival and progression although the mechanisms are unknown. In
the present study, the RALBP1 mRNA and protein level in cells
transfected with miR-143-3p mimics and RALBP1 siRNA were
downregulated, while notably upregulated subsequent to transfection
with miR-143-3p inhibitor, when compared with scramble control.
Additionally, flow cytometric analysis was used to study the
molecular mechanism underlying the observed viability-reduced and
apoptosis-promoting effect of miR-143-3p, the results suggested the
viability of cells was inhibited following transfection with
miR-143-3p mimics and RALBP1 siRNA, while notably promoted
subsequent to transfection with miR-143-3p inhibitor. Apoptosis of
cells were promoted following transfection with miR-143-3p mimics
and RALBP1 siRNA, while notably inhibited subsequent to
transfection with miR-143-3p inhibitor. In conclusion, these
findings provide support that downregulation of miR-143-3p caused
an increased expression of RALBP1, which may be a molecular
mechanism of tumorigenesis of ovarian cancer. miR-143-3p and RALBP1
show promise as therapeutic targets in the management of ovarian
cancer.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kaku T, Ogawa S, Kawano Y, Ohishi Y,
Kobayashi H, Hirakawa T and Nakano H: Histological classification
of ovarian cancer. Med Electron Microsc. 36:9–17. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bodemann BO and White MA: Ral GTPases and
cancer: Linchpin support of the tumorigenic platform. Nat Rev
Cancer. 8:133–140. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lim KH, Baines AT, Fiordalisi JJ,
Shipitsin M, Feig LA, Cox AD, Der CJ and Counter CM: Activation of
RalA is critical for Ras-induced tumorigenesis of human cells.
Cancer Cell. 7:533–545. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smith SC, Oxford G, Baras AS, Owens C,
Havaleshko D, Brautigan DL, Safo MK and Theodorescu D: Expression
of ral GTPases, their effectors, and activators in human bladder
cancer. Clin Cancer Res. 13:3803–3813. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Singhal SS, Awasthi YC and Awasthi S:
Regression of melanoma in a murine model by RLIP76 depletion.
Cancer Res. 66:2354–2360. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang H, Lu Z, Luo JQ, Wolfman A and
Foster DA: Ras mediates the activation of phospholipase D by v-Src.
J Biol Chem. 270:6006–6009. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reinhart BJ, Weinstein EG, Rhoades MW,
Bartel B and Bartel DP: MicroRNAs in plants. Genes Dev.
16:1616–1626. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim VN and Nam JW: Genomics of microRNA.
Trends Genet. 22:165–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shahab SW, Matyunina LV, Mezencev R,
Walker LD, Bowen NJ, Benigno BB and McDonald JF: Evidence for the
complexity of microRNA-mediated regulation in ovarian cancer: A
systems approach. PLoS One. 6:e225082011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Permuth-Wey J, Kim D, Tsai YY, Lin HY,
Chen YA, Barnholtz-Sloan J, Birrer MJ, Bloom G, Chanock SJ, Chen Z,
et al: Ovarian Cancer Association Consortium: LIN28B polymorphisms
influence susceptibility to epithelial ovarian cancer. Cancer Res.
71:3896–3903. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim
JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hudson ME, Pozdnyakova I, Haines K, Mor G
and Snyder M: Identification of differentially expressed proteins
in ovarian cancer using high-density protein microarrays. Proc Natl
Acad Sci USA. 104:17494–17499. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nakagawa M, Tsuzuki S, Honma K, Taguchi O
and Seto M: Synergistic effect of Bcl2, Myc and Ccnd1 transforms
mouse primary B cells into malignant cells. Haematologica.
96:1318–1326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takagi T, Iio A, Nakagawa Y, Naoe T,
Tanigawa N and Akao Y: Decreased expression of microRNA-143 and
−145 in human gastric cancers. Oncology. 77:12–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li X, Luo F, Li Q, Xu M, Feng D, Zhang G
and Wu W: Identification of new aberrantly expressed miRNAs in
intestinal-type gastric cancer and its clinical significance. Oncol
Rep. 26:1431–1439. 2011.PubMed/NCBI
|
|
21
|
Keely PJ, Rusyn EV, Cox AD and Parise LV:
R-Ras signals through specific integrin alpha cytoplasmic domains
to promote migration and invasion of breast epithelial cells. J
Cell Biol. 145:1077–1088. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Holly SP, Larson MK and Parise LV: The
unique N-terminus of R-ras is required for Rac activation and
precise regulation of cell migration. Mol Biol Cell. 16:2458–2469.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Radhakrishna H and Donaldson JG:
ADP-ribosylation factor 6 regulates a novel plasma membrane
recycling pathway. J Cell Biol. 139:49–61. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jullien-Flores V, Mahé Y, Mirey G,
Leprince C, Meunier-Bisceuil B, Sorkin A and Camonis JH: RLIP76, an
effector of the GTPase Ral, interacts with the AP2 complex:
Involvement of the Ral pathway in receptor endocytosis. J Cell Sci.
113:2837–2844. 2000.PubMed/NCBI
|
|
25
|
Paleotti O, Macia E, Luton F, Klein S,
Partisani M, Chardin P, Kirchhausen T and Franco M: The small
G-protein Arf6GTP recruits the AP-2 adaptor complex to membranes. J
Biol Chem. 280:21661–21666. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ceresa BP and Schmid SL: Regulation of
signal transduction by endocytosis. Curr Opin Cell Biol.
12:204–210. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Awasthi S, Singhal SS, Awasthi YC, Martin
B, Woo JH, Cunningham CC and Frankel AE: RLIP76 and cancer. Clin
Cancer Res. 14:4372–4377. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jullien-Flores V, Dorseuil O, Romero F,
Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G and
Camonis JH: Bridging Ral GTPase to Rho pathways. RLIP76, a Ral
effector with CDC42/Rac GTPase-activating protein activity. J Biol
Chem. 270:22473–22477. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Awasthi S, Cheng JZ, Singhal SS, Pandya U,
Sharma R, Singh SV, Zimniak P and Awasthi YC: Functional reassembly
of ATP-dependent xenobiotic transport by the N- and C-terminal
domains of RLIP76 and identification of ATP binding sequences.
Biochemistry. 40:4159–4168. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Goldfinger LE, Ptak C, Jeffery ED,
Shabanowitz J, Hunt DF and Ginsberg MH: RLIP76 (RalBP1) is an R-Ras
effector that mediates adhesion-dependent Rac activation and cell
migration. J Cell Biol. 174:877–888. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kashatus DF, Lim KH, Brady DC, Pershing
NL, Cox AD and Counter CM: RALA and RALBP1 regulate mitochondrial
fission at mitosis. Nat Cell Biol. 13:1108–1115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nakashima S, Morinaka K, Koyama S, Ikeda
M, Kishida M, Okawa K, Iwamatsu A, Kishida S and Kikuchi A: Small G
protein Ral and its downstream molecules regulate endocytosis of
EGF and insulin receptors. EMBO J. 18:3629–3642. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Singhal SS, Roth C, Leake K, Singhal J,
Yadav S and Awasthi S: Regression of prostate cancer xenografts by
RLIP76 depletion. Biochem Pharmacol. 77:1074–1083. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Singhal SS, Singhal J, Yadav S, Dwivedi S,
Boor PJ, Awasthi YC and Awasthi S: Regression of lung and colon
cancer xenografts by depleting or inhibiting RLIP76 (Ral-binding
protein 1). Cancer Res. 67:4382–4389. 2007. View Article : Google Scholar : PubMed/NCBI
|