Introduction
The mortality rate of pancreatic cancer (PC) is
high, and the 5-year survival rate is less than 6% in the United
States (1–3) with an average survival period of 5 to
6 months. Thus, early diagnosis of PC is very important. PC cell
metastasis negatively impacts radiation and chemotherapy (4,5). The
early diagnosis of PC is difficult due to a lack of clinical
symptoms and reliable diagnostic markers. Previous studies have
found aberrant overexpression of various mucins in various
epithelial malignancies, including pancreatic and breast cancer
(6–8). For example, mucin 1 (MUC1), MUC4 and
MUC5AC are highly expressed in human PC tissues and are associated
with disease progression and poor prognosis (9,10).
Using northern blot analysis and PCR, Andrianifahanana et al
(11) found that MUC4 mRNA was not
expressed in normal pancreatic tissues or in chronic pancreatitis
tissue but was highly expressed in PC tissue. Therefore, MUC4 plays
an important role in pancreatic tumor development and metastasis
and may be a specific tumor marker for the diagnosis and therapy of
PC (12,13).
Mimeault et al found that silencing of the
MUC4 gene can inhibit the growth and metastasis of PC (14). Effective MUC4 gene knockdown in PC
has contributed to the elucidation of pancreatic tumor development
and metastasis, and may be valuable in new therapeutic approaches.
Recently, RNA interference (RNAi) has become widely used 3,15–17 due
to a more specific, effective and complete inhibition of gene
expression compared to other methods. Short hairpin RNA (shRNA) for
a specific gene can be integrated into the host chromosome using a
lentivirus vector to stably express a small interfering RNA
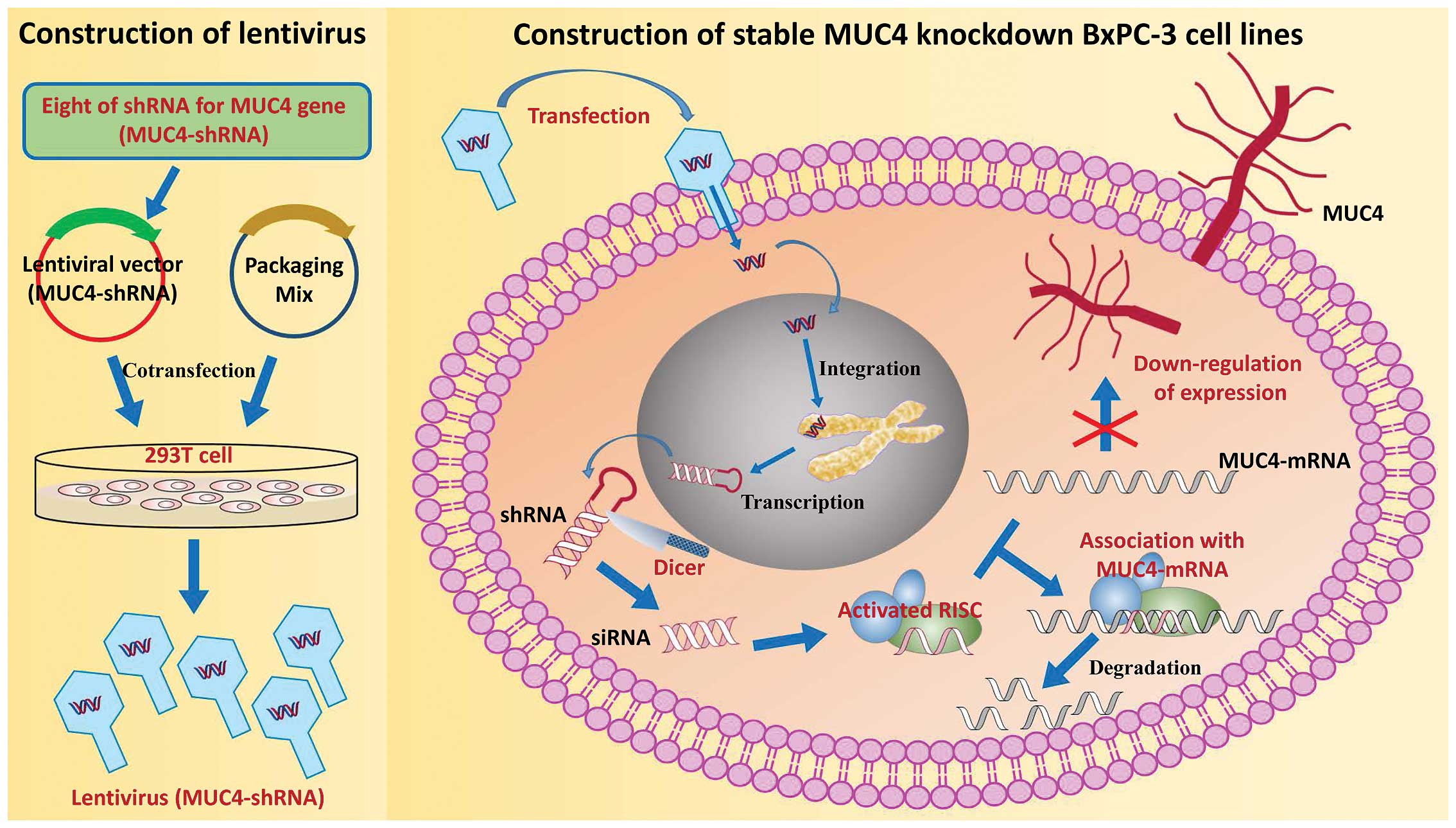
(siRNA), which can inhibit long-term gene expression. Here, we used
BxPC-3 cells as an in vitro model of PC. Eight shRNAs for
the MUC4 gene (MUC4-shRNA) were designed and integrated into
lentiviral vectors. The MUC4-shRNA with a high interference
efficiency was identified and used for stable MUC4 knockdown in the
BxPC-3 cell line (Fig. 1).
Subsequently, cell proliferation, migration, protein expression,
and tumor formation in vivo were assessed.
Materials and methods
Chemicals, reagents, and cell
lines
The lentiviral expression vector was purchased from
the Zimmer Medical International Trading Co., Ltd. (Shanghai,
China). DH5α Escherichia coli (E. coli) was obtained
from the Chinese Academy of Sciences (Shanghai, China). The
restriction endonucleases BbsI and BamHI, T4 ligase
and Taq enzyme were purchased from Takara (Dalian, China).
Roswell Park Memorial Institute (RPMI)-1640 culture medium, fetal
bovine serum (FBS), trypsin, Lipofectamine 2000 and TRIzol reagent
were purchased from Gibco (Invitrogen Life Technologies, Carlsbad,
CA, USA). A plasmid DNA extraction kit was obtained from Qiagen
(Shanghai, China). A reverse transcription kit was purchased from
Promega (Fitchburg, WI, USA). All oligonucleotides, including PCR
primers, were purchased from Shanghai YingJun Biotechnology Co.,
Ltd. (Shanghai, China). A Cell Counting Kit-8 (CCK-8) was purchased
from Dojindo Laboratories (Kumamoto, Japan). A Boyden chamber for
the migration assay was obtained from Corning Costar (Rochester,
NY, USA). Matrigel (0.3 mg/ml) was purchased from BD Biosciences
(Bedford, MA, USA).
BxPC-3 cells, a human PC cell line, was purchased
from the Shanghai Cell Bank of the Chinese Academy of Sciences.
These cells were grown in RPMI-1640 media supplemented with 10%
fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin at
37°C in a humidified atmosphere containing 95% air and 5%
CO2.
shRNA synthesis, vector construction
and verification
The eight MUC4-siRNAs, and a negative control (with
a scrambled sequence that did not match any known gene) were
selected based on the full-length cDNA of human MUC4 mRNA (gene
bank no. NM_004532) using Ambion siRNA design software. A siRNA
against GAPDH was included as a positive control to verify
transfection reliability, RNA extraction and gene expression
quantification. The TTCAAGAGA sequence was used in the loop of all
constructs to avoid premature termination. The sequences of eight
shRNAs coresponding to MUC4-siRNA are listed in Table I. PCR products were annealed using
the following conditions: 95°C for 5 min, 85°C for 4 min, 75°C for
5 min, 70°C for 5 min and preservation at 4°C. After the annealing
process, 20 µl of shRNA template (10 µl of forward primer and 10 µl
of reverse primer), 10 µl of annealing buffer (10X) and 70 µl of
double distilled water (ddH2O) were mixed at 100°C for
ligation.
 | Table I.Sequences of the MUC4-shRNA
primers. |
Table I.
Sequences of the MUC4-shRNA
primers.
| No. |
| 5-3-Primer
sequences |
|---|
| A139 | F |
GATCCGGCCTGTGAATTACTGCTACAATGGTACCATTGTAGCAGTAATTCACAGGTTTTTG |
|
| R |
AATTCAAAAACCTGTGAATTACTGCTACAATGGTACCATTGTAGCAGTAATTCACAGGCCG |
| A140 | F |
GATCCGGCCCTGTGAATTACTGCTACAAGGTACCTTGTAGCAGTAATTCACAGGGTTTTTG |
|
| R |
AATTCAAAAACCCTGTGAATTACTGCTACAAGGTACCTTGTAGCAGTAATTCACAGGGCCG |
| A141 | F |
GATCCGGGGGAACAACTTCAGTCCAACTGGTACCAGTTGGACTGAAGTTGTTCCCTTTTTG |
|
| R |
AATTCAAAAAGGGAACAACTTCAGTCCAACTGGTACCAGTTGGACTGAAGTTGTTCCCCCG |
| A142 | F |
GATCCGGCAATGGTGGTCGTGTGATTGAGGTACCTCAATCACACGACCACCATTGTTTTTG |
|
| R |
AATTCAAAAACAATGGTGGTCGTGTGATTGAGGTACCTCAATCACACGACCACCATTGCCG |
| A143 | F |
GATCCGGGCCCTGATAGATTCCTGAATTGGTACCAATTCAGGAATCTATCAGGGCTTTTTG |
|
| R |
AATTCAAAAAGCCCTGATAGATTCCTGAATTGGTACCAATTCAGGAATCTATCAGGGCCCG |
| A144 | F |
GATCCGGCGCCCTGATAGATTCCTGAATGGTACCATTCAGGAATCTATCAGGGCGTTTTTG |
|
| R |
AATTCAAAAACGCCCTGATAGATTCCTGAATGGTACCATTCAGGAATCTATCAGGGCGCCG |
| A145 | F |
GATCCGGAGTCCTGCCCTGTGAATTACTGGTACCAGTAATTCACAGGGCAGGACTTTTTTG |
|
| R |
AATTCAAAAAAGTCCTGCCCTGTGAATTACTGGTACCAGTAATTCACAGGGCAGGACTCCG |
| A146 | F |
GATCCGGGGAGATGGCTATTTCGAAAACGGTACCGTTTTCGAAATAGCCATCTCCTTTTTG |
|
| R |
AATTCAAAAAGGAGATGGCTATTTCGAAAACGGTACCGTTTTCGAAATAGCCATCTCCCCG |
shRNA verification
After agarose electrophoresis purification, shRNA
was inserted into the pUETP/td-tomoto/puro vector at the
BamHI and BbsI sites. The resulting vector was
inoculated into competent DH5α E. coli, selected using
kanamycin resistance, and verified with sequencing using Shanghai
3D Biopharm Bio-technology. A total of eight constructs, referred
to as pUETP/td-tomoto/puro-shRNA-1-8 (A139-A146) were evaluated in
cultured BxPC-3 cells. The experiment also included
pUETP/td-tomoto/puro-shNC (shNC) as a negative control and
pUETP/td-tomoto/puro-shPC (shPC) as a positive control.
Quantitative real-time PCR analysis
(qPCR)
Two microliters of total RNA were reverse
transcribed with Premix Ex Taq™ (Takara) using random
primers. MUC4 first-strand cDNA was amplified with specific primers
as follows: forward, 5-GGAG AGGTATCGCCCTGATAGATT-3 and reverse,
5-ACGGTA GTTGGGCCTTTCTTC-3. The primers used for housekeeping gene,
glyceraldehyde-phosphate dehydrogenase (GAPDH), were as follows:
forward, 5-CATGAGAAGTATG ACAACAGCCT-3 and reverse,
5-AGTCCTTCCACGATAC CAAAGT-3. We evaluated the specificity of the
primers using GenBank. The PCR products were separated on a 1.5%
agarose gel and analyzed using an imaging system from Bio-Rad
Laboratories, Inc. (Hercules, CA, USA).
Western blot (WB) analysis
Whole cell proteins were prepared from
5×106 cells after transfection for 96 h using WB lysis
buffer. For WB, we used an anti-MUC4 mouse monoclonal antibody
(8G7) and an HRP-linked anti-rabbit IgG secondary antibody (Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA). We used the BCA
protein assay kit to detect the protein concentration in the
supernatant (Takara, Otsu, Japan). The protein bands were
visualized using an enhanced chemiluminescence system (Bio-Rad
Laboratories).
Experiments in the BxPC-3 cells
After seeding in 6-well plates at a density of
1×105 cells/well, BxPC-3 cells were transfected with
A139-A146, sh-NC, or sh-PC. After incubation for 96 h, MUC4 mRNA
was examined using qPCR. Subsequently, we counted the number of
cells at 48, 72 and 96 h after transfection using a colorimetric
CCK-8 assay at 450 nm. Migration was examined using a 24-well, 8-µm
pore transparent insert containing a gelatin-coated polycarbonate
membrane filter. Cell migration was quantified by counting the
cells that migrated to the lower chamber using crystal violet
staining and an optical microscope. Tumor formation and growth were
examined in nude mice.
Statistical analyses
The quantitative results are expressed as the mean
value ± SD. All statistical analyses were carried out by one-way
analysis of variance (ANOVA) using SPSS version 13.0 (SPSS, Inc.,
Chicago, IL, USA). A P-value of <0.05 was considered to indicate
a statistically significant difference.
Results
Construction and verification of
MUC4-shRNA lentiviral vector
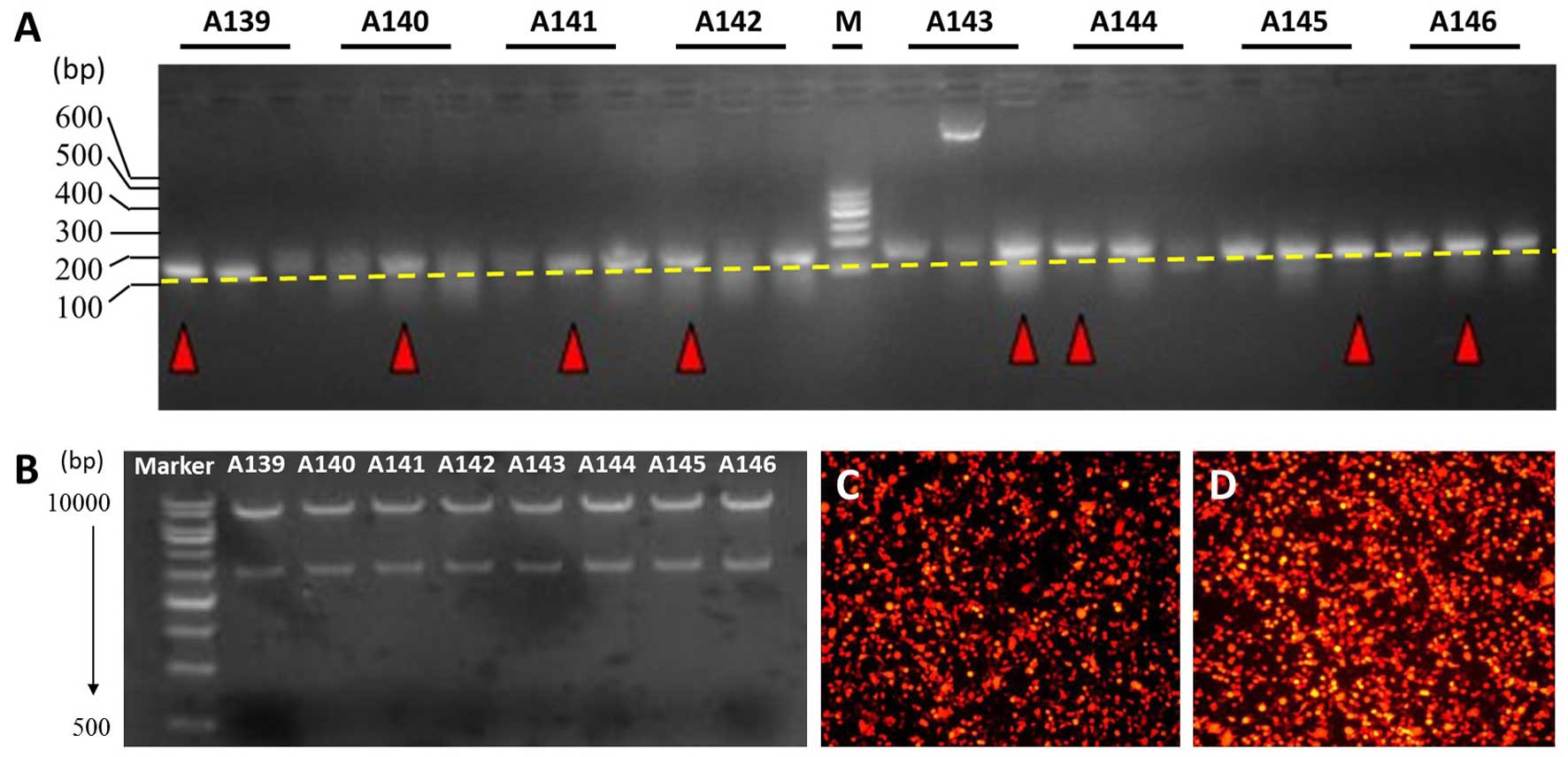
Eight pairs of MUC4-shRNA were ligated into the
lentiviral vector using T4 DNA ligase. A KpnI single enzyme
digestion was performed, and the fragments were examined by agarose
gel electrophoresis. A 2.95+7.5 kb fragment confirmed that the
shRNA size conformed to the design (Fig. 2B). A transfection experiment on 293T
cells showed that more than 80% of the cells produced red
fluorescent protein, as observed with fluorescence microscopy
(Fig. 2D).
Construction of MUC4 knockdown in
BxPC-3 cells
The BxPC-3 cell lines were transfected with eight
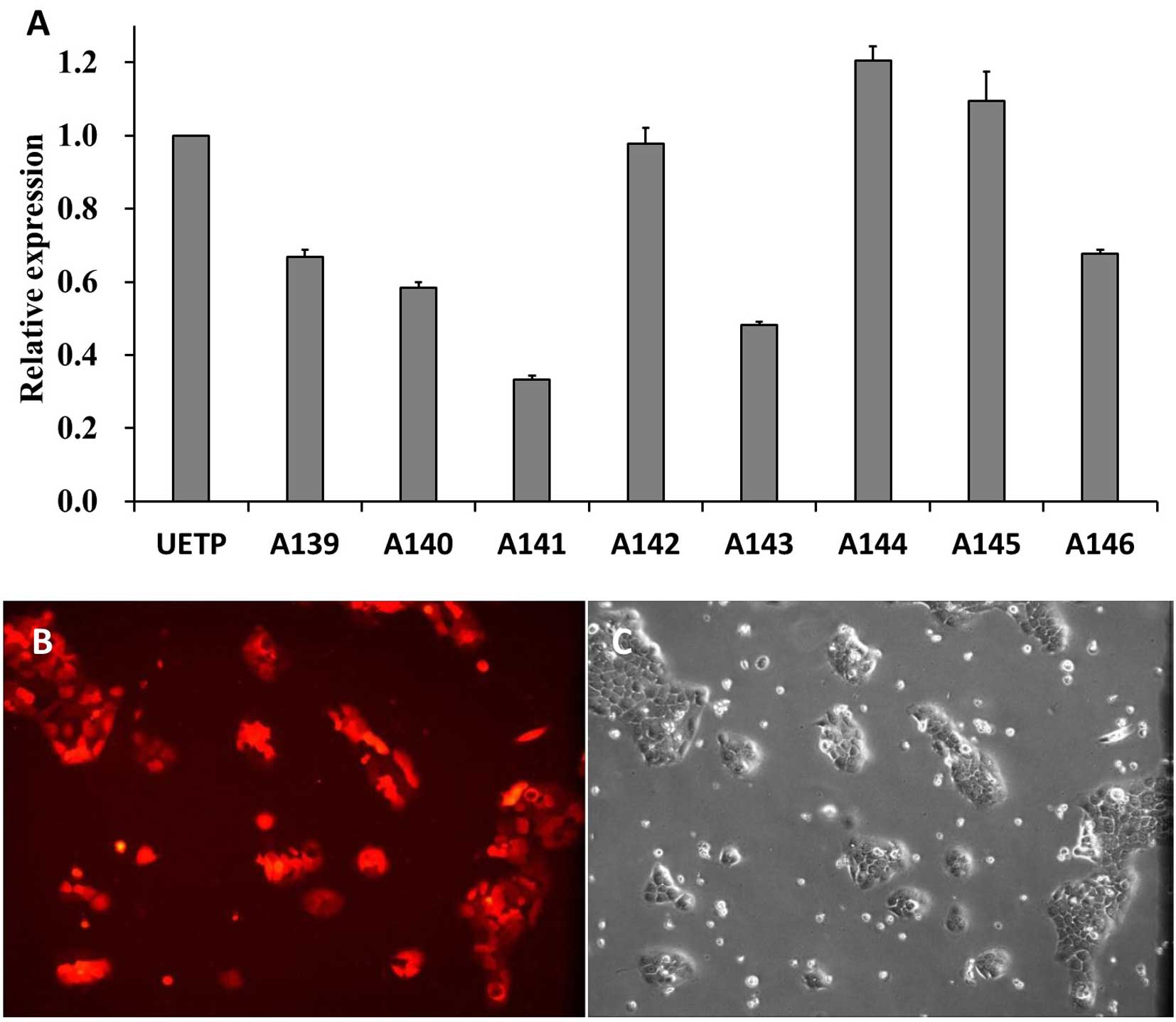
pairs of lentiviral vectors based on MUC4-shRNA. The qPCR results
showed that expression of MUC4 mRNA in the BxPC-3 cells was
significantly decreased by all eight shRNAs (P<0.05 vs. the NC;
Fig. 3A) at 96 h after
transfection. A141 had the strongest suppressive effect on MUC4
mRNA expression among the eight shRNAs and was used for further
experiments. To obtain MUC4 knockdown in BxPC-3 cells, the BxPC-3
cell line was transfected with shRNA-A141, and cultured for nearly
20 days until the cell fluorescence ratio reached 100% (Fig. 3B).
Proliferation, migration and protein
expression of MUC4 following knockdown in BxPC-3 cells
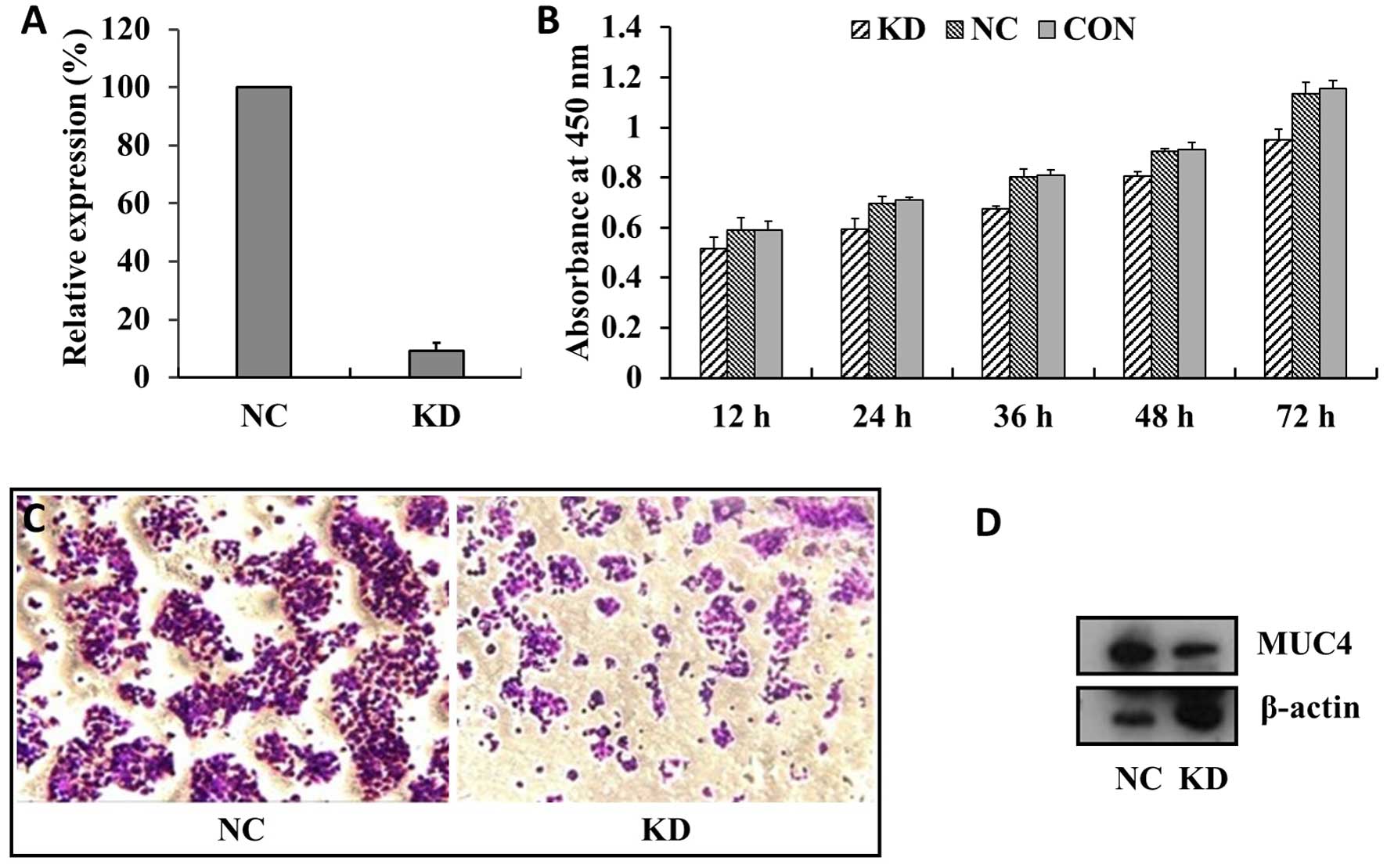
After stable transfection with shRNA-A141 (KD
group), qPCR and western blot analysis were performed. The results
showed that the efficiency of MUC4 mRNA interference was ~91%
(Fig. 4A). At the same time, MUC4
knockdown in the BXPC-3 cells (KD) showed reduced growth compared
with cells transfected with empty or scrambled RNAi vectors (NC and
CON groups), and the number of floating cells in the cell culture
increased (Fig. 4B). Transwell
migration assays showed that the number of cells in the
polycarbonate membrane for the KD group were significantly lower
than that of the NC group (P<0.01) (Fig. 4C). In fact, the expression level of
MUC4 in the KD group was also significantly reduced, compared with
the NC group (P<0.01) (Fig. 4D).
The results showed that MUC4 protein knockdown in the BxPC-3 cells
was noteworthy.
Tumor formation in vivo after MUC4
knockdown
BxPC-3 cells of the KD, NC and CON groups were
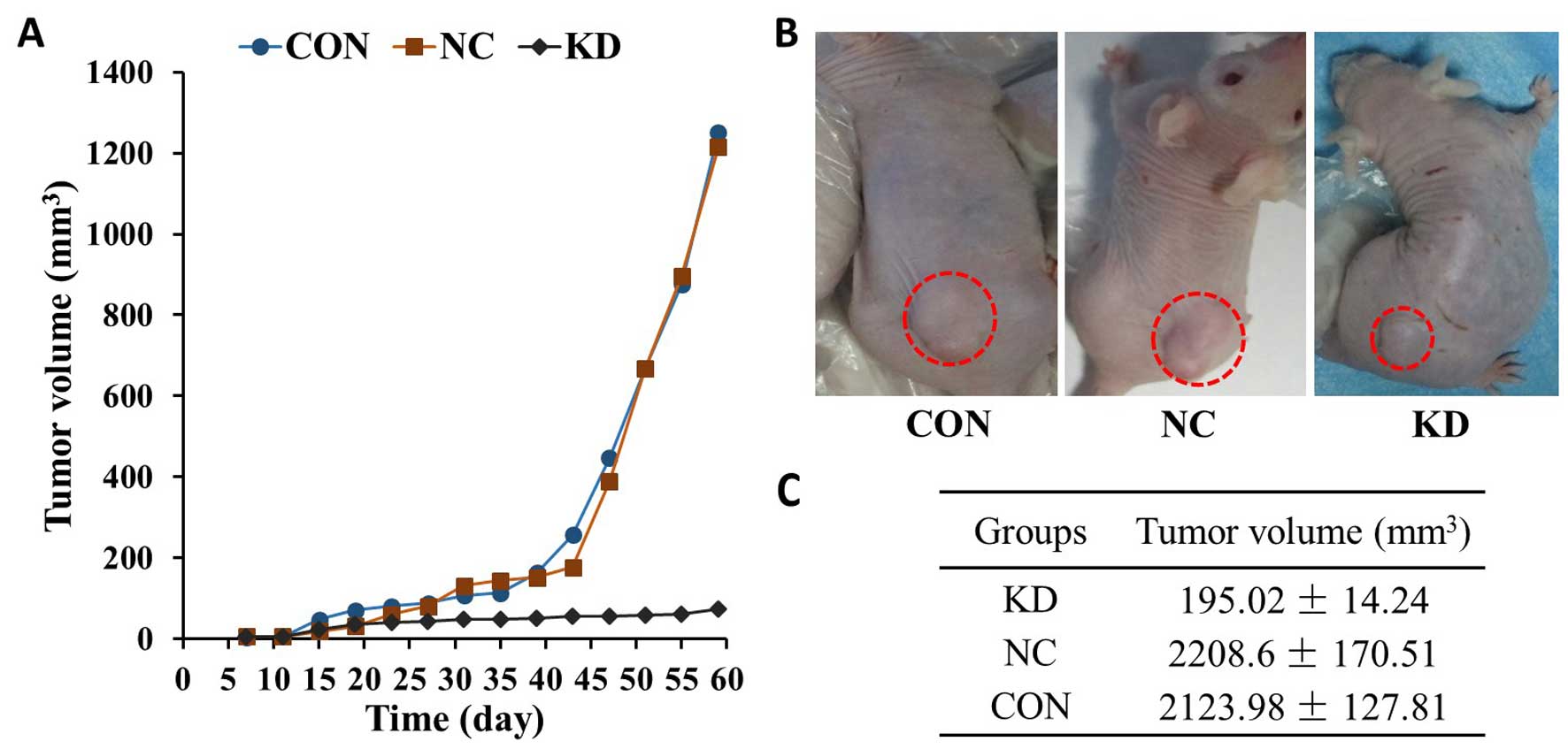
transplanted into nude mice. Tumors formed within 7–10 days. Lower
tumor growth rates were observed for tumors derived from the KD
group, whereas the transplanted tumors derived from the other
groups grew rapidly (Fig. 5A). At 8
weeks after cell inoculation, the tumor volume reached 195.02±14.24
mm2 in the KD group, which was less than a tenth of that
in the NC and CON groups (Fig. 5C).
Throughout the experiment, no significant differences were observed
in the activity and eating behavior of the KD cell-transplanted
nude mice. In the NC and CON groups, the activity of the nude mice
gradually reduced with tumor enlargement.
Discussion
In the present study, MUC4 gene sequences of human
PC BxPC-3 cells were used to design eight shRNAs (Table I). Typically, shRNAs are designed
based on the principle that the first base in the shRNA sequence
must be a G. If the first base is not a G, a G must be added to the
sense strand to ensure transcription by RNA polymerase.
Furthermore, 5–6 Ts must be included at the 3 end of the shRNA to
ensure termination by RNA polymerase. However, more than 3 As or 3
Ts must appear in the sense strand and antisense strand to avoid
early termination of shRNA transcription. Restriction enzyme sites
must be at the ends of the two complementary nucleotide sequences,
and the GC content should be between 36 and 52%.
A lentiviral vector carrying the MUC4 gene was
successfully constructed and eight pairs of shRNA were successfully
ligated into the lentiviral vector pUETP. Positive clones were
identified and chosen using colony PCR (Fig. 2A). PCR positive products had 2.95
and 7.5 kb fragments, as determined by agarose gel electrophoresis,
after KpnI single enzyme digestion (Fig. 2B). Recombinant lentiviral vector
plasmid, auxiliary plasmid and transit transfection reagents were
transfected into the 293T cells. More than 80% of the cells
produced red fluorescent protein, as determined by fluorescence
microscopy, which confirmed that the lentiviral vectors were
efficiently transferred into the 293T cells (Fig. 2D). Moreover, the viral titer was
measured to confirm the lentiviral vector concentration necessary
for subsequent experiments.
The expression of MUC4 mRNA in BxPC-3 cells was
evaluated directly before and 96 h after transfection with the
eight pairs of lentiviral vectors and was used to evaluate the
interference efficiency. qPCR, as performed previously, was also
used to analyze the interference efficiency (18,19).
The results showed that MUC4 mRNA in the A141-transfected group
(MUC4-shRNA transfection KD group) had a significantly lower
expression level when compared with A139, A140, A142, A143, A144,
A145, A146 and the UETP groups. After stable transfection with
shRNA-A141 in the BxPC-3 cells, the MUC4 mRNA knockdown was ~91%
(Fig. 4A, P<0.01), which was
significantly higher than that found in other studies (20,21).
These results suggest that our shRNAs designed against the MUC4
gene sequences effectively blocked MUC4 gene expression in the
human PC BxPC-3 cells. Furthermore, MUC4 protein expression was
detected by western blot analysis in the NC (UETP) and KD (A141)
groups. MUC4 protein expression in the KD group was significantly
reduced compared with the NC group (P<0.01). Our knockdown of
MUC4 protein expression in the BxPC-3 cells was marked.
MUC4 is a high-molecular-weight glycol protein
related to the growth, metastasis and angiogenesis of PC (21–24).
Thus, we examined the cell growth and active state of the BxPC-3
cells in our study (Figs. 4 and
5). Compared with the NC group and
CON group, the KD group had a lower cell growth rate and an
increased number of floating cells. These results are consistent
with other studies (14,20) suggesting that the MUC4-shRNA used in
our study could promote the apoptosis of BxPC-3 cells. Moreover, we
also found that cell proliferation in the KD group was
significantly suppressed compared with the proliferation in the NC
and CON groups, as determined by a CCK-8 cell proliferation test.
We found that cell migration in the KD group was significantly
lower than the migration in the NC group, as determined using a
Transwell chamber test. These results are similar to previous
results showing that MUC4 knockdown reduces the migration ability
of PC cells (24) Moreover, BxPC-3
cells of the KD, NC and CON groups were transplanted into nude
mice. Lower tumor growth rates were observed for tumors derived
from the KD group, whereas the transplanted tumors derived from the
other groups grew rapidly (Fig. 5).
At 8 weeks after cell inoculation, the tumor volume in the KD group
was less than a tenth of that in the NC and CON groups.
In conclusion, we demonstrated for the first time
that MUC4 promotes the migration and proliferation of BxPC-3 cells.
These results indicate that inhibition of MUC4 expression may be an
effective means for limiting metastasis and invasion of PC cells.
Thus, MUC4 may be a new potential target for the treatment of
PC.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China, no. 81271643.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Frank TS, Sun X, Zhang Y, Yang J, Fisher
WE, Gingras MC and Li M: Genomic profiling guides the choice of
molecular targeted therapy of PC. Cancer Lett. 363:1–6. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rachagani S, Macha MA, Heimann N,
Seshacharyulu P, Haridas D, Chugh S and Batra SK: Clinical
implications of miRNAs in the pathogenesis, diagnosis and therapy
of PC. Adv Drug Deliv Rev. 81:16–33. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Matsuno S, Egawa S, Fukuyama S, Motoi F,
Sunamura M, Isaji S, Imaizumi T, Okada S, Kato H, Suda K, et al: PC
Registry in Japan: 20 years of experience. Pancreas. 28:219–230.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sultana A, Smith C Tudur, Cunningham D,
Starling N, Neoptolemos JP and Ghaneh P: Meta-analyses of
chemotherapy for locally advanced and metastatic PC: Results of
secondary end points analyses. Br J Cancer. 99:6–13. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kufe DW: Mucins in cancer: Function,
prognosis and therapy. Nat Rev Cancer. 9:874–885. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ponnusamy MP, Seshacharyulu P, Lakshmanan
I, Vaz AP, Chugh S and Batra SK: Emerging role of mucins in
epithelial to mesenchymal transition. Curr Cancer Drug Targets.
13:945–956. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kaur S, Kumar S, Momi N, Sasson AR and
Batra SK: Mucins in PC and its microenvironment. Nat Rev
Gastroenterol Hepatol. 10:607–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Torres MP, Chakraborty S, Souchek J and
Batra SK: Mucin-based targeted PC therapy. Curr Pharm Des.
18:2472–2481. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rachagani S, Torres MP, Kumar S, Haridas
D, Baine M, Macha MA, Kaur S, Ponnusamy MP, Dey P, Seshacharyulu P,
et al: Mucin (Muc) expression during PC progression in spontaneous
mouse model: Potential implications for diagnosis and therapy. J
Hematol Oncol. 5:682012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Andrianifahanana M, Moniaux N, Schmied BM,
Ringel J, Friess H, Hollingsworth MA, Büchler MW, Aubert JP and
Batra SK: Mucin (MUC) gene expression in human pancreatic
adenocarcinoma and chronic pancreatitis: a potential role of MUC4
as a tumor marker of diagnostic significance. Clin Cancer Res.
7:4033–4040. 2001.PubMed/NCBI
|
|
12
|
Wu SC, Chen YJ, Lin YJ, Wu TH and Wang YM:
Development of a mucin4-targeting SPIO contrast agent for effective
detection of pancreatic tumor cells in vitro and in vivo. J Med
Chem. 56:9100–9109. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jhala N, Jhala D, Vickers SM, Eltoum I,
Batra SK, Manne U, Eloubeidi M, Jones JJ and Grizzle WE: Biomarkers
in diagnosis of pancreatic carcinoma in fine-needle aspirates: A
translational research application. Am J Clin Pathol. 126:572–579.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mimeault M, Johansson SL, Senapati S, Momi
N, Chakraborty S and Batra SK: MUC4 down-regulation reverses
chemoresistance of PC stem/progenitor cells and their progenies.
Cancer Lett. 295:69–84. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ambesajir A, Kaushik A, Kaushik JJ and
Petros ST: RNA interference: A futuristic tool and its therapeutic
applications. Saudi J Biol Sci. 19:395–403. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pecot CV, Calin GA, Coleman RL,
Lopez-Berestein G and Sood AK: RNA interference in the clinic:
Challenges and future directions. Nat Rev Cancer. 11:59–67. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Agrawal N, Dasaradhi PV, Mohmmed A,
Malhotra P, Bhatnagar RK and Mukherjee SK: RNA interference:
Biology, mechanism, and applications. Microbiol Mol Biol Rev.
67:657–685. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang C, Fu J, Wang Y, Bao Z and Zhao H:
Identification of suitable reference genes for gene expression
normalization in the quantitative real-time PCR analysis of sweet
osmanthus (Osmanthus fragrans Lour.). PLoS One. 10:e01363552015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dijkstra JR, van Kempen LC, Nagtegaal ID
and Bustin SA: Critical appraisal of quantitative PCR results in
colorectal cancer research: Can we rely on published qPCR results?
Mol Oncol. 8:813–818. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chaturvedi P, Singh AP, Moniaux N,
Senapati S, Chakraborty S, Meza JL and Batra SK: MUC4 mucin
potentiates pancreatic tumor cell proliferation, survival, and
invasive properties and interferes with its interaction to
extracellular matrix proteins. Mol Cancer Res. 5:309–320. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Singh AP, Moniaux N, Chauhan SC, Meza JL
and Batra SK: Inhibition of MUC4 expression suppresses pancreatic
tumor cell growth and metastasis. Cancer Res. 64:622–630. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhi X, Tao J, Xie K, Zhu Y, Li Z, Tang J,
Wang W, Xu H, Zhang J and Xu Z: MUC4-induced nuclear translocation
of β-catenin: A novel mechanism for growth, metastasis and
angiogenesis in PC. Cancer Lett. 346:104–113. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rachagani S, Macha MA, Ponnusamy MP,
Haridas D, Kaur S, Jain M and Batra SK: MUC4 potentiates invasion
and metastasis of PC cells through stabilization of fibroblast
growth factor receptor 1. Carcinogenesis. 33:1953–1964. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lahdaoui F, Delpu Y, Vincent A, Renaud F,
Messager M, Duchêne B, Leteurtre E, Mariette C, Torrisani J,
Jonckheere N, et al: miR-219-1-3p is a negative regulator of the
mucin MUC4 expression and is a tumor suppressor in PC. Oncogene.
34:780–788. 2015. View Article : Google Scholar : PubMed/NCBI
|